BMP-Induced MicroRNA-101 Expression Regulates Vascular Smooth Muscle Cell Migration
Abstract
:1. Introduction
2. Results
2.1. miR-101 Affects Functions of VSMCs
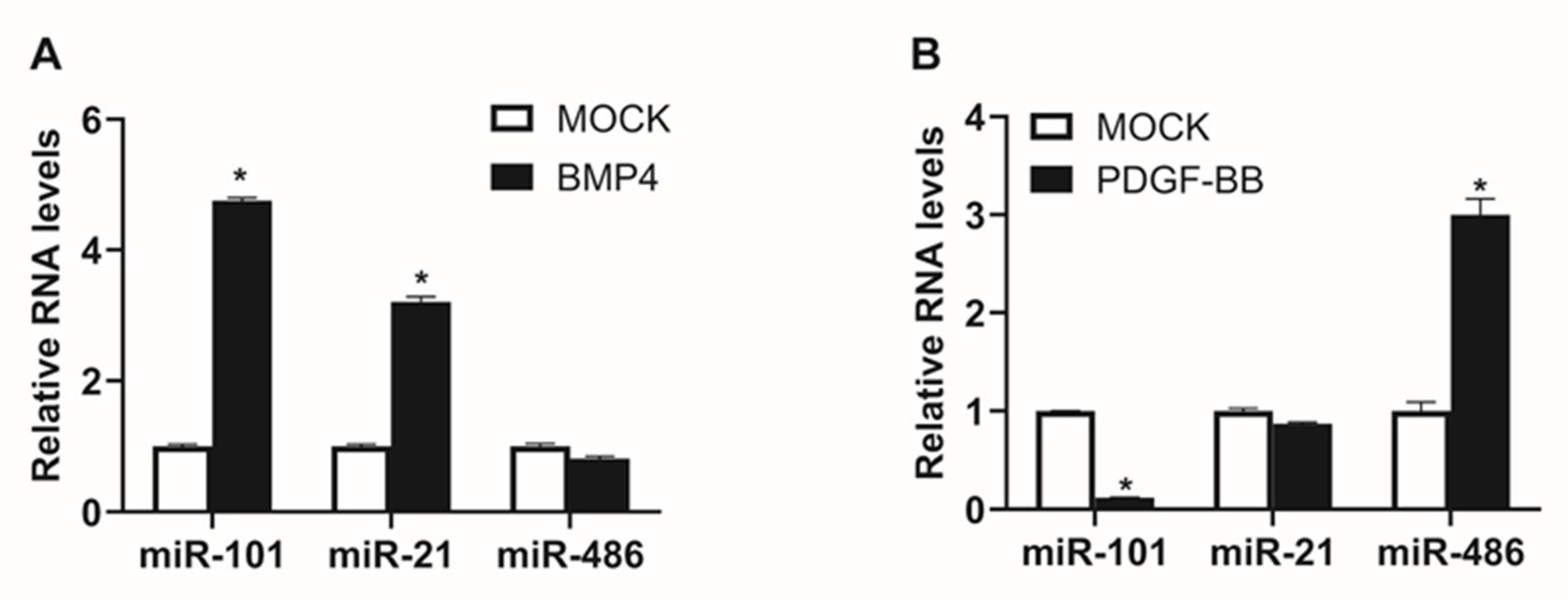
2.2. Expression of miR-101 is Induced by BMP Signaling
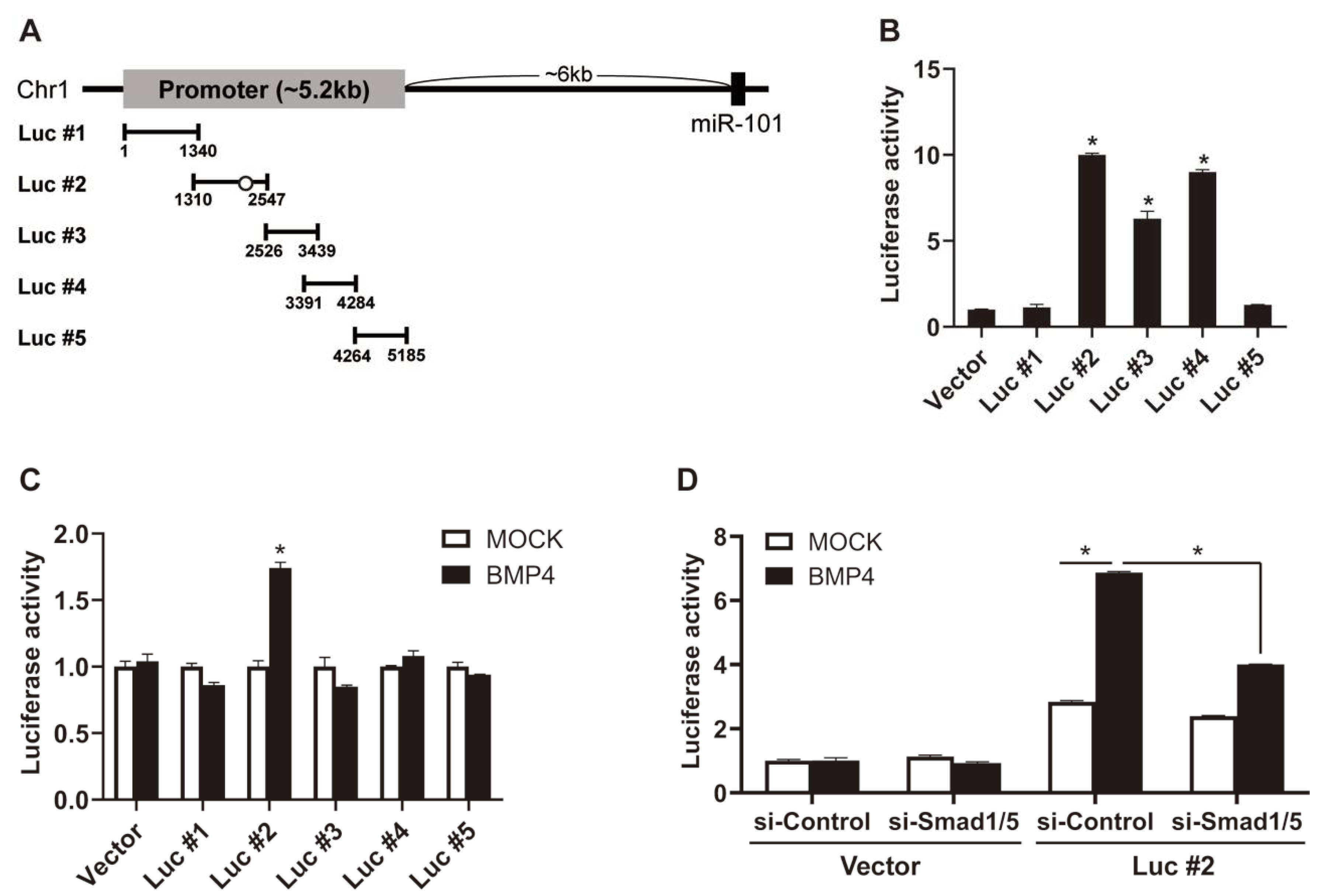
2.3. BMP Signaling Induces miR-101 Expression in a Smad-Dependent Manner
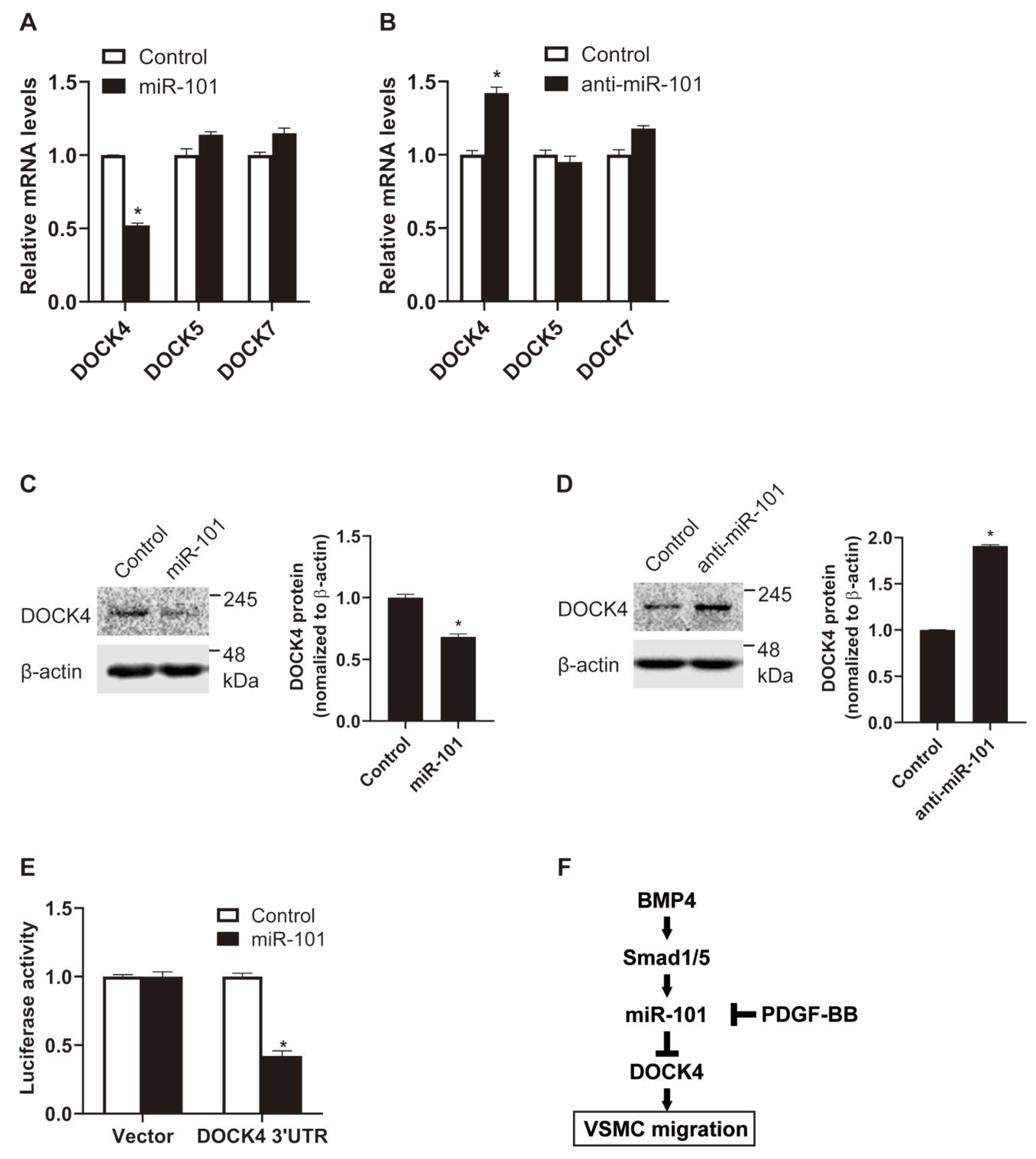
2.4. DOCK4 Is a Novel Target of miR-101
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Reverse Transcription Quatitative PCR (qRT-PCR)
4.3. miRNA Mimics and Anti-miRNA Oligonucleotides
4.4. RNA Interference
4.5. Immunoblotting
4.6. Cell Proliferation Assay
4.7. In Vitro Scratch Wound Assay
4.8. Luciferase Reporter Constructs
4.9. Luciferase Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Small, E.M.; Frost, R.J.; Olson, E.N. MicroRNAs add a new dimension to cardiovascular disease. Circulation 2010, 121, 1022–1032. [Google Scholar] [CrossRef] [Green Version]
- Boettger, T.; Beetz, N.; Kostin, S.; Schneider, J.; Kruger, M.; Hein, L.; Braun, T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J. Clin. Investig. 2009, 119, 2634–2647. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Liu, X.; Yang, J.; Lin, Y.; Xu, D.Z.; Lu, Q.; Deitch, E.A.; Huo, Y.; Delphin, E.S.; Zhang, C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ. Res. 2009, 105, 158–166. [Google Scholar] [CrossRef]
- Ji, R.; Cheng, Y.; Yue, J.; Yang, J.; Liu, X.; Chen, H.; Dean, D.B.; Zhang, C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ. Res. 2007, 100, 1579–1588. [Google Scholar] [CrossRef]
- Xin, M.; Small, E.M.; Sutherland, L.B.; Qi, X.; McAnally, J.; Plato, C.F.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009, 23, 2166–2178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, R.; Mani, R.S.; Russo, N.; Scanlon, C.S.; Tsodikov, A.; Jing, X.; Cao, Q.; Palanisamy, N.; Metwally, T.; Inglehart, R.C.; et al. The tumor suppressor gene rap1GAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene 2011, 30, 4339–4349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, H.M.; Jeon, H.S.; Lee, S.Y.; Jeong, K.J.; Park, S.Y.; Lee, H.Y.; Lee, J.U.; Kim, J.H.; Kwon, S.J.; Choi, E.; et al. microRNA-101 inhibits lung cancer invasion through the regulation of enhancer of zeste homolog 2. Exp. Ther. Med. 2011, 2, 963–967. [Google Scholar] [CrossRef] [Green Version]
- Kottakis, F.; Polytarchou, C.; Foltopoulou, P.; Sanidas, I.; Kampranis, S.C.; Tsichlis, P.N. FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol. Cell 2011, 43, 285–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, Q.; Shen, F.; Wu, J.; Zhang, W.; Wang, J.; Zhang, L. MiR-101, downregulated in retinoblastoma, functions as a tumor suppressor in human retinoblastoma cells by targeting EZH2. Oncol. Rep. 2014, 32, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Lv, P.; Zhang, P.; Li, X.; Chen, Y. Micro ribonucleic acid (RNA)-101 inhibits cell proliferation and invasion of lung cancer by regulating cyclooxygenase-2. Thorac. Cancer 2015, 6, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Zheng, C.; Bai, E.; Yang, K. miR-101 inhibits glioma cell invasion via the downregulation of COX-2. Oncol. Lett. 2016, 12, 2538–2544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smits, M.; Nilsson, J.; Mir, S.E.; van der Stoop, P.M.; Hulleman, E.; Niers, J.M.; de Witt Hamer, P.C.; Marquez, V.E.; Cloos, J.; Krichevsky, A.M.; et al. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget 2010, 1, 710–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strillacci, A.; Griffoni, C.; Sansone, P.; Paterini, P.; Piazzi, G.; Lazzarini, G.; Spisni, E.; Pantaleo, M.A.; Biasco, G.; Tomasi, V. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp. Cell Res. 2009, 315, 1439–1447. [Google Scholar] [CrossRef]
- Varambally, S.; Cao, Q.; Mani, R.S.; Shankar, S.; Wang, X.; Ateeq, B.; Laxman, B.; Cao, X.; Jing, X.; Ramnarayanan, K.; et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008, 322, 1695–1699. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Schulz, R.; Edmunds, S.; Kruger, E.; Markert, E.; Gaedcke, J.; Cormet-Boyaka, E.; Ghadimi, M.; Beissbarth, T.; Levine, A.J.; et al. MicroRNA-101 Suppresses Tumor Cell Proliferation by Acting as an Endogenous Proteasome Inhibitor via Targeting the Proteasome Assembly Factor POMP. Mol. Cell 2015, 59, 243–257. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhuang, L.; Rong, H.; Guo, Y.; Ling, X.; Wang, R.; Yu, X.; Zhang, W. MicroRNA-101 inhibits proliferation of pulmonary microvascular endothelial cells in a rat model of hepatopulmonary syndrome by targeting the JAK2/STAT3 signaling pathway. Mol. Med. Rep. 2015, 12, 8261–8267. [Google Scholar] [CrossRef] [Green Version]
- Waldo, K.L.; Hutson, M.R.; Ward, C.C.; Zdanowicz, M.; Stadt, H.A.; Kumiski, D.; Abu-Issa, R.; Kirby, M.L. Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev. Biol. 2005, 281, 78–90. [Google Scholar] [CrossRef] [Green Version]
- Owens, G.K.; Kumar, M.S.; Wamhoff, B.R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004, 84, 767–801. [Google Scholar] [CrossRef]
- Beamish, J.A.; He, P.; Kottke-Marchant, K.; Marchant, R.E. Molecular regulation of contractile smooth muscle cell phenotype: Implications for vascular tissue engineering. Tissue Eng. Part B Rev. 2010, 16, 467–491. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.; Davis-Dusenbery, B.N.; Nguyen, P.H.; Lal, A.; Lieberman, J.; Van Aelst, L.; Lagna, G.; Hata, A. Bone morphogenetic protein 4 promotes vascular smooth muscle contractility by activating microRNA-21 (miR-21), which down-regulates expression of family of dedicator of cytokinesis (DOCK) proteins. J. Biol. Chem. 2012, 287, 3976–3986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.; Louie, J.; Weisman, A.; Sheu-Gruttadauria, J.; Davis-Dusenbery, B.N.; Lagna, G.; Hata, A. Inhibition of microRNA-302 (miR-302) by bone morphogenetic protein 4 (BMP4) facilitates the BMP signaling pathway. J. Biol. Chem. 2012, 287, 38656–38664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Hata, A.; Kang, H. Down-regulation of miR-96 by bone morphogenetic protein signaling is critical for vascular smooth muscle cell phenotype modulation. J. Cell. Biochem. 2014, 115, 889–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Heo, J.; Kang, H. miR-92b-3p-TSC1 axis is critical for mTOR signaling-mediated vascular smooth muscle cell proliferation induced by hypoxia. Cell Death Differ. 2019, 26, 1782–1795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Kang, H. Hypoxia Promotes Vascular Smooth Muscle Cell Proliferation through microRNA-Mediated Suppression of Cyclin-Dependent Kinase Inhibitors. Cells 2019, 8, 802. [Google Scholar] [CrossRef] [Green Version]
- Seong, M.; Kang, H. Hypoxia-induced miR-1260b regulates vascular smooth muscle cell proliferation by targeting GDF11. BMB Rep. 2020, 53, 206–211. [Google Scholar] [CrossRef]
- Heo, J.; Yang, H.C.; Rhee, W.J.; Kang, H. Vascular Smooth Muscle Cell-Derived Exosomal MicroRNAs Regulate Endothelial Cell Migration Under PDGF Stimulation. Cells 2020, 9, 639. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Fan, W.; Wang, X.; Ke, X.; Wu, G.; Hu, C. MicroRNA-101 mediates the suppressive effect of laminar shear stress on mTOR expression in vascular endothelial cells. Biochem. Biophys. Res. Commun. 2012, 427, 138–142. [Google Scholar] [CrossRef]
- Lagna, G.; Ku, M.M.; Nguyen, P.H.; Neuman, N.A.; Davis, B.N.; Hata, A. Control of phenotypic plasticity of smooth muscle cells by bone morphogenetic protein signaling through the myocardin-related transcription factors. J. Biol. Chem. 2007, 282, 37244–37255. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.; Hata, A. MicroRNA regulation of smooth muscle gene expression and phenotype. Curr. Opin. Hematol. 2012, 19, 224–231. [Google Scholar] [CrossRef]
- Davis, B.N.; Hilyard, A.C.; Nguyen, P.H.; Lagna, G.; Hata, A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol. Cell 2010, 39, 373–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurin, M.; Cote, J.F. Insights into the biological functions of Dock family guanine nucleotide exchange factors. Genes Dev. 2014, 28, 533–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivey, K.N.; Srivastava, D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell 2010, 7, 36–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, N.; Kang, H. BMP-Induced MicroRNA-101 Expression Regulates Vascular Smooth Muscle Cell Migration. Int. J. Mol. Sci. 2020, 21, 4764. https://doi.org/10.3390/ijms21134764
Park N, Kang H. BMP-Induced MicroRNA-101 Expression Regulates Vascular Smooth Muscle Cell Migration. International Journal of Molecular Sciences. 2020; 21(13):4764. https://doi.org/10.3390/ijms21134764
Chicago/Turabian StylePark, Nanju, and Hara Kang. 2020. "BMP-Induced MicroRNA-101 Expression Regulates Vascular Smooth Muscle Cell Migration" International Journal of Molecular Sciences 21, no. 13: 4764. https://doi.org/10.3390/ijms21134764



