Cell Fate Determination of Lymphatic Endothelial Cells
Abstract
1. Introduction
2. Origins of Lymphatic Endothelial Cells
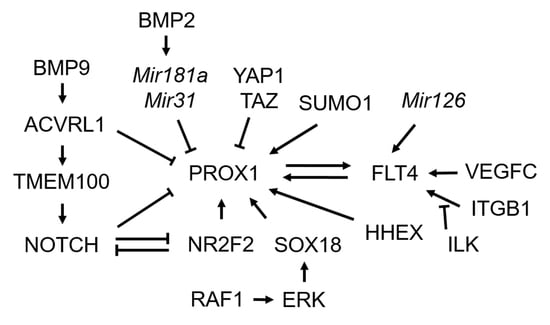
3. Specification of Lymphatic Endothelial Cells
3.1. Transcription Factor Prospero Homeobox 1 (PROX1)
3.2. Transcriptional Regulators of PROX1
3.3. Post-Transcriptional Regulators of PROX1 and Post-Translational Modification for PROX1
3.4. FMS-Like Tyrosine Kinase 4 (FLT4)/Vascular Endothelial Growth Factor Receptor 3 (VEGFR3) Signaling
3.5. NOTCH Signaling
3.6. Bone Morphogenetic Protein (BMP) Signaling
3.7. Transmembrane Protein 100 (TMEM100)
4. Human Diseases Associated with Genes for the Cell Fate Determination of Lymphatic Endothelial Cells
5. Conclusions
Funding
Conflicts of Interest
Abbreviations
| BMP | Bone morphogenetic protein |
| E | Embryonic day |
| HLTRS | Hypotrichosis-lymphedema-telangiectasia-renal defect syndrome |
| HLTS | Hypotrichosis-lymphedema-telangiectasia syndrome |
| LMPHM1 | Lymphatic malformation-1 |
| LMPHM4 | Lymphatic malformation-4 |
| MiRNA | MicroRNA |
| NICD | NOTCH intracellular domain |
| R-SMAD | Receptor-regulated SMAD |
| RAF1S259A | RAF1 S259A mutant |
| SNP | Single-nucleotide polymorphism |
| TGF-β | Transforming growth factor-β |
References
- Kesler, C.T.; Liao, S.; Munn, L.L.; Padera, T.P. Lymphatic vessels in health and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2013, 5, 111–124. [Google Scholar] [CrossRef]
- Alitalo, K.; Tammela, T.; Petrova, T.V. Lymphangiogenesis in development and human disease. Nature 2005, 438, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Coso, S.; Bovay, E.; Petrova, T.V. Pressing the right buttons: Signaling in lymphangiogenesis. Blood 2014, 123, 2614–2624. [Google Scholar] [CrossRef]
- Escobedo, N.; Oliver, G. Lymphangiogenesis: Origin, Specification, and Cell Fate Determination. Annu. Rev. Cell Dev. Biol. 2016, 32, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.R.; Lee, E.Y.; Shaikh, R. Lymphatics. In Pediatric Body MRI; Springer: Berlin/Heidelberg, Germany, 2020; pp. 113–124. [Google Scholar]
- Srinivasan, R.S.; Dillard, M.E.; Lagutin, O.V.; Lin, F.-J.; Tsai, S.; Tsai, M.-J.; Samokhvalov, I.M.; Oliver, G. Lineage tracing demonstrates the venous origin of the mammalian lymphatic vasculature. Genes Dev. 2007, 21, 2422–2432. [Google Scholar] [CrossRef] [PubMed]
- Wigle, J.T.; Oliver, G. Prox1 function is required for the development of the murine lymphatic system. Cell 1999, 98, 769–778. [Google Scholar] [CrossRef]
- François, M.; Short, K.; Secker, G.A.; Combes, A.; Schwarz, Q.; Davidson, T.L.; Smyth, I.; Hong, Y.K.; Harvey, N.L.; Koopman, P. Segmental territories along the cardinal veins generate lymph sacs via a ballooning mechanism during embryonic lymphangiogenesis in mice. Dev. Biol. 2012, 364, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; García-Verdugo, J.M.; Soriano-Navarro, M.; Srinivasan, R.S.; Scallan, J.P.; Singh, M.K.; Epstein, J.A.; Oliver, G. Lymphatic endothelial progenitors bud from the cardinal vein and intersomitic vessels in mammalian embryos. Blood 2012, 120, 2340–2348. [Google Scholar] [CrossRef]
- Hägerling, R.; Pollmann, C.; Andreas, M.; Schmidt, C.; Nurmi, H.; Adams, R.H.; Alitalo, K.; Andresen, V.; Schulte-Merker, S.; Kiefer, F. A novel multistep mechanism for initial lymphangiogenesis in mouse embryos based on ultramicroscopy. EMBO J. 2013, 32, 629–644. [Google Scholar] [CrossRef]
- Schulte-Merker, S.; Sabine, A.; Petrova, T.V. Lymphatic vascular morphogenesis in development, physiology, and disease. J. Cell Biol. 2011, 193, 607–618. [Google Scholar] [CrossRef]
- Karkkainen, M.J.; Haiko, P.; Sainio, K.; Partanen, J.; Taipale, J.; Petrova, T.V.; Jeltsch, M.; Jackson, D.G.; Talikka, M.; Rauvala, H.; et al. Vascular endothelial growth factor C is required for sprouting of the first lymphatic vessels from embryonic veins. Nat. Immunol. 2004, 5, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Kazenwadel, J.; Harvey, N.L. Lymphatic endothelial progenitor cells: Origins and roles in lymphangiogenesis. Curr. Opin. Immunol. 2018, 53, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Stanczuk, L.; Martinez-Corral, I.; Ulvmar, M.H.; Zhang, Y.; Lavina, B.; Fruttiger, M.; Adams, R.H.; Saur, D.; Betsholtz, C.; Ortega, S.; et al. cKit Lineage Hemogenic Endothelium-Derived Cells Contribute to Mesenteric Lymphatic Vessels. Cell Rep. 2015, 10, 1708–1721. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Corral, I.; Ulvmar, M.H.; Stanczuk, L.; Tatin, F.; Kizhatil, K.; John, S.W.; Alitalo, K.; Ortega, S.; Makinen, T. Nonvenous origin of dermal lymphatic vasculature. Circ. Res. 2015, 116, 1649–1654. [Google Scholar] [CrossRef] [PubMed]
- Klotz, L.; Norman, S.; Vieira, J.M.; Masters, M.; Rohling, M.; Dube, K.N.; Bollini, S.; Matsuzaki, F.; Carr, C.A.; Riley, P.R. Cardiac lymphatics are heterogeneous in origin and respond to injury. Nature 2015, 522, 62–67. [Google Scholar] [CrossRef]
- Sabin, F.R. On the origin of the lymphatics system from the veins and the development of the lymph hearts and the thoracic duct in the pig. Am. J. Anat. 1902, 1, 367–389. [Google Scholar] [CrossRef]
- Huntington, G.S.; McClure, C.F.W. The anatomy and development of the jugular lymph sac in the domestic cat (Felis domestica). Am. J. Anat. 1910, 10, 177–312. [Google Scholar] [CrossRef]
- Zinovieva, R.D.; Duncan, M.K.; Johnson, T.R.; Torres, R.; Polymeropoulos, M.H.; Tomarev, S.I. Structure and chromosomal localization of the human homeobox gene Prox 1. Genomics 1996, 35, 517–522. [Google Scholar] [CrossRef][Green Version]
- Hassan, B.; Li, L.; Bremer, K.A.; Chang, W.; Pinsonneault, J.; Vaessin, H. Prospero is a panneural transcription factor that modulates homeodomain protein activity. Proc. Natl. Acad. Sci. USA 1997, 94, 10991–10996. [Google Scholar] [CrossRef]
- Wigle, J.T.; Harvey, N.; Detmar, M.; Lagutina, I.; Grosveld, G.; Gunn, M.D.; Jackson, D.G.; Oliver, G. An essential role for Prox1 in the induction of the lymphatic endothelial cell phenotype. EMBO J. 2002, 21, 1505–1513. [Google Scholar] [CrossRef]
- Johnson, N.C.; Dillard, M.E.; Baluk, P.; McDonald, D.M.; Harvey, N.L.; Frase, S.L.; Oliver, G. Lymphatic endothelial cell identity is reversible and its maintenance requires Prox1 activity. Genes Dev. 2008, 22, 3282–3291. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.K.; Harvey, N.; Noh, Y.H.; Schacht, V.; Hirakawa, S.; Detmar, M.; Oliver, G. Prox1 is a master control gene in the program specifying lymphatic endothelial cell fate. Dev. Dyn. 2002, 225, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kang, J.; Yoo, J.; Ganesan, S.K.; Cook, S.C.; Aguilar, B.; Ramu, S.; Lee, J.; Hong, Y.K. Prox1 physically and functionally interacts with COUP-TFII to specify lymphatic endothelial cell fate. Blood 2009, 113, 1856–1859. [Google Scholar] [CrossRef]
- Srinivasan, R.S.; Escobedo, N.; Yang, Y.; Interiano, A.; Dillard, M.E.; Finkelstein, D.; Mukatira, S.; Gil, H.J.; Nurmi, H.; Alitalo, K.; et al. The Prox1-Vegfr3 feedback loop maintains the identity and the number of lymphatic endothelial cell progenitors. Genes Dev. 2014, 28, 2175–2187. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Cruz, M.; Bourdeau, A.; Dumont, D.J. Cell-cell interactions influence vascular reprogramming by Prox1 during embryonic development. PLoS ONE 2013, 8, e52197. [Google Scholar] [CrossRef] [PubMed]
- Petrova, T.V.; Makinen, T.; Makela, T.P.; Saarela, J.; Virtanen, I.; Ferrell, R.E.; Finegold, D.N.; Kerjaschki, D.; Yla-Herttuala, S.; Alitalo, K. Lymphatic endothelial reprogramming of vascular endothelial cells by the Prox-1 homeobox transcription factor. EMBO J. 2002, 21, 4593–4599. [Google Scholar] [CrossRef]
- François, M.; Caprini, A.; Hosking, B.; Orsenigo, F.; Wilhelm, D.; Browne, C.; Paavonen, K.; Karnezis, T.; Shayan, R.; Downes, M.; et al. Sox18 induces development of the lymphatic vasculature in mice. Nature 2008, 456, 643–647. [Google Scholar] [CrossRef]
- Gauvrit, S.; Villasenor, A.; Strilic, B.; Kitchen, P.; Collins, M.M.; Marin-Juez, R.; Guenther, S.; Maischein, H.M.; Fukuda, N.; Canham, M.A.; et al. HHEX is a transcriptional regulator of the VEGFC/FLT4/PROX1 signaling axis during vascular development. Nat. Commun. 2018, 9, 2704. [Google Scholar] [CrossRef]
- Cho, H.; Kim, J.; Ahn, J.H.; Hong, Y.K.; Makinen, T.; Lim, D.S.; Koh, G.Y. YAP and TAZ Negatively Regulate Prox1 During Developmental and Pathologic Lymphangiogenesis. Circ. Res. 2019, 124, 225–242. [Google Scholar] [CrossRef]
- Hosking, B.M.; Muscat, G.E.; Koopman, P.A.; Dowhan, D.H.; Dunn, T.L. Trans-activation and DNA-binding properties of the transcription factor, Sox-18. Nucleic Acids Res. 1995, 23, 2626–2628. [Google Scholar] [CrossRef][Green Version]
- Deng, Y.; Atri, D.; Eichmann, A.; Simons, M. Endothelial ERK signaling controls lymphatic fate specification. J. Clin. Investig. 2013, 123, 1202–1215. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Simons, M. Lymphatic fate determination: Playing RAF with ERK. Cell Cycle 2013, 12, 1157–1158. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.S.; Geng, X.; Yang, Y.; Wang, Y.; Mukatira, S.; Studer, M.; Porto, M.P.; Lagutin, O.; Oliver, G. The nuclear hormone receptor Coup-TFII is required for the initiation and early maintenance of Prox1 expression in lymphatic endothelial cells. Genes Dev. 2010, 24, 696–707. [Google Scholar] [CrossRef]
- Pereira, F.A.; Qiu, Y.; Zhou, G.; Tsai, M.J.; Tsai, S.Y. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999, 13, 1037–1049. [Google Scholar] [CrossRef]
- You, L.R.; Lin, F.J.; Lee, C.T.; DeMayo, F.J.; Tsai, M.J.; Tsai, S.Y. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature 2005, 435, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Yoshimatsu, Y.; Morishita, Y.; Miyazono, K.; Watabe, T. COUP-TFII regulates the functions of Prox1 in lymphatic endothelial cells through direct interaction. Genes Cells 2009, 14, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Martinez Barbera, J.P.; Clements, M.; Thomas, P.; Rodriguez, T.; Meloy, D.; Kioussis, D.; Beddington, R.S. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development 2000, 127, 2433–2445. [Google Scholar] [PubMed]
- Panciera, T.; Azzolin, L.; Cordenonsi, M.; Piccolo, S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 2017, 18, 758–770. [Google Scholar] [CrossRef]
- Kazenwadel, J.; Michael, M.Z.; Harvey, N.L. Prox1 expression is negatively regulated by miR-181 in endothelial cells. Blood 2010, 116, 2395–2401. [Google Scholar] [CrossRef]
- Pedrioli, D.M.; Karpanen, T.; Dabouras, V.; Jurisic, G.; van de Hoek, G.; Shin, J.W.; Marino, D.; Kälin, R.E.; Leidel, S.; Cinelli, P.; et al. miR-31 functions as a negative regulator of lymphatic vascular lineage-specific differentiation in vitro and vascular development in vivo. Mol. Cell. Biol. 2010, 30, 3620–3634. [Google Scholar] [CrossRef]
- Pan, M.R.; Chang, T.M.; Chang, H.C.; Su, J.L.; Wang, H.W.; Hung, W.C. Sumoylation of Prox1 controls its ability to induce VEGFR3 expression and lymphatic phenotypes in endothelial cells. J. Cell Sci. 2009, 122, 3358–3364. [Google Scholar] [CrossRef] [PubMed]
- Dumont, D.J.; Jussila, L.; Taipale, J.; Lymboussaki, A.; Mustonen, T.; Pajusola, K.; Breitman, M.; Alitalo, K. Cardiovascular failure in mouse embryos deficient in VEGF receptor-3. Science 1998, 282, 946–949. [Google Scholar] [CrossRef] [PubMed]
- Kaipainen, A.; Korhonen, J.; Mustonen, T.; van Hinsbergh, V.W.; Fang, G.H.; Dumont, D.; Breitman, M.; Alitalo, K. Expression of the fms-like tyrosine kinase 4 gene becomes restricted to lymphatic endothelium during development. Proc. Natl. Acad. Sci. USA 1995, 92, 3566–3570. [Google Scholar] [CrossRef] [PubMed]
- Mäkinen, T.; Veikkola, T.; Mustjoki, S.; Karpanen, T.; Catimel, B.; Nice, E.C.; Wise, L.; Mercer, A.; Kowalski, H.; Kerjaschki, D.; et al. Isolated lymphatic endothelial cells transduce growth, survival and migratory signals via the VEGF-C/D receptor VEGFR-3. EMBO J. 2001, 20, 4762–4773. [Google Scholar] [CrossRef]
- Salameh, A.; Galvagni, F.; Bardelli, M.; Bussolino, F.; Oliviero, S. Direct recruitment of CRK and GRB2 to VEGFR-3 induces proliferation, migration, and survival of endothelial cells through the activation of ERK, AKT, and JNK pathways. Blood 2005, 106, 3423–3431. [Google Scholar] [CrossRef]
- Wang, J.F.; Zhang, X.F.; Groopman, J.E. Stimulation of beta 1 integrin induces tyrosine phosphorylation of vascular endothelial growth factor receptor-3 and modulates cell migration. J. Biol. Chem. 2001, 276, 41950–41957. [Google Scholar] [CrossRef]
- Zhang, X.; Groopman, J.E.; Wang, J.F. Extracellular matrix regulates endothelial functions through interaction of VEGFR-3 and integrin alpha5beta1. J. Cell. Physiol. 2005, 202, 205–214. [Google Scholar] [CrossRef]
- Galvagni, F.; Pennacchini, S.; Salameh, A.; Rocchigiani, M.; Neri, F.; Orlandini, M.; Petraglia, F.; Gotta, S.; Sardone, G.L.; Matteucci, G.; et al. Endothelial cell adhesion to the extracellular matrix induces c-Src-dependent VEGFR-3 phosphorylation without the activation of the receptor intrinsic kinase activity. Circ. Res. 2010, 106, 1839–1848. [Google Scholar] [CrossRef]
- Urner, S.; Planas-Paz, L.; Hilger, L.S.; Henning, C.; Branopolski, A.; Kelly-Goss, M.; Stanczuk, L.; Pitter, B.; Montanez, E.; Peirce, S.M.; et al. Identification of ILK as a critical regulator of VEGFR3 signalling and lymphatic vascular growth. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Kontarakis, Z.; Rossi, A.; Ramas, S.; Dellinger, M.T.; Stainier, D.Y.R. Mir-126 is a conserved modulator of lymphatic development. Dev. Biol. 2018, 437, 120–130. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef]
- Bentley, K.; Chakravartula, S. The temporal basis of angiogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20150522. [Google Scholar] [CrossRef] [PubMed]
- Villa, N.; Walker, L.; Lindsell, C.E.; Gasson, J.; Iruela-Arispe, M.L.; Weinmaster, G. Vascular expression of Notch pathway receptors and ligands is restricted to arterial vessels. Mech. Dev. 2001, 108, 161–164. [Google Scholar] [CrossRef]
- Guruharsha, K.G.; Kankel, M.W.; Artavanis-Tsakonas, S. The Notch signalling system: Recent insights into the complexity of a conserved pathway. Nat. Rev. Genet. 2012, 13, 654–666. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Yoo, J.; Lee, S.; Tang, W.; Aguilar, B.; Ramu, S.; Choi, I.; Otu, H.H.; Shin, J.W.; Dotto, G.P.; et al. An exquisite cross-control mechanism among endothelial cell fate regulators directs the plasticity and heterogeneity of lymphatic endothelial cells. Blood 2010, 116, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Qin, J.; Cheng, C.M.; Tsai, M.J.; Tsai, S.Y. COUP-TFII is a major regulator of cell cycle and Notch signaling pathways. Mol. Endocrinol. 2012, 26, 1268–1277. [Google Scholar] [CrossRef]
- Murtomaki, A.; Uh, M.K.; Choi, Y.K.; Kitajewski, C.; Borisenko, V.; Kitajewski, J.; Shawber, C.J. Notch1 functions as a negative regulator of lymphatic endothelial cell differentiation in the venous endothelium. Development 2013, 140, 2365–2376. [Google Scholar] [CrossRef]
- Fatima, A.; Culver, A.; Culver, F.; Liu, T.; Dietz, W.H.; Thomson, B.R.; Hadjantonakis, A.-K.; Quaggin, S.E.; Kume, T. Murine Notch1 is required for lymphatic vascular morphogenesis during development. Dev. Dyn. 2014, 243, 957–964. [Google Scholar] [CrossRef]
- Choi, D.; Park, E.; Jung, E.; Seong, Y.J.; Yoo, J.; Lee, E.; Hong, M.; Lee, S.; Ishida, H.; Burford, J.; et al. Laminar flow downregulates Notch activity to promote lymphatic sprouting. J. Clin. Investig. 2017, 127, 1225–1240. [Google Scholar] [CrossRef]
- Beets, K.; Staring, M.W.; Criem, N.; Maas, E.; Schellinx, N.; de Sousa Lopes, S.M.C.; Umans, L.; Zwijsen, A. BMP-SMAD signalling output is highly regionalized in cardiovascular and lymphatic endothelial networks. BMC Dev. Biol. 2016, 16, 34. [Google Scholar] [CrossRef]
- Dunworth, W.P.; Cardona-Costa, J.; Bozkulak, E.C.; Kim, J.D.; Meadows, S.; Fischer, J.C.; Wang, Y.; Cleaver, O.; Qyang, Y.; Ober, E.A.; et al. Bone morphogenetic protein 2 signaling negatively modulates lymphatic development in vertebrate embryos. Circ. Res. 2014, 114, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Levet, S.; Ciais, D.; Merdzhanova, G.; Mallet, C.; Zimmers, T.A.; Lee, S.-J.; Navarro, F.P.; Texier, I.; Feige, J.-J.; Bailly, S.; et al. Bone morphogenetic protein 9 (BMP9) controls lymphatic vessel maturation and valve formation. Blood 2013, 122, 598–607. [Google Scholar] [CrossRef]
- Yoshimatsu, Y.; Lee, Y.G.; Akatsu, Y.; Taguchi, L.; Suzuki, H.I.; Cunha, S.I.; Maruyama, K.; Suzuki, Y.; Yamazaki, T.; Katsura, A.; et al. Bone morphogenetic protein-9 inhibits lymphatic vessel formation via activin receptor-like kinase 1 during development and cancer progression. Proc. Natl. Acad. Sci. USA 2013, 110, 18940–18945. [Google Scholar] [CrossRef]
- Moon, E.-H.; Kim, M.-J.; Ko, K.S.; Kim, Y.S.; Seo, J.; Oh, S.P.; Lee, Y.J. Generation of mice with a conditional and reporter allele for Tmem100. Genesis 2010, 48, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Somekawa, S.; Imagawa, K.; Hayashi, H.; Sakabe, M.; Ioka, T.; Sato, G.E.; Inada, K.; Iwamoto, T.; Mori, T.; Uemura, S.; et al. Tmem100, an ALK1 receptor signaling-dependent gene essential for arterial endothelium differentiation and vascular morphogenesis. Proc. Natl. Acad. Sci. USA 2012, 109, 12064–12069. [Google Scholar] [CrossRef] [PubMed]
- Moon, E.-H.; Kim, Y.S.; Seo, J.; Lee, S.; Lee, Y.J.; Oh, S.P. Essential role for TMEM100 in vascular integrity but limited contributions to the pathogenesis of hereditary haemorrhagic telangiectasia. Cardiovasc. Res. 2015, 105, 353–360. [Google Scholar] [CrossRef]
- Tachida, Y.; Izumi, N.; Sakurai, T.; Kobayashi, H. Mutual interaction between endothelial cells and mural cells enhances BMP9 signaling in endothelial cells. Biol. Open 2017, 6, 370–380. [Google Scholar] [CrossRef]
- Moon, E.H.; Kim, Y.H.; Vu, P.N.; Yoo, H.; Hong, K.; Lee, Y.J.; Oh, S.P. TMEM100 is a key factor for specification of lymphatic endothelial progenitors. Angiogenesis 2020, 23, 339–355. [Google Scholar] [CrossRef]
- Irrthum, A.; Devriendt, K.; Chitayat, D.; Matthijs, G.; Glade, C.; Steijlen, P.M.; Fryns, J.P.; Van Steensel, M.A.; Vikkula, M. Mutations in the transcription factor gene SOX18 underlie recessive and dominant forms of hypotrichosis-lymphedema-telangiectasia. Am. J. Hum. Genet. 2003, 72, 1470–1478. [Google Scholar] [CrossRef]
- Moalem, S.; Brouillard, P.; Kuypers, D.; Legius, E.; Harvey, E.; Taylor, G.; Francois, M.; Vikkula, M.; Chitayat, D. Hypotrichosis-lymphedema-telangiectasia-renal defect associated with a truncating mutation in the SOX18 gene. Clin. Genet. 2015, 87, 378–382. [Google Scholar] [CrossRef]
- Pandit, B.; Sarkozy, A.; Pennacchio, L.A.; Carta, C.; Oishi, K.; Martinelli, S.; Pogna, E.A.; Schackwitz, W.; Ustaszewska, A.; Landstrom, A.; et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat. Genet. 2007, 39, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Razzaque, M.A.; Nishizawa, T.; Komoike, Y.; Yagi, H.; Furutani, M.; Amo, R.; Kamisago, M.; Momma, K.; Katayama, H.; Nakagawa, M.; et al. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat. Genet. 2007, 39, 1013–1017. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, M.; Mehler, E.L.; Goldberg, R.; Zampino, G.; Brunner, H.G.; Kremer, H.; van der Burgt, I.; Crosby, A.H.; Ion, A.; Jeffery, S.; et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 2001, 29, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Carta, C.; Pantaleoni, F.; Bocchinfuso, G.; Stella, L.; Vasta, I.; Sarkozy, A.; Digilio, C.; Palleschi, A.; Pizzuti, A.; Grammatico, P.; et al. Germline missense mutations affecting KRAS Isoform B are associated with a severe Noonan syndrome phenotype. Am. J. Hum. Genet. 2006, 79, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Schubbert, S.; Zenker, M.; Rowe, S.L.; Böll, S.; Klein, C.; Bollag, G.; van der Burgt, I.; Musante, L.; Kalscheuer, V.; Wehner, L.E.; et al. Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 2006, 38, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.E.; Araki, T.; Swanson, K.D.; Montgomery, K.T.; Schiripo, T.A.; Joshi, V.A.; Li, L.; Yassin, Y.; Tamburino, A.M.; Neel, B.G.; et al. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat. Genet. 2007, 39, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, M.; Pennacchio, L.A.; Zhao, C.; Yadav, K.K.; Fodale, V.; Sarkozy, A.; Pandit, B.; Oishi, K.; Martinelli, S.; Schackwitz, W.; et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat. Genet. 2007, 39, 75–79. [Google Scholar] [CrossRef]
- Lanning, P.; Similä, S.; Suramo, I.; Paavilainen, T. Lymphatic abnormalities in Noonan’s syndrome. Pediatr. Radiol. 1978, 7, 106–109. [Google Scholar] [CrossRef]
- Baltaxe, H.A.; Lee, J.G.; Ehlers, K.H.; Engle, M.A. Pulmonary lymphangiectasia demonstrated by lymphangiography in 2 patients with Noonan’s syndrome. Radiology 1975, 115, 149–153. [Google Scholar] [CrossRef]
- Ghalamkarpour, A.; Holnthoner, W.; Saharinen, P.; Boon, L.M.; Mulliken, J.B.; Alitalo, K.; Vikkula, M. Recessive primary congenital lymphoedema caused by a VEGFR3 mutation. J. Med. Genet. 2009, 46, 399–404. [Google Scholar] [CrossRef]
- Gordon, K.; Schulte, D.; Brice, G.; Simpson, M.A.; Roukens, M.G.; van Impel, A.; Connell, F.; Kalidas, K.; Jeffery, S.; Mortimer, P.S.; et al. Mutation in vascular endothelial growth factor-C, a ligand for vascular endothelial growth factor receptor-3, is associated with autosomal dominant milroy-like primary lymphedema. Circ. Res. 2013, 112, 956–960. [Google Scholar] [CrossRef] [PubMed]
- Online Mendelian Inheritance in Man, OMIM®. McKusick-Nathans Institute of Genetic Medicine: Johns Hopkins University (Baltimore, MD). Available online: https://omim.org/ (accessed on 8 June 2020).
- Mendola, A.; Schlögel, M.J.; Ghalamkarpour, A.; Irrthum, A.; Nguyen, H.L.; Fastré, E.; Bygum, A.; van der Vleuten, C.; Fagerberg, C.; Baselga, E.; et al. Mutations in the VEGFR3 signaling pathway explain 36% of familial lymphedema. Mol. Syndromol. 2013, 4, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Harvey, N.L.; Srinivasan, R.S.; Dillard, M.E.; Johnson, N.C.; Witte, M.H.; Boyd, K.; Sleeman, M.W.; Oliver, G. Lymphatic vascular defects promoted by Prox1 haploinsufficiency cause adult-onset obesity. Nat. Genet. 2005, 37, 1072–1081. [Google Scholar] [CrossRef] [PubMed]
- Horra, A.; Salazar, J.; Ferré, R.; Vallvé, J.C.; Guardiola, M.; Rosales, R.; Masana, L.; Ribalta, J. Prox-1 and FOXC2 gene expression in adipose tissue: A potential contributory role of the lymphatic system to familial combined hyperlipidaemia. Atherosclerosis 2009, 206, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Yoo, Y.J.; Ju, Y.S.; Lee, S.; Cho, S.I.; Sung, J.; Kim, J.I.; Seo, J.S. Combined linkage and association analyses identify a novel locus for obesity near PROX1 in Asians. Obesity (Silver Spring) 2013, 21, 2405–2412. [Google Scholar] [CrossRef] [PubMed]
- Kretowski, A.; Adamska, E.; Maliszewska, K.; Wawrusiewicz-Kurylonek, N.; Citko, A.; Goscik, J.; Bauer, W.; Wilk, J.; Golonko, A.; Waszczeniuk, M.; et al. The rs340874 PROX1 type 2 diabetes mellitus risk variant is associated with visceral fat accumulation and alterations in postprandial glucose and lipid metabolism. Genes Nutr. 2015, 10, 4. [Google Scholar] [CrossRef]
- Adamska-Patruno, E.; Godzien, J.; Ciborowski, M.; Samczuk, P.; Bauer, W.; Siewko, K.; Gorska, M.; Barbas, C.; Kretowski, A. The Type 2 Diabetes Susceptibility PROX1 Gene Variants Are Associated with Postprandial Plasma Metabolites Profile in Non-Diabetic Men. Nutrients 2019, 11, 882. [Google Scholar] [CrossRef]
- Norden, P.R.; Kume, T. The Role of Lymphatic Vascular Function in Metabolic Disorders. Front. Physiol. 2020, 11, 404. [Google Scholar] [CrossRef]
- Franceschini, N.; Almasy, L.; MacCluer, J.W.; Göring, H.H.; Cole, S.A.; Diego, V.P.; Laston, S.; Howard, B.V.; Lee, E.T.; Best, L.G.; et al. Diabetes-specific genetic effects on obesity traits in American Indian populations: The Strong Heart Family Study. BMC Med. Genet. 2008, 9, 90. [Google Scholar] [CrossRef]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U.; Wheeler, E.; Glazer, N.L.; Bouatia-Naji, N.; Gloyn, A.L.; et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef]
- Lecompte, S.; Pasquetti, G.; Hermant, X.; Grenier-Boley, B.; Gonzalez-Gross, M.; De Henauw, S.; Molnar, D.; Stehle, P.; Béghin, L.; Moreno, L.A.; et al. Genetic and molecular insights into the role of PROX1 in glucose metabolism. Diabetes 2013, 62, 1738–1745. [Google Scholar] [CrossRef] [PubMed]
- Hamet, P.; Haloui, M.; Harvey, F.; Marois-Blanchet, F.C.; Sylvestre, M.P.; Tahir, M.R.; Simon, P.H.; Kanzki, B.S.; Raelson, J.; Long, C.; et al. PROX1 gene CC genotype as a major determinant of early onset of type 2 diabetes in slavic study participants from Action in Diabetes and Vascular Disease: Preterax and Diamicron MR Controlled Evaluation study. J. Hypertens. 2017, 35 Suppl 1, S24–S32. [Google Scholar] [CrossRef]
- Burgio, G. Redefining mouse transgenesis with CRISPR/Cas9 genome editing technology. Genome Biol. 2018, 19, 27. [Google Scholar] [CrossRef] [PubMed]
- Wojnowski, L.; Stancato, L.F.; Zimmer, A.M.; Hahn, H.; Beck, T.W.; Larner, A.C.; Rapp, U.R.; Zimmer, A. Craf-1 protein kinase is essential for mouse development. Mech. Dev. 1998, 76, 141–149. [Google Scholar] [CrossRef]
- Mikula, M.; Schreiber, M.; Husak, Z.; Kucerova, L.; Rüth, J.; Wieser, R.; Zatloukal, K.; Beug, H.; Wagner, E.F.; Baccarini, M. Embryonic lethality and fetal liver apoptosis in mice lacking the c-raf-1 gene. EMBO J. 2001, 20, 1952–1962. [Google Scholar] [CrossRef] [PubMed]

| Gene | Roles in Lymphangiogenesis | Viability and Gross Morphology of Knockout (KO) Embryos | Human Diseases 1 |
|---|---|---|---|
| Prox1 | Specification and maintenance of lymphatic endothelial cells | KO mice [7,21] die ~E14.5; lymphedema; lack of lymphatics cKO mice (Tek-cre) 2 [6] lymphedema; compromised lymphangiogenesis cKO mice (CAGGCreER, E8.5~E10.5, E12.5 and E13.5) 3 [22] lymphedema; blood-filled lymphatics OE mice (tie1 tTA:tetOS prox1) 4 [26] lymphedema; anemia | Human SNP rs1704198 located in the proximity of PROX1 associated with a larger waist circumference Human SNP rs340874 located in the 5′-UTR of PROX1 associated with fasting glycemia and type 2 diabetes |
| Sox18 | Activation of Prox1 expression | KO mice [28] die ~E14.5; lymphedema; lack of lymphatics | Hypotrichosis-lymphedema-telangiectasia syndrome (OMIM #607823) Hypotrichosis-lymphedema-telangiectasia-renal defect syndrome (OMIM #137940) |
| Raf1 | Activation of Sox18 and Prox1 expression through ERK signaling | KO mice [96,97] die after E11.5 (until E16.5); growth retardation; defects in several organs including the skin, eyelids, lung, placenta, and liver OE mice (VE-cadherin-tTA/RAF1S259A) 4 [32] die at E15.5; lymphedema; enlarged lymphatics; heart defects; induction of Sox18 and Prox1 expression | Noonan syndrome 5 (OMIM #611553) LEOPARD syndrome 2 (OMIM #611554) Cardiomyopathy, dilated, 1NN (OMIM #615916) |
| Nr2f2 | Activation of Prox1 expression Inhibition of NOTCH signaling | KO mice [35] die before E11.5; heart defects; angiogenesis defects cKO mice (Tek-cre) [6,34,36] die at E11.5; compromised lymphangiogenesis; ectopic expression of Notch1 | 46, XX sex reversal 5 (OMIM #618901) Congenital heart defects, multiple types, 4 (OMIM #615779) |
| Hhex | Activation of Prox1 expression | KO mice [29,38] die after E11.5; pericardial edema; blood-filled lymphatics; growth retardation; vascular patterning defects cKO mice (Tek-cre) [29] die after E11.5; pericardial edema; growth retardation; vascular patterning defects; blood-filled lymphatics; lymphedema; defects in lymphatic vessels cKO mice (Prox1-CreER, E10.5~E12.5) [29] blood-filled lymphatics; lymphedema; defects in lymphatic vessels | |
| Yap1 and Taz | Inhibition of Prox1 expression | Double cKO mice (Prox1-CreER, E11.5 and E13.5) [30] lymphedema; defects in lymphatic vessels | YAP1: Coloboma, ocular, with or without hearing impairment, cleft lip/palate, and/or mental retardation (OMIM #120433) TAZ: Barth syndrome (OMIM #302060) |
| Vegfc | Ligand for FLT4 Budding-off of lymphatic endothelial cells | KO mice [10,12,25] die after E15.5; lymphedema; failure of the budding-off of lymphatic endothelial cells from the cardinal vein | Lymphatic malformation 4 (OMIM #615907) |
| Flt4 | Receptor for VEGFC Budding-off of lymphatic endothelial cells | KO mice [43] die after E10.5; severe cardiovascular defects; yolk sac vasculature defects; pericardial edema; growth retardation | Lymphatic malformation 1 (OMIM #153100) Congenital heart defects, multiple types, 7 (OMIM #618780) Hemangioma, capillary infantile, somatic (OMIM #602089) |
| Ilk | Inhibition of the interaction between β1 integrin and FLT4 | cKO mice (Kdr-cre) [50] die after E13.5; lymphedema; head bleeding; enlarged lymphatics; lymphatic and blood vascular sprouting defects | |
| Mir126 | Control of FLT4 signaling | KO mice 1 [51] No obvious defects KO mice 2 [52] partial embryonic lethality; edema; hemorrhage; growth retardation Mir126-/-; Flt4+/- [51] Die before birth; lymphedema at E14.5 | |
| Notch1 | Inhibition of Prox1 and Nr2f2 expression | cKO mice (Prox1-CreER, E9.75) [58] mild lymphedema; bold-filled lymphatics; enlarged lymphatic sacs cKO mice (Prox1-CreER, E10.5) [59] enlarged lymphatic vessels OE mice (Prox1-CreER, E10.5) [58] numerous small and disorganized lymphatic sac-like structures | Adams-Oliver syndrome 5 (OMIM #616028) Aortic valve disease 1 (OMIM #109730) |
| Bmp9 | Downregulation of Prox1 expression through ACVRL1 | KO mice [63,64] enlarged lymphatic vessels; defective lymphatic valve formation | Telangiectasia, hereditary hemorrhagic, type 5 (OMIM #615506) |
| Tmem100 | Inhibition of NOTCH signaling | cKO mice (ROSA26-CreER, E10.5) [69] die around E16.5; lymphedema, blood-filled lymphatic vessels; lymphatic vessel dilation OE mice (Tek-cre) [69] die around E15.5; lymphedema, small size and number of lymphatic vessels |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.J. Cell Fate Determination of Lymphatic Endothelial Cells. Int. J. Mol. Sci. 2020, 21, 4790. https://doi.org/10.3390/ijms21134790
Lee YJ. Cell Fate Determination of Lymphatic Endothelial Cells. International Journal of Molecular Sciences. 2020; 21(13):4790. https://doi.org/10.3390/ijms21134790
Chicago/Turabian StyleLee, Young Jae. 2020. "Cell Fate Determination of Lymphatic Endothelial Cells" International Journal of Molecular Sciences 21, no. 13: 4790. https://doi.org/10.3390/ijms21134790
APA StyleLee, Y. J. (2020). Cell Fate Determination of Lymphatic Endothelial Cells. International Journal of Molecular Sciences, 21(13), 4790. https://doi.org/10.3390/ijms21134790