How Do Yeast Cells Contend with Prions?
Abstract
1. Introduction
2. Prion Variants
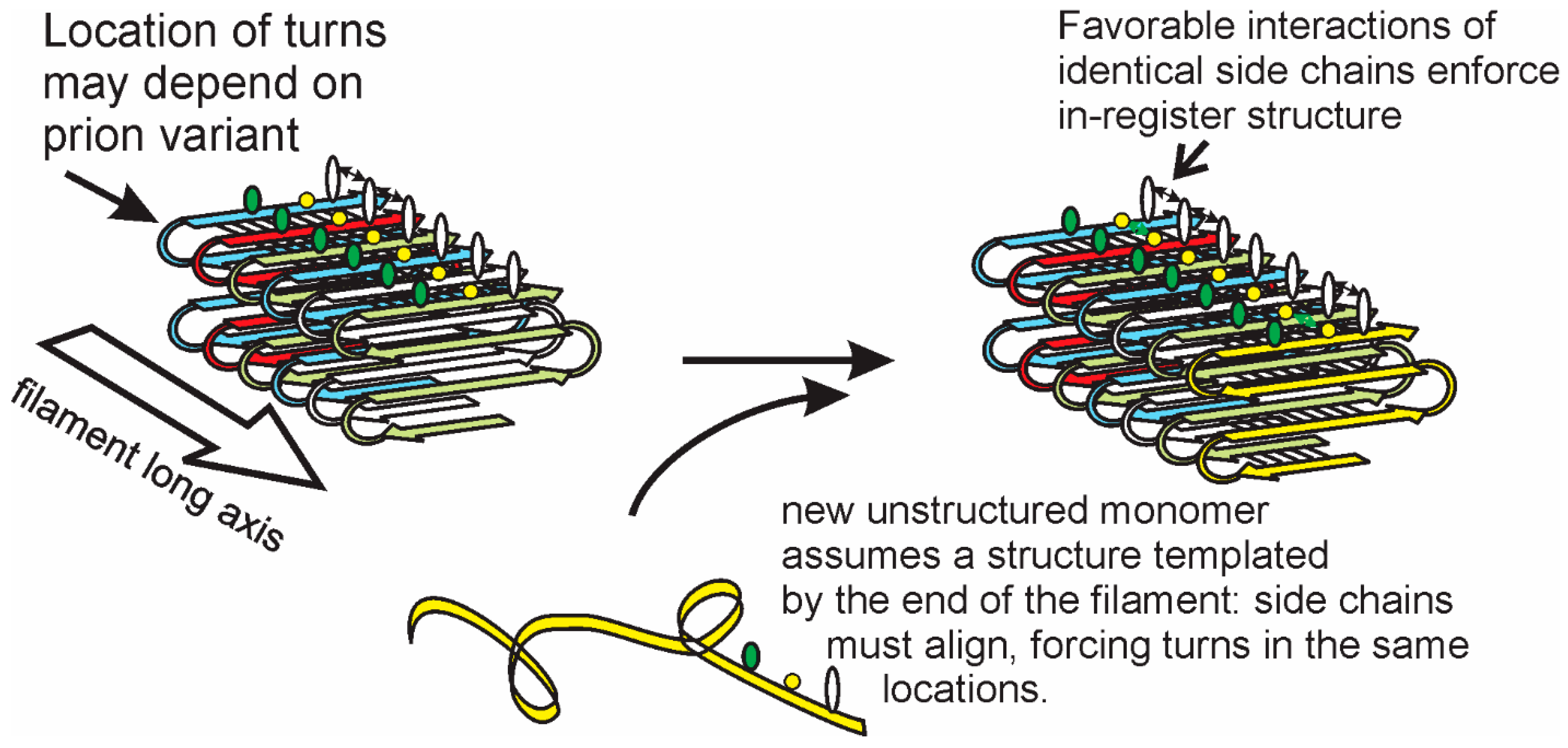
3. Structures of Yeast/Fungal Prion Amyloids: Explanation of Templating
4. Biology of Yeast/Fungal Prions
5. Prions Evolve on Two Levels
6. Chaperones and Prions
7. Anti-Prion Systems
7.1. Ribosome-Associated Complex
7.2. Cur1p Cures [URE3] without Visibly Collecting Ure2p Amyloid Aggregates
7.3. Normal Levels of Btn2p and Cur1p Cure Most [URE3] Variants Arising in Their Absence
7.4. Hsp104 Curing Activity Works at Normal Chaperone Levels
7.5. Siw14p, Inositol Polyphosphates, and [PSI+] Prion Propagation
7.6. Nonsense-Mediated mRNA Decay Factors Upf1,2,3 Cure some [PSI+] Variants
7.7. Sis1p Prevents Toxicity of [PSI+] Variants that Are Mild in Normal Cells
7.8. Lug1p Lets [URE3] Grow
8. Perspectives and Prospects
- >>
- What exactly do Hsp90s and their co-chaperones do to/for prions?
- >>
- What are the relations among the various anti-prion components? Are they co-operating in some systematic way or are they just different systems that happen to have similar effects?
- >>
- Which anti-prion systems are inducible (like Hsp104) and which are constitutive?
- >>
- Are there mammalian anti-prion systems?
- >>
- What are the detailed mechanisms of the yeast anti-prion systems? This applies to all of the systems discussed here, but particularly Lug1p, inositol polyphosphates, and Cur1p.
Funding
Conflicts of Interest
References
- Jaunmuktane, Z.; Mead, S.; Ellis, M.; Wadsworth, J.D.; Nicoll, A.J.; Kenny, J.; Launchbury, F.; Linehan, J.; Richard-Loendt, A.; Walker, A.S.; et al. Evidence for human transmission of amyloid-β pathology and cerebral amyloid angiopathy. Nature 2015, 525, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Morales-Scheihing, D.; Salvadores, N.; Moreno-Gonzales, I.; Gonzales, C.; Taylor-Presse, K.; Mendez, N.; Shahnawaz, M.; Gaber, A.O.; Sabek, O.M.; et al. Inductionn of IAPP amyloid deposition and associated diabetic abnormalities by a prion-like mechanism. J. Exp. Med. 2017, 214, 2591–2610. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kwon, S.-H.; Kam, T.-I.; Panicker, N.; Karuppagounder, S.S.; Lee, S.; Lee, J.H.; Kim, W.R.; Kook, M.; Foss, C.A.; et al. Transneuronal propagation of pathologic α-synuclein from the gut to the brain models Parkinson’s disease. Neuron 2019, 103, 627–641. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Challis, C.; Jain, N.; Moiseyenko, A.; Ladinsky, M.S.; Shastri, G.G.; Thron, T.; Needham, B.D.; Horvath, I.; Debelius, J.W.; et al. A gut bacterial amyloid promotes α-synuclein aggregation and motor impairment in mice. Elife 2020, 9, e53111. [Google Scholar] [CrossRef] [PubMed]
- Cox, B.S. PSI, a cytoplasmic suppressor of super-suppressor in yeast. Heredity 1965, 20, 505–521. [Google Scholar] [CrossRef]
- Lacroute, F. Non-Mendelian mutation allowing ureidosuccinic acid uptake in yeast. J. Bacteriol. 1971, 106, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B. [URE3] as an altered URE2 protein: Evidence for a prion analog in S. cerevisiae. Science 1994, 264, 566–569. [Google Scholar] [CrossRef]
- Liebman, S.W.; Chernoff, Y.O. Prions in yeast. Genetics 2012, 191, 1041–1072. [Google Scholar] [CrossRef]
- Wickner, R.B.; Shewmaker, F.; Bateman, D.A.; Edskes, H.E.; Gorkovskiy, A.; Dayani, Y.; Bezsonov, E.E. Yeast prions: Structure, biology and prion-handling systems. Microbiol. Mol. Biol. Rev. 2015, 79, 1–17. [Google Scholar] [CrossRef]
- Cooper, T.G. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: Connecting the dots. FEMS Microbiol. Rev. 2002, 26, 223–238. [Google Scholar] [CrossRef]
- Masison, D.C.; Wickner, R.B. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 1995, 270, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Edskes, H.K.; VGray, T.; Wickner, R.B. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl. Acad. Sci. USA 1999, 96, 1498–1503. [Google Scholar] [CrossRef]
- Taylor, K.L.; Cheng, N.; Williams, R.W.; Steven, A.C.; Wickner, R.B. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 1999, 283, 1339–1343. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, A.; Baxa, U.; Wickner, R.B. Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J. 2005, 24, 3082–3092. [Google Scholar] [CrossRef] [PubMed]
- Schlumpberger, M.; Prusiner, S.B.; Herskowitz, I. Induction of distinct [URE3] yeast prion strains. Mol. Cell. Biol. 2001, 21, 7035–7046. [Google Scholar] [CrossRef]
- Stansfield, I.; Jones, K.M.; Kushnirov, V.V.; Dagkesamanskaya, A.R.; Poznyakovski, A.I.; Paushkin, S.V.; Nierras, C.R.; Cox, B.S.; Ter-Avanesyan, M.D.; Tuite, M.F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995, 14, 4365–4373. [Google Scholar] [CrossRef] [PubMed]
- Frolova, L.; LeGoff, X.; Rasmussen, H.H.; Cheperegin, S.; Drugeon, G.; Kress, M.; Arman, I.; Haenni, A.-L.; Celis, J.E.; Philippe, M.; et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 1994, 372, 701–703. [Google Scholar] [CrossRef] [PubMed]
- Paushkin, S.V.; Kushnirov, V.V.; Smirnov, V.N.; Ter-Avanesyan, M.D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996, 15, 3127–3134. [Google Scholar] [CrossRef]
- King, C.-Y.; Tittmann, P.; Gross, H.; Gebert, R.; Aebi, M.; Wuthrich, K. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. USA 1997, 94, 6618–6622. [Google Scholar] [CrossRef]
- Glover, J.R.; Kowal, A.S.; Shirmer, E.C.; Patino, M.M.; Liu, J.-J.; Lindquist, S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 1997, 89, 811–819. [Google Scholar] [CrossRef]
- King, C.-Y.; Diaz-Avalos, R. Protein-only transmission of three yeast prion strains. Nature 2004, 428, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Chien, P.; Naber, N.; Cooke, R.; Weissman, J.S. Conformational variations in an infectious protein determine prion strain differences. Nature 2004, 428, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Chernoff, Y.O.; Kushnirov, V.V.; Inge-Vechtomov, S.G.; Liebman, S.W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 1996, 144, 1375–1386. [Google Scholar]
- Sondheimer, N.; Lindquist, S. Rnq1: An epigenetic modifier of protein function in yeast. Mol. Cell 2000, 5, 163–172. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Bradley, M.E.; Hong, J.Y.; Liebman, S.W. Prions affect the appearance of other prions: The story of [PIN]. Cell 2001, 106, 171–182. [Google Scholar] [CrossRef]
- Coustou, V.; Deleu, C.; Saupe, S.; Begueret, J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. USA 1997, 94, 9773–9778. [Google Scholar] [CrossRef]
- Saupe, S.J. The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. Sem. Cell Dev. Biol. 2011, 22, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Bradley, M.E.; Zhou, P.; Chernoff, Y.O.; Liebman, S.W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997, 147, 507–519. [Google Scholar]
- Patel, B.K.; Gavin-Smyth, J.; Liebman, S.W. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat. Cell Biol. 2009, 11, 344–349. [Google Scholar] [CrossRef]
- Du, Z.; Park, K.-W.; Yu, H.; Fan, Q.; Li, L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 2008, 40, 460–465. [Google Scholar] [CrossRef]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, G.; Shimazu, N.; Tanaka, M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 2012, 336, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Roberts, B.T.; Wickner, R.B. A class of prions that propagate via covalent auto-activation. Genes Dev. 2003, 17, 2083–2087. [Google Scholar] [CrossRef]
- Chakravarty, A.K.; Smejkal, T.; Itakura, A.K.; Garcia, D.M.; Jarosz, D.F. A non-amyloid prion particle that activates a heritable gene expression program. Mol. Cell 2020, 77, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Chernova, T.A.; Kiktev, D.A.; Romanyuk, A.V.; Shanks, J.R.; Laur, O.; Ali, M.; Ghosh, A.; Kim, D.; Yang, Z.; Mang, M.; et al. Yeast short-lived actin-associated protein forms a metastable prion in response to thermal stress. Cell Rep. 2017, 18, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Taneja, V.; Maddelein, M.L.; Talarek, N.; Saupe, S.J.; Liebman, S.W. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol. Cell 2007, 27, 67–77. [Google Scholar] [CrossRef][Green Version]
- Sengupta, S.; Maji, S.K.; Ghosh, S.K. Evidence fo a prion-like transmission of p53 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2017, 37, e00118-17. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Galkin, A.P.; Lewitin, E.; Chernova, T.A.; Newnam, G.P.; Belenkiy, S.M. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol. Microbiol. 2000, 35, 865–876. [Google Scholar] [CrossRef]
- Santoso, A.; Chien, P.; Osherovich, L.Z.; Weissman, J.S. Molecular basis of a yeast prion species barrier. Cell 2000, 100, 277–288. [Google Scholar] [CrossRef]
- Nakayashiki, T.; Ebihara, K.; Bannai, H.; Nakamura, Y. Yeast [PSI+] “prions” that are crosstransmissible and susceptible beyond a species barrier through a quasi-prion state. Mol. Cell 2001, 7, 1121–1130. [Google Scholar] [CrossRef]
- Bradley, M.E.; Edskes, H.K.; Hong, J.Y.; Wickner, R.B.; Liebman, S.W. Interactions among prions and prion “strains” in yeast. Proc. Natl. Acad. Sci. USA 2002, 99 (Suppl. 4), 16392–16399. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Bruce, K.L.; Newnam, G.P.; Gyoneva, S.; Romanyuk, A.V.; Chernoff, Y.O. Genetic and epigenetic control of the efficiency and fidelity of cross-species prion transmission. Mol. Microbiol. 2010, 76, 1483–1499. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, J.Y.; Liao, T.Y.; Lee, H.C.; King, C.Y. Inter-allelic prion propagation reveals conformational relationships among a multitude of [PSI] strains. PLoS Genet 2011, 7, e1002297. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Newnam, G.P.; Chernoff, Y.O. Prion species barrier between the closely related yeast proteins is detected despite coaggregation. Proc. Natl. Acad. Sci. USA 2007, 104, 2791–2796. [Google Scholar] [CrossRef]
- Edskes, H.K.; McCann, L.M.; Hebert, A.M.; Wickner, R.B. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics 2009, 181, 1159–1167. [Google Scholar] [CrossRef]
- McGlinchey, R.; Kryndushkin, D.; Wickner, R.B. Suicidal [PSI+] is a lethal yeast prion. Proc. Natl. Acad. Sci. USA 2011, 108, 5337–5341. [Google Scholar] [CrossRef]
- Bateman, D.A.; Wickner, R.B. [PSI+] prion transmission barriers protect Saccharomyces cerevisiae from infection: Intraspecies ‘species barriers’. Genetics 2012, 190, 569–579. [Google Scholar] [CrossRef]
- Wickner, R.B.; Beszonov, E.; Bateman, D.A. Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc. Natl. Acad. Sci. USA 2014, 111, E2711–E2720. [Google Scholar] [CrossRef]
- Wickner, R.B.; Son, M.; Edskes, B.K. Prion variants of yeast are numerous, mutable, and segregate on growth, affecting prion pathogenesis, transmission barriers and sensitivity to anti-prioin systems. Viruses 2019, 11, 238. [Google Scholar] [CrossRef]
- Killian, A.N.; Miller, S.C.; Hines, J.K. Impact of amyloid polymorphism on prion-chaperone interactions in yeast. Viruses 2019, 11, 349. [Google Scholar] [CrossRef]
- Sharma, J.; Liebman, S.W. [PSI+] prion variant establishment in yeast. Mol. Microbiol. 2012, 86, 866–881. [Google Scholar] [CrossRef]
- Bateman, D.; Wickner, R.B. The [PSI+] prion exists as a dynamic cloud of variants. PLoS Genet 2013, 9, e1003257. [Google Scholar] [CrossRef] [PubMed]
- Dergalev, A.A.; Alexandrovg, A.; Ivannikov, R.I.; Ter-Avanesyan, M.D.; Kushnirov, V.V. Yeast Sup35 prion structure: Two types, four parts, many variants. Int. J. Mol. Sci. 2019, 20, 2633. [Google Scholar] [CrossRef] [PubMed]
- Tycko, R.; Wickner, R.B. Molecular structures of amyloid and prion fibrils: Consensus vs. controversy. Acc. Chem. Res. 2013, 46, 1487–1496. [Google Scholar] [CrossRef] [PubMed]
- Benzinger, T.L.; Gregory, D.M.; Burkoth, T.S.; Miller-Auer, H.; Lynn, D.G.; Botto, R.E.; Meredith, S.C. Propagating structure of Alzheimer’s beta-amyloid(10-35) is parallel beta-sheet with residues in exact register. Proc. Natl. Acad. Sci. USA 1998, 95, 13407–13412. [Google Scholar] [CrossRef] [PubMed]
- Shewmaker, F.; Wickner, R.B.; Tycko, R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl. Acad. Sci. USA 2006, 103, 19754–19759. [Google Scholar] [CrossRef] [PubMed]
- Baxa, U.; Wickner, R.B.; Steven, A.C.; Anderson, D.; Marekov, L.; Yau, W.-M.; Tycko, R. Characterization of β-sheet structure in Ure2p1-89 yeast prion fibrils by solid state nuclear magnetic resonance. Biochemistry 2007, 46, 13149–13162. [Google Scholar] [CrossRef]
- Wickner, R.B.; Dyda, F.; Tycko, R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register β-sheet structure. Proc. Natl. Acad. Sci. USA 2008, 105, 2403–2408. [Google Scholar] [CrossRef]
- Shewmaker, F.; Kryndushkin, D.; Chen, B.; Tycko, R.; Wickner, R.B. Two prion variants of Sup35p have in-register β-sheet structures, independent of hydration. Biochemistry 2009, 48, 5074–5082. [Google Scholar] [CrossRef]
- Gorkovskiy, A.; Thurber, K.R.; Tycko, R.; Wickner, R.B. Locating folds of the in-register parallel β-sheet of the Sup35p prion domain infectious amyloid. Proc. Natl. Acad. Sci. USA 2014, 111, E4615–E4622. [Google Scholar] [CrossRef]
- Ohhashi, Y.; Yamaguchi, Y.; Kurahashi, H.; Kamatari, Y.O.; Sugiyama, S.; Uluca, B.; Piechatzek, T.; Komi, Y.; Shida, T.; Muller, H.; et al. Molecular basis for diversification of yeast prion strain conformation. Proc. Natl. Acad. Sci. USA 2018, 115, 2389–2394. [Google Scholar] [CrossRef]
- Ngo, S.; Chiang, V.; Guo, Z. Quantitative analysis of spin exchange interactions to identify β strand and turn regions in Ure2 prion domain fibrils with site-directed spin labeling. J. Struct. Biol. 2012, 180, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.; Gu, L.; Guo, Z. Hierarchical organization in the amyloid core of yeast prion protein Ure2. J. Biol. Chem. 2011, 286, 29691–29699. [Google Scholar] [CrossRef] [PubMed]
- Baxa, U.; Taylor, K.L.; Wall, J.S.; Simon, M.N.; Cheng, N.; Wickner, R.B.; Steven, A. Architecture of Ure2p prion filaments: The N-terminal domain forms a central core fiber. J. Biol. Chem. 2003, 278, 43717–43727. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Avalos, R.; King, C.Y.; Wall, J.S.; Simon, M.; Caspar, D.L.D. Strain-specific morphologies of yeast prion amyloids. Proc. Natl. Acad. Sci. USA 2005, 102, 10165–10170. [Google Scholar] [CrossRef]
- Chen, B.; Thurber, K.R.; Shewmaker, F.; Wickner, R.B.; Tycko, R. Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc. Natl. Acad. Sci. USA 2009, 106, 14339–14344. [Google Scholar] [CrossRef]
- Ross, E.D.; Baxa, U.; Wickner, R.B. Scrambled prion domains form prions and amyloid. Mol. Cell. Biol. 2004, 24, 7206–7213. [Google Scholar] [CrossRef]
- Ross, E.D.; Edskes, H.K.; Terry, M.J.; Wickner, R.B. Primary sequence independence for prion formation. Proc. Natl. Acad. Sci. USA 2005, 102, 12825–12830. [Google Scholar] [CrossRef]
- Ross, E.D.; Minton, A.P.; Wickner, R.B. Prion domains: Sequences, structures and interactions. Nat. Cell Biol. 2005, 7, 1039–1044. [Google Scholar] [CrossRef]
- Wong, S.-H.; King, C.-Y. Amino acid proximities in two Sup35 prion strains revealed by chemical cross-linking. J. Biol. Chem. 2015, 290, 25062–25071. [Google Scholar] [CrossRef]
- Wickner, R.B.; Edskes, H.K.; Shewmaker, F.; Nakayashiki, T. Prions of fungi: Inherited structures and biological roles. Nat. Rev. Microbiol. 2007, 5, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Schwimmer, C.; Masison, D.C. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 2002, 22, 3590–3598. [Google Scholar] [CrossRef] [PubMed]
- Franzmann, T.M.; Jahnel, M.; Pozniakovsky, A.; Mahamid, J.; Holehouse, A.S.; Nuske, E.; Richter, D.; Baumeister, W.; Grill, S.W.; Pappu, R.V.; et al. Phase separation of a yeast protein promotes cellular fitness. Science 2018, 359, eaao5654. [Google Scholar] [CrossRef]
- Nakayashiki, T.; Kurtzman, C.P.; Edskes, H.K.; Wickner, R.B. Yeast prions [URE3] and [PSI+] are diseases. Proc. Natl. Acad. Sci. USA 2005, 102, 10575–10580. [Google Scholar] [CrossRef] [PubMed]
- Futcher, A.B.; Cox, B.S. Maintenance of the 2 m circle plasmid in populations of Saccharomyces cerevisiae. J. Bacteriol. 1983, 154, 612–622. [Google Scholar] [CrossRef]
- Mead, D.J.; Gardner, D.C.J.; Oliver, S.G. The yeast 2 plasmid: Strategies for the survival of a selfish DNA. Mol. Gen. Genet. 1986, 205, 417–421. [Google Scholar] [CrossRef]
- Futcher, B.; Reid, E.; Hickey, D.A. Maintenance of the 2 micron circle plasmid of Saccharomyces cerevisiae by sexual transmission: An example of selfish DNA. Genetics 1988, 118, 411–415. [Google Scholar]
- Kelly, A.C.; Shewmaker, F.P.; Kryndushkin, D.; Wickner, R.B. Sex, prions and plasmids in yeast. Proc. Natl. Acad. Sci. USA 2012, 109, E2683–E2690. [Google Scholar] [CrossRef]
- Debets, A.J.; Dalstra, H.J.; Slakhorst, M.; Koopmanschap, B.; Hoekstra, R.F.; Saupe, S.J. High natural prevalence of a fungal prion. Proc. Natl. Acad. Sci. USA 2012, 109, 10432–10437. [Google Scholar] [CrossRef]
- Ritter, C.; Maddelein, M.L.; Siemer, A.B.; Luhrs, T.; Ernst, M.; Meier, B.H.; Saupe, S.J.; Riek, R. Correlation of structural elements and infectivity of the HET-s prion. Nature 2005, 435, 844–848. [Google Scholar] [CrossRef]
- Wasmer, C.; Lange, A.; van Melckebeke, H.; Siemer, A.B.; Riek, R.; Meier, B.H. Amyloid fibrils of the HET-s(218-279) prion form a beta solenoid with a triangular hydrophobic core. Science 2008, 319, 1523–1526. [Google Scholar] [CrossRef] [PubMed]
- Vishveshwara, N.; Bradley, M.E.; Liebman, S.W. Sequestration of essential proteins causes prion associated toxicity in yeast. Mol. Microbiol. 2009, 73, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Treusch, S.; Lindquist, S. An intrinsically disordered yeast prion arrests the cell cycle by sequestering a spindle pole body component. J. Cell Biol. 2012, 197, 369–379. [Google Scholar] [CrossRef] [PubMed]
- King, C.Y. Supporting the structural basis of prion strains: Induction and identification of [PSI] variants. J Mol. Biol. 2001, 307, 1247–1260. [Google Scholar] [CrossRef]
- Shewmaker, F.; Mull, L.; Nakayashiki, T.; Masison, D.C.; Wickner, R.B. Ure2p function is enhanced by its prion domain in Saccharomyces cerevisiae. Genetics 2007, 176, 1557–1565. [Google Scholar] [CrossRef]
- Hosoda, N.; Kobayashii, T.; Uchida, N.; Funakoshi, Y.; Kikuchi, Y.; Hoshino, S.; Katada, T. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 2003, 278, 38287–38291. [Google Scholar] [CrossRef]
- Wickner, R.B.; Kelly, A.C. Prions are affected by evolution at two levels. Cell. Mol. Life Sci. 2016, 73, 1131–1144. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Lindquist, S.L.; Ono, B.-I.; Inge-Vechtomov, S.G.; Liebman, S.W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 1995, 268, 880–884. [Google Scholar] [CrossRef]
- Glover, J.R.; Lindquist, S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 1998, 94, 73–82. [Google Scholar] [CrossRef]
- Jung, G.; Jones, G.; Wegrzyn, R.D.; Masison, D.C. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 2000, 156, 559–570. [Google Scholar]
- Jung, G.; Jones, G.; Masison, D.C. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc. Natl. Acad. Sci. USA 2002, 99, 9936–9941. [Google Scholar] [CrossRef]
- Ness, F.; Ferreira, P.; Cox, B.S.; Tuite, M.F. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 2002, 22, 5593–5605. [Google Scholar] [CrossRef]
- Inoue, Y.; Taguchi, H.; Kishimoto, A.; Yoshida, M. Hsp104 binds to yeast Sup35 prion fiber but needs other factor(s) to sever it. J. Biol. Chem. 2004, 279, 52319–52323. [Google Scholar] [CrossRef] [PubMed]
- Higurashi, T.; Hines, J.K.; Sahi, C.; Aron, R.; Craig, E.A. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. USA 2008, 105, 16596–16601. [Google Scholar] [CrossRef]
- Reidy, M.; Miot, M.; Masison, D.C. Prokaryotic chaperones support yeast prions and thermotolerance and define disaggregation machinery interactions. Genetics 2012, 192, 185–193. [Google Scholar] [CrossRef]
- Winkler, J.; Tyedmers, J.; Bukau, B.; Mogk, A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 2012, 198, 387–404. [Google Scholar] [CrossRef]
- Troisi, E.M.; Rockman, M.E.; Nguyen, P.P.; Oliver, E.E.; Hines, J.K. Swa2, the yeast homolog of mammalian auxilin, is specifically required for the propagation of the prion variant [URE3-1]. Mol. Microbiol. 2015, 97, 926–941. [Google Scholar] [CrossRef]
- Lum, R.; Tkach, J.M.; Vierling, E.; Glover, J.R. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 2004, 279, 29139–29146. [Google Scholar] [CrossRef] [PubMed]
- Hung, G.C.; Masison, D.C. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics 2006, 173, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Tessarz, P.; Mogk, A.; Bukau, B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol. Microbiol. 2008, 68, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Kummer, E.; Szlachcic, A.; Franke, K.B.; Ungelenk, S.; Bukau, B.; Mogk, A. Bacterial and yeast AAA+ disaggregases ClpB and Hsp104 operate through conserved mechanism involving cooperation with Hsp70. J. Mol. Biol. 2016, 428, 4378–4391. [Google Scholar] [CrossRef]
- Jones, G.; Song, Y.; Chung, S.; Masison, D.C. Propagation of yeast [PSI+] prion impaired by factors that regulate Hsp70 substrate binding. Mol. Cell. Biol. 2004, 24, 3928–3937. [Google Scholar] [CrossRef][Green Version]
- Sharma, D.; Masison, D.C. Hsp70 structure, function, regulation and influence on yeast prions. Prot. Pept. Lett. 2009, 16, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Chernoff, Y.O.; Ono, B.-I. Dosage-dependent modifiers of PSI-dependent omnipotent suppression in yeast. In Protein Synthesis and Targeting in Yeast; Brown, A.J.P., Tuite, M.F., McCarthy, J.E.G., Eds.; Springer: Berlin, Germany, 1992; pp. 101–107. [Google Scholar]
- Reidy, M.; Masison, D.C. Sti1 regulation of Hsp70 and Hsp90 is critical for curing of Saccharomyces cerevisiae [PSI+] prions by Hsp104. Mol. Cell. Biol. 2010, 30, 3542–3552. [Google Scholar] [CrossRef] [PubMed]
- Moosavi, B.; Wongwigkam, J.; Tuite, M.F. Hsp70/Hsp90 co-chaperones are required for efficient Hsp104-mediated elimination of the yeast [PSI+] prion but not for prion propagation. Yeast 2010, 27, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Helsen, C.W.; Glover, J.R. Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104). J. Biol. Chem. 2012, 287, 542–556. [Google Scholar] [CrossRef]
- Tuite, M.F.; Mundy, C.R.; Cox, B.S. Agents that cause a high frequency of genetic change from [psi+] to [psi-] in Saccharomyces cerevisiae. Genetics 1981, 98, 691–711. [Google Scholar]
- Newnam, G.P.; Birchmore, J.L.; Chernoff, Y.O. Destabilization and recovery of a yeast prion after mild heat shock. J. Mol. Biol. 2011, 408, 432–448. [Google Scholar] [CrossRef]
- Howie, R.L.; Jay-Garcia, L.M.; Kiktev, D.A.; Faber, Q.L.; Murphy, M.; Rees, K.A.; Sachwani, N.; Chernoff, Y.O. Role of the cell asymmetry apparatus and ribosome-associated chaperones in the destabilization of a Saccharomyces cerevisiae prion by heat shock. Genetics 2019, 212, 757–771. [Google Scholar] [CrossRef]
- Aguilaniu, H.; Gustafsson, L.; Gigoulet, M.; Nystrom, T. Assymetric inheritance of oxidatively damaged proteins during cytokinesis. Science 2003, 299, 1751–1753. [Google Scholar] [CrossRef]
- Liu, B.; Larsson, L.; Caballero, A.; Hao, X.; Oling, D.; Grantham, J.; Nystrom, T. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 2010, 140, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, D.L.; Dobson, C.M.; Rachubinski, R.A. Chaperone proteins select and maintain [PIN+] prion conformations in Saccharomyces cerevisiae. J. Biol. Chem. 2013, 288, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Gaur, D.; Gupta, A.; Puri, A.; Sharma, D. Hsp90-associated immunophilin homolog Cpr7 is required for the mitotic stability of [URE3] prion in Saccharomyces cerevisiae. PLoS Genet 2015, 11, e1005567. [Google Scholar] [CrossRef] [PubMed]
- Oliver, E.E.; Troisi, E.M.; Hines, J.K. Prion-specific Hsp40 function: The role of the auxilin homolog Swa2. Prion 2017, 11, 174–185. [Google Scholar] [CrossRef]
- Berger, S.E.; Nolte, A.N.; Kamiya, E.; Hines, J.K. Three J-proteins impact Hsp104-mediated variant-specific protein elimination: A new critical role for a low-complexity domain. Curr. Genet. 2019, 66, 51–58. [Google Scholar] [CrossRef]
- Reidy, M.; Sharma, R.; Shastry, S.; Roberts, B.L.; Albino-Flores, I.; Wickner, S.; Masison, D.C. Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines. PLoS Genet 2014, 10, e1004720. [Google Scholar] [CrossRef]
- Hines, J.K.; Li, X.; Du, Z.; Higurashi, T.; Li, L.; Craig, E.A. [SWI], the prion formed by the chromatin remodeling factor Swi1, is highly sensitive to alterations in Hsp70 chaperone system activity. PLoS Genet 2011, 7, e1001309. [Google Scholar] [CrossRef]
- Kirkland, P.A.; Reidy, M.; Masison, D.C. Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics 2011, 188, 565–577. [Google Scholar] [CrossRef]
- Astor, M.T.; Kamiya, E.; Sporn, Z.A.; Berger, S.E.; Hines, J.K. Variant-specific and reciprocal Hsp40 functions in Hsp104-mediated prion elimination. Mol. Microbiol. 2018, 109, 41–62. [Google Scholar] [CrossRef]
- Nelson, R.J.; Ziegilhoffer, T.; Nicolet, C.; Werner-Washburne, M.; Craig, E.A. The translation machinery and 70 kDal heat shock protein cooperate in protein synthesis. Cell 1992, 71, 97–105. [Google Scholar] [CrossRef]
- Pfund, C.; Lopez-Hoyo, N.; Ziegelhoffer, T.; Schilke, B.A.; Lopez-Buesa, P.; Walter, W.A.; Wiedmann, M.; Craig, E.A. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 1998, 17, 3981–3989. [Google Scholar] [CrossRef]
- Koplin, A.; Preissler, S.; Ilina, Y.; Kock, M.; Scior, A.; Erhardt, M.; Deuerling, E. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 2010, 189, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Chernoff, Y.O.; Kiktev, D.A. Dual role of ribosome-associated chaperones in prion formation and propagation. Curr. Genet. 2016, 62, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Chernoff, Y.O.; Newnam, G.P.; Kumar, J.; Allen, K.; Zink, A.D. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone Ssb in formation, stability and toxicity of the [PSI+] prion. Mol. Cell. Biol. 1999, 19, 8103–8112. [Google Scholar] [CrossRef] [PubMed]
- Kiktev, D.A.; Melomed, M.M.; Lu, C.D.; Newnam, G.P.; Chernoff, Y.O. Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol. Microbiol. 2015, 96, 621–632. [Google Scholar] [CrossRef]
- Amor, A.J.; Castanzo, D.T.; Delany, S.P.; Selechnik, D.M.; van Ooy, A.; Cameron, D.M. The ribosome-associated complex antagonizes prion formation in yeast. Prion 2015, 9, 144–164. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Kryndushkin, D.; Boguta, M.; Smirnov, V.N.; Ter-Avanesyan, M.D. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 2000, 10, 1443–1446. [Google Scholar] [CrossRef]
- Cachinska, A.; Szczesniak, B.; Kochneva-Pervukhova, N.V.; Kushnirov, V.V.; Ter-Avanesyan, M.D.; Boguta, M. Ssb1 chaperone is a [PSI+] prion-curing factor. Curr. Genet. 2001, 39, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Kryndushkin, D.; Shewmaker, F.; Wickner, R.B. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 2008, 27, 2725–2735. [Google Scholar] [CrossRef]
- Kanneganti, V.; Kama, R.; Gerst, J.E. Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast. Mol. Biol. Cell 2011, 22, 1648–1663. [Google Scholar] [CrossRef]
- Malinovska, L.; Kroschwald, S.; Munder, M.C.; Richter, D.; Alberti, S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spaciotemporal distribution of protein aggregates. Mol. Biol. Cell 2012, 23, 3041–3056. [Google Scholar] [CrossRef] [PubMed]
- Kryndushkin, D.; Ihrke, G.; Piermartiri, T.C.; Shewmaker, F. A yeast model of optineurin proteinopathy reveals a unique aggregation pattern associated with cellular toxicity. Mol. Microbiol. 2012, 86, 1531–1547. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.T.; Grousi, T.; Shatz, O.; Jawed, A.; Ruger-Herreros, C.; Semmelink, M.; Zahn, R.; Richter, K.; Bukau, B.; Mogk, A. Cellular sequestrases maintain basal Hsp70 capacity ensuring balanced proteostasis. Nat. Commun. 2019, 10, 4851. [Google Scholar] [CrossRef]
- Kama, R.; Robinson, M.; Gerst, J.E. Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast. Mol. Cell. Biol. 2007, 27, 605–621. [Google Scholar] [CrossRef] [PubMed]
- Olenick, M.A.; Holzbaur, E.L.F. Dynein activator and adaptor at a glance. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef]
- Specht, S.; Miller, S.B.M.; Mogk, A.; Bukau, B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J. Cell Biol. 2011, 195, 617–629. [Google Scholar] [CrossRef]
- Miller, S.B.; Ho, C.T.; Winkler, J.; Khokhrina, M.; Neuner, A.; Mohamed, M.Y.; Guilbride, D.L.; Richter, K.; Lisby, M.; Scheibel, E.; et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 2015, 34, 778–797. [Google Scholar] [CrossRef]
- Lopez, N.; Aron, R.; Craig, E.A. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+]. Mol. Biol. Cell 2003, 14, 1172–1181. [Google Scholar] [CrossRef]
- Barbitoff, Y.A.; Matveenko, A.G.; Moskalnko, S.E.; Zemlyanko, O.M.; Newnam, G.P.; Patel, A.; Chernova, T.A.; Chernoff, Y.O.; Zhouravleva, G.A. To CURe or not to CURe? Differential effects of the chaperone sorting factor Cur1 on yeast prions are mediated by the chaperone Sis1. Mol. Microbiol. 2017, 105, 242–257. [Google Scholar] [CrossRef]
- Gorkovskiy, A.; Reidy, M.; Masison, D.C.; Wickner, R.B. Hsp104 at normal levels cures many [PSI+] variants in a process promoted by Sti1p, Hsp90 and Sis1p. Proc. Natl. Acad. Sci. USA 2017, 114, E4193–E4202. [Google Scholar] [CrossRef]
- Wickner, R.B.; Kelly, A.C.; Bezsonov, E.E.; Edskes, H.E. Prion propagation is controlled by inositol polyphosphates. Proc. Natl. Acad. Sci. USA 2017, 114, E8402–E8410. [Google Scholar] [CrossRef] [PubMed]
- Steidle, E.A.; Chong, L.S.; Wu, M.; Crooke, E.; Fiedler, D.; Resnick, A.C.; Rolfes, R.J. A novel inositol pyrophosphate phosphatase in Saccharomyces cerevisiae: Siw14 protein selectively cleaves the β-phosphate from 5-diphosphoinositol pentakisphosphate (5PP-IP5). J. Biol. Chem. 2016, 291, 6772–6783. [Google Scholar] [CrossRef]
- Tsui, M.M.; York, J.D. Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv. Biol. Regul. 2010, 50, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Worley, J.; Luo, X.; Capaldi, A.P. Inositol pyrophosphates regulate cell growth and the environmental stress response by activating the HDAC Rpd3L. Cell Rep. 2013, 3, 1476–1482. [Google Scholar] [CrossRef] [PubMed]
- Steidle, E.A.; Morrisette, V.A.; Fujimaki, K.; Chong, L.; Resnick, A.C.; Capaldi, A.P.; Rolfes, R.J. The InsP7 phosphatase Siw14 regulates inositol pyrophosphate levels to control localization of the general stress response transcription factor Msn2. J. Biol. Chem. 2020, 295, 2043–2056. [Google Scholar] [CrossRef]
- Wu, M.; Chong, L.S.; Perlman, D.H.; Resnick, A.C.; Fiedler, D. Inositol polyphosphates intersect with signaling and metabolic networks via two distinct mechanisms. Proc. Natl. Acad. Sci. USA 2016, 113, E6757–E6765. [Google Scholar] [CrossRef]
- Son, M.; Wickner, R.B. Nonsense-mediated mRNA decay factors cure most [PSI+] prion variants. Proc. Natl. Acad. Sci. USA 2018, 115, E1184–E1193. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Jacobson, A. Nonsense-mediated mRNA decay: Degradation of defective transcripts is only part of the story. Annu. Rev. Genet. 2015, 49, 339–366. [Google Scholar] [CrossRef] [PubMed]
- Czaplinski, K.; Ruiz-Echevarria, M.J.; Paushkin, S.V.; Han, X.; Weng, Y.; Perlick, H.A.; Dietz, H.C.; Ter-Avanesyan, M.D.; Peltz, S.W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998, 12, 1665–1677. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.M.; Nguyen, P.P.; Patel, M.J.; Sporn, Z.A.; Hines, J.K. Functional diversification of Hsp40: Distinct J-protein functional requirements for two prions allow for chaperone-dependent prion selection. PLoS Genet 2014, 10, e41004510. [Google Scholar] [CrossRef][Green Version]
- Edskes, H.E.; Mukhamedova, M.; Edskes, B.K.; Wickner, R.B. Hermes transposon mutagenesis shows [URE3] prion pathology prevented by a ubiquitin-targeting protein: Evidence for carbon/nitrogen assimilation cross-talk and a second function for Ure2p. Genetics 2018, 209, 789–800. [Google Scholar] [PubMed]
- Seol, J.H.; Shevchenko, A.; Shevchenko, A.; Deshales, R.J. Skip1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 2001, 3, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Sarikas, A.; Hartmann, T.; Pan, Z.Q. The cullin protein family. Genome Biol. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, S.; Maytham, E.G.; Pota, H.; Grizenkova, J.; Molou, E.; Uphill, J.; Hummerich, H.; Whitfield, J.; Alpers, M.P.; Mead, S.; et al. HECTD2 is associated with susceptibility to mouse and human prion disease. PLoS Genet. 2009, 5, e1000383. [Google Scholar] [CrossRef]
- Shimura, H.; Hattori, N.; Kubo, S.; Mizuno, Y.; Asakawa, S.; Minoshima, S.; Shimizu, N.; Iwai, K.; Chiba, T.; Tanaka, K.; et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 2000, 25, 302–305. [Google Scholar] [CrossRef]
- Erjavec, N.; Larsson, L.; Grantham, J.; Nystrom, T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007, 21, 2410–2421. [Google Scholar] [CrossRef]
- Johnston, J.A.; Ward, C.L.; Kopito, R.R. Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 1999, 143, 1883–1898. [Google Scholar] [CrossRef]
- Mays, C.E.; Armijo, E.; Morales, R.; Telling, G.C.; Pandita, T.K.; Hunt, C.R.; Soto, C. Prion disease is accelerated in mice lacking stress-induced heat shock protein 70 (HSP70). J. Biol. Chem. 2019, 294, 13619–13628. [Google Scholar] [CrossRef]

| Prion | Prion Protein | Prion Phenotype | Normal Protein Function | Reference |
|---|---|---|---|---|
| [URE3] | Ure2p | Derepressed genes for using poor N-sources in presence of a good N-source; slow growth | repression of genes for utilizing poor N-sources in presence of a good N-source | [7] |
| [PSI+] | Sup35p | Readthrough of termination codons; slow growth; death | translation termination | [7] |
| [PIN+] or [RNQ+] | Rnq1p | Rare generation (by cross-seeding) of [PSI+] or [URE3] | none known | [28] |
| [OCT+] | Cyc8p | Slow growth; impaired mating and sporulatiion | transcription repressor subunit | [29] |
| [SWI+] | Swi1p | Poor growth on raffinose, galactose or glycerol | chromatin remodeling subunit | [30] |
| [MOT+] | Mot3p | Inappropriate derepression of anaerobic genes; colony polymorphisms | transcription regulator | [31] |
| [MOD+] | Mod5p | Partial azole-resistance; slow growth | tRNA isopentenyltransferase | [32] |
| [BETA] | Prb1p | Active protease B (non-amyloid prion) * | Active protease B (this is a functional prion) | [33] |
| [SMAUG+] | Vts1p | Increased mRNA decay * | stimulates mRNA degradation | [34] |
| [Het-s] | HET-s | Heterokaryon incompatibility | Heterokaryon incompatibility (this is a functional prion) | [26] |
| [LSB+] | Lsb2p | Transient Pin activity ([PSI+] prion generation) | Inhibitor of actin filament nucleation | [35] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wickner, R.B.; Edskes, H.K.; Son, M.; Wu, S.; Niznikiewicz, M. How Do Yeast Cells Contend with Prions? Int. J. Mol. Sci. 2020, 21, 4742. https://doi.org/10.3390/ijms21134742
Wickner RB, Edskes HK, Son M, Wu S, Niznikiewicz M. How Do Yeast Cells Contend with Prions? International Journal of Molecular Sciences. 2020; 21(13):4742. https://doi.org/10.3390/ijms21134742
Chicago/Turabian StyleWickner, Reed B., Herman K. Edskes, Moonil Son, Songsong Wu, and Madaleine Niznikiewicz. 2020. "How Do Yeast Cells Contend with Prions?" International Journal of Molecular Sciences 21, no. 13: 4742. https://doi.org/10.3390/ijms21134742
APA StyleWickner, R. B., Edskes, H. K., Son, M., Wu, S., & Niznikiewicz, M. (2020). How Do Yeast Cells Contend with Prions? International Journal of Molecular Sciences, 21(13), 4742. https://doi.org/10.3390/ijms21134742





