Toxicity of TiO2 Nanoparticles: Validation of Alternative Models
Abstract
1. Introduction
2. Results
2.1. TiO2 Nanoparticles Characterization
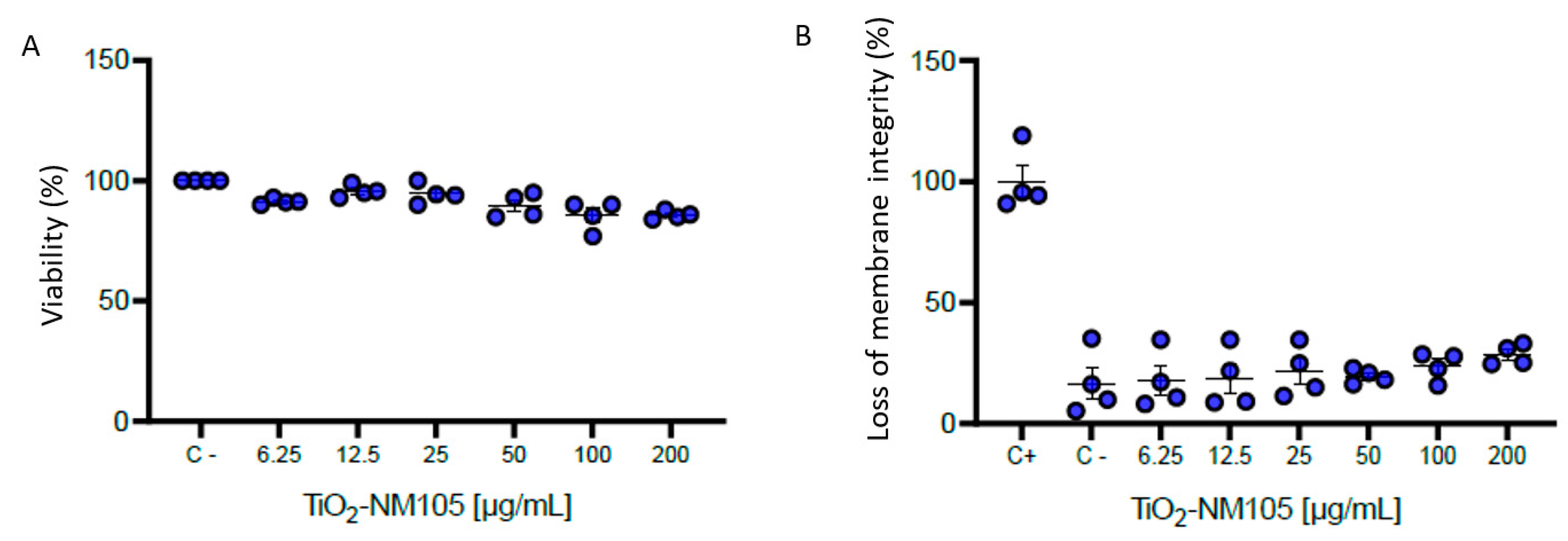
2.2. In Vitro Cytotoxicity Study
2.3. Transcriptomic Analysis of Dysregulated Genes Following In Vivo, In Vitro Submerged, and ALI Vitrocell Cloud ® Exposure (ALI) to NM-105 TiO2 NP
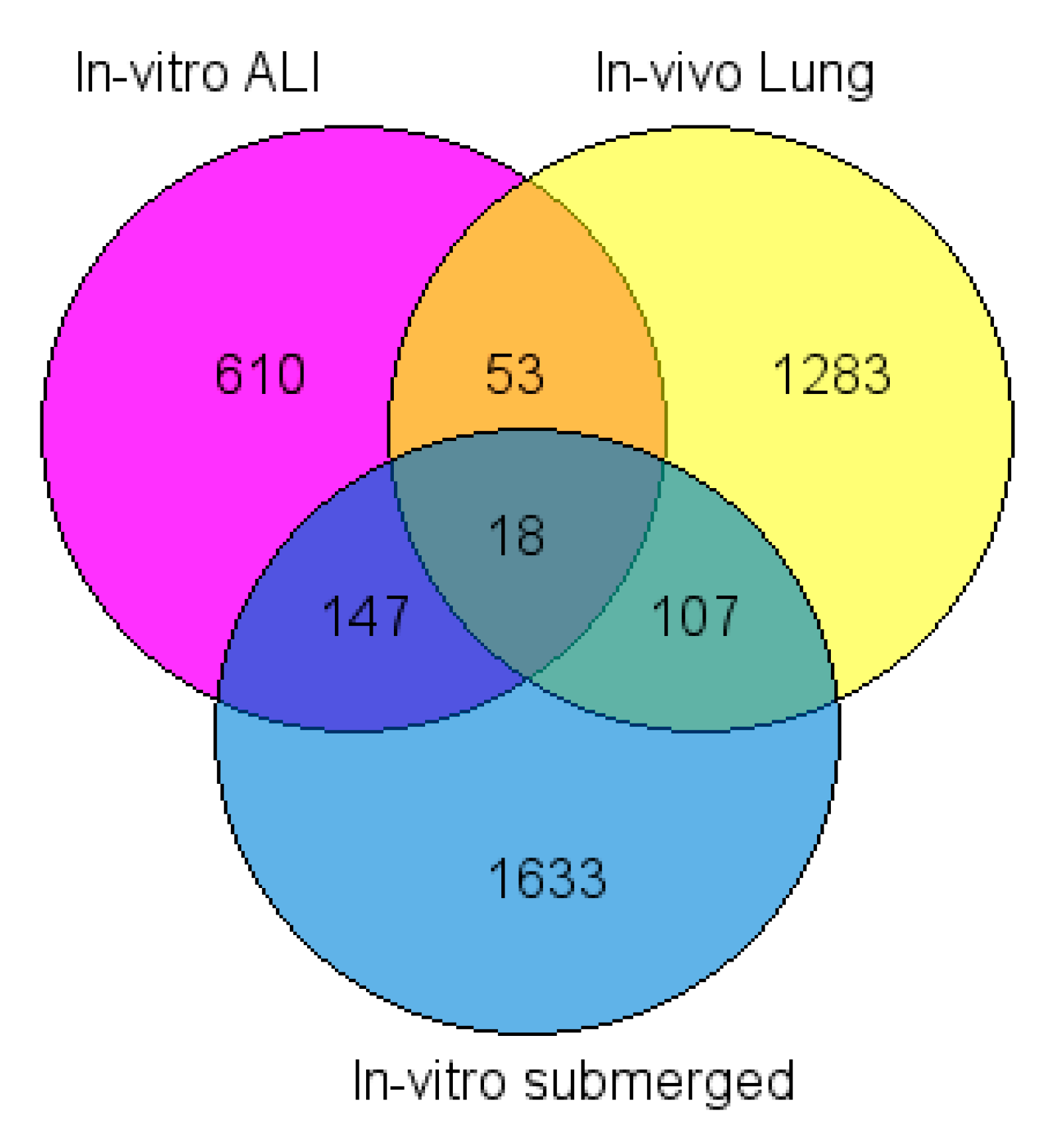
2.4. Common Dysregulated Genes between In Vivo, In Vitro, and ALI Exposures to TiO2 NP
2.5. Comparison of Functional Annotations of Dysregulated Genes In Vivo, In Vitro, and ALI after Exposition to TiO2 NP
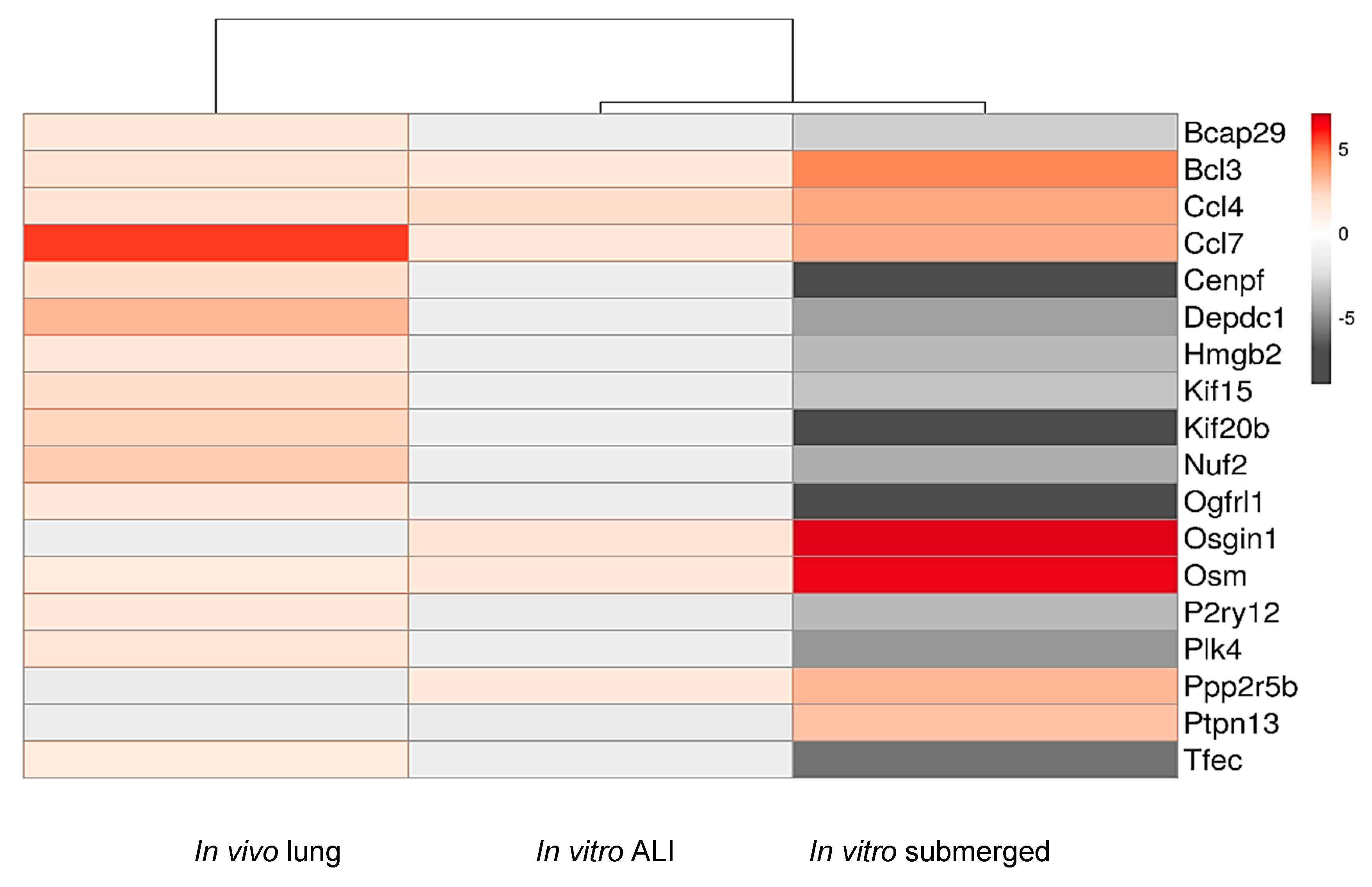
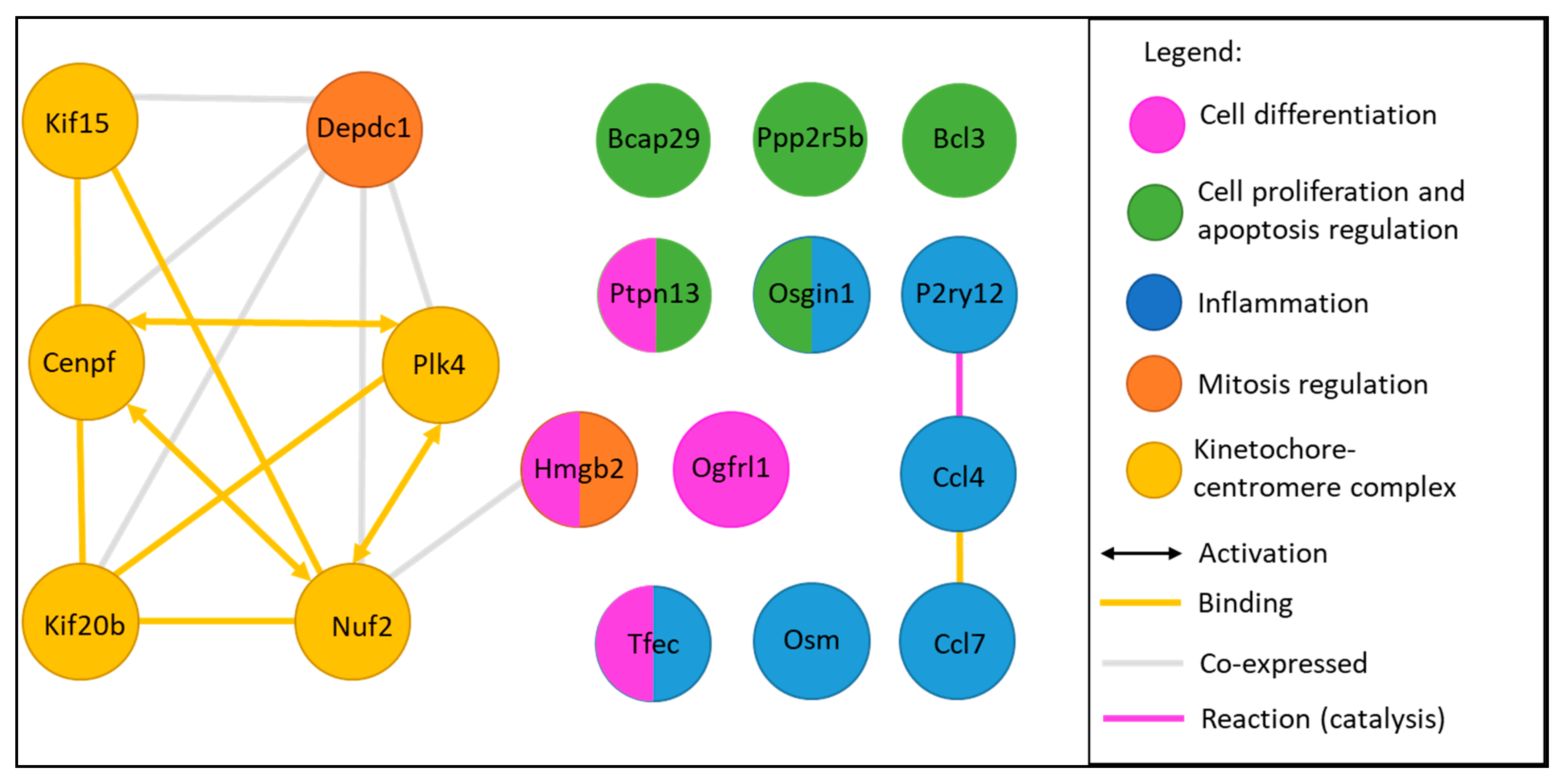
2.6. Functional Analysis of Common Dysregulated Genes between In Vivo, In Vitro, and ALI Expositions to TiO2 NP
3. Discussion
3.1. Methodology
3.2. Viability of NR8383 Cells Exposed to TiO2
3.3. Number of DEGs in Each Exposure Method
3.4. Transcriptomic Study
4. Materials and Methods
4.1. TiO2 NP Characterization
4.2. Cell Culture
4.3. In Vitro Cytotoxicity Study
4.4. In Vivo Exposure
4.5. Exposure at the Air–liquid Interface
4.6. Transcriptomic Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jaroenworaluck, A.; Sunsaneeyametha, W.; Kosachan, N.; Stevens, R. Characteristics of silica-coated TiO2 and its UV absorption for sunscreen cosmetic applications. Surf. Interface Anal. 2006, 38, 473–477. [Google Scholar] [CrossRef]
- Weir, A.; Westerhoff, P.; Fabricius, L.; Hristovski, K.; von Goetz, N. Titanium Dioxide Nanoparticles in Food and Personal Care Products. Environ. Sci. Technol. 2012, 46, 2242–2250. [Google Scholar] [CrossRef] [PubMed]
- Ahn, T.-K.; Lee, D.H.; Kim, T.; Jang, G.C.; Choi, S.; Oh, J.B.; Ye, G.; Lee, S. Modification of Titanium Implant and Titanium Dioxide for Bone Tissue Engineering. In Novel Biomaterials for Regenerative Medicine; Chun, H.J., Park, K., Kim, C.-H., Khang, G., Eds.; Springer: Singapore, 2018; Volume 1077, pp. 355–368. ISBN 9789811309465. [Google Scholar]
- Shi, H.; Magaye, R.; Castranova, V.; Zhao, J. Titanium dioxide nanoparticles: A review of current toxicological data. Part. Fibre Toxicol. 2013, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Ziental, D.; Czarczynska-Goslinska, B.; Mlynarczyk, D.T.; Glowacka-Sobotta, A.; Stanisz, B.; Goslinski, T.; Sobotta, L. Titanium Dioxide Nanoparticles: Prospects and Applications in Medicine. Nanomaterials 2020, 10, 387. [Google Scholar] [CrossRef] [PubMed]
- Howarter, J.A.; Youngblood, J.P. Self-Cleaning and Next Generation Anti-Fog Surfaces and Coatings. Macromol. Rapid Commun. 2008, 29, 455–466. [Google Scholar] [CrossRef]
- Li, S.-X.; Lin, X.; Zheng, F.-Y.; Liang, W.; Zhong, Y.; Cai, J. Constituting Fully Integrated Visual Analysis System for Cu(II) on TiO2 /Cellulose Paper. Anal. Chem. 2014, 86, 7079–7083. [Google Scholar] [CrossRef]
- Markowska-Szczupak, A.; Ulfig, K.; Grzmil, B.; Morawski, A. A preliminary study on antifungal effect of TiO2-based paints in natural indoor light. Pol. J. Chem. Technol. 2010, 12, 53–57. [Google Scholar] [CrossRef]
- Rathee, D.; Arya, S.K.; Kumar, M. Analysis of TiO2 for microelectronic applications: Effect of deposition methods on their electrical properties. Front. Optoelectron. China 2011, 4, 349–358. [Google Scholar] [CrossRef]
- Wu, M.J.; Bak, T.; O’Doherty, P.J.; Moffitt, M.C.; Nowotny, J.; Bailey, T.D.; Kersaitis, C. Photocatalysis of Titanium Dioxide for Water Disinfection: Challenges and Future Perspectives. Int. J. Photochem. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Armand, L.; Tarantini, A.; Beal, D.; Biola-Clier, M.; Bobyk, L.; Sorieul, S.; Pernet-Gallay, K.; Marie-Desvergne, C.; Lynch, I.; Herlin-Boime, N.; et al. Long-term exposure of A549 cells to titanium dioxide nanoparticles induces DNA damage and sensitizes cells towards genotoxic agents. Nanotoxicology 2016, 10, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Johnston, H.J.; Hutchison, G.R.; Christensen, F.M.; Peters, S.; Hankin, S.; Stone, V. Identification of the mechanisms that drive the toxicity of TiO(2)particulates: The contribution of physicochemical characteristics. Part. Fibre Toxicol. 2009, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Landsiedel, R.; Ma-Hock, L.; Hofmann, T.; Wiemann, M.; Strauss, V.; Treumann, S.; Wohlleben, W.; Gröters, S.; Wiench, K.; van Ravenzwaay, B. Application of short-term inhalation studies to assess the inhalation toxicity of nanomaterials. Part. Fibre Toxicol. 2014, 11, 16. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yan, J.; Ding, W.; Chen, Y.; Pack, L.M.; Chen, T. Genotoxicity and gene expression analyses of liver and lung tissues of mice treated with titanium dioxide nanoparticles. Mutagenesis 2017, 32, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Gong, X.; Duan, Y.; Li, N.; Hu, R.; Liu, H.; Hong, M.; Zhou, M.; Wang, L.; Wang, H.; et al. Hepatocyte apoptosis and its molecular mechanisms in mice caused by titanium dioxide nanoparticles. J. Hazard. Mater. 2010, 183, 874–880. [Google Scholar] [CrossRef]
- Cui, Y.; Liu, H.; Ze, Y.; Zengli, Z.; Hu, Y.; Cheng, Z.; Cheng, J.; Hu, R.; Gao, G.; Wang, L.; et al. Gene expression in liver injury caused by long-term exposure to titanium dioxide nanoparticles in mice. Toxicol. Sci. Off. J. Soc. Toxicol. 2012, 128, 171–185. [Google Scholar] [CrossRef]
- Lenz, A.G.; Karg, E.; Lentner, B.; Dittrich, V.; Brandenberger, C.; Rothen-Rutishauser, B.; Schulz, H.; Ferron, G.A.; Schmid, O. A Dose-Controlled System for Air-Liquid Interface Cell Exposure and Application to Zinc Oxide Nanoparticles. Part. Fibre Toxicol. 2009, 6, 32. [Google Scholar] [CrossRef]
- Lenz, A.-G.; Karg, E.; Brendel, E.; Hinze-Heyn, H.; Maier, K.L.; Eickelberg, O.; Stoeger, T.; Schmid, O. Inflammatory and Oxidative Stress Responses of an Alveolar Epithelial Cell Line to Airborne Zinc Oxide Nanoparticles at the Air-Liquid Interface: A Comparison with Conventional, Submerged Cell-Culture Conditions. Biomed Res. Int. 2013, 2013, 652632. [Google Scholar] [CrossRef]
- Lenz, A.-G.; Stoeger, T.; Cei, D.; Schmidmeir, M.; Semren, N.; Burgstaller, G.; Lentner, B.; Eickelberg, O.; Meiners, S.; Schmid, O. Efficient Bioactive Delivery of Aerosolized Drugs to Human Pulmonary Epithelial Cells Cultured in Air-Liquid Interface Conditions. Am. J. Respir. Cell Mol. Biol. 2014, 51, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Eidi, H.; Joubert, O.; Némos, C.; Grandemange, S.; Mograbi, B.; Foliguet, B.; Tournebize, J.; Maincent, P.; Le Faou, A.; Aboukhamis, I.; et al. Drug delivery by polymeric nanoparticles induces autophagy in macrophages. Int. J. Pharm. 2012, 422, 495–503. [Google Scholar] [CrossRef]
- Hussain, S.; Vanoirbeek, J.A.J.; Hoet, P.H.M. Interactions of nanomaterials with the immune system. WIREs Nanomed. Nanobiotechnol. 2012, 4, 169–183. [Google Scholar] [CrossRef]
- Chézeau, L.; Sébillaud, S.; Safar, R.; Seidel, C.; Dembélé, D.; Lorcin, M.; Langlais, C.; Grossmann, S.; Nunge, H.; Michaux, S.; et al. Short- and long-term gene expression profiles induced by inhaled TiO2 nanostructured aerosol in rat lung. Toxicol. Appl. Pharmacol. 2018, 356, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Volkov, V.A.; Grissom, P.M.; Arzhanik, V.K.; Zaytsev, A.V.; Renganathan, K.; McClure-Begley, T.; Old, W.M.; Ahn, N.; McIntosh, J.R. Centromere protein F includes two sites that couple efficiently to depolymerizing microtubules. J. Cell Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- McCleland, M.L.; Gardner, R.D.; Kallio, M.J.; Daum, J.R.; Gorbsky, G.J.; Burke, D.J.; Stukenberg, P.T. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003, 17, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Tanenbaum, M.E.; Macůrek, L.; Janssen, A.; Geers, E.F.; Alvarez-Fernández, M.; Medema, R.H. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr. Biol. CB 2009, 19, 1703–1711. [Google Scholar] [CrossRef]
- Mi, Y.; Zhang, C.; Bu, Y.; Zhang, Y.; He, L.; Li, H.; Zhu, H.; Li, Y.; Lei, Y.; Zhu, J. DEPDC1 is a novel cell cycle related gene that regulates mitotic progression. BMB Rep. 2015, 48, 413–418. [Google Scholar] [CrossRef]
- Pallier, C.; Scaffidi, P.; Chopineau-Proust, S.; Agresti, A.; Nordmann, P.; Bianchi, M.E.; Marechal, V. Association of Chromatin Proteins High Mobility Group Box (HMGB) 1 and HMGB2 with Mitotic Chromosomes. Mol. Biol. Cell 2003, 14, 3414–3426. [Google Scholar] [CrossRef]
- Wang, W.; Jiang, H.; Zhu, H.; Zhang, H.; Gong, J.; Zhang, L.; Ding, Q. Overexpression of high mobility group box 1 and 2 is associated with the progression and angiogenesis of human bladder carcinoma. Oncol. Lett. 2013, 5, 884–888. [Google Scholar] [CrossRef]
- Kwon, J.-H.; Kim, J.; Park, J.Y.; Hong, S.M.; Park, C.W.; Hong, S.J.; Park, S.Y.; Choi, Y.J.; Do, I.-G.; Joh, J.-W.; et al. Overexpression of High-Mobility Group Box 2 Is Associated with Tumor Aggressiveness and Prognosis of Hepatocellular Carcinoma. Clin. Cancer Res. 2010. [Google Scholar] [CrossRef]
- Hechler, B.; Gachet, C. P2 receptors and platelet function. Purinergic Signal. 2011, 7, 293–303. [Google Scholar] [CrossRef]
- Zhu, C.; Kros, J.M.; van der Weiden, M.; Zheng, P.; Cheng, C.; Mustafa, D.A.M. Expression site of P2RY12 in residential microglial cells in astrocytomas correlates with M1 and M2 marker expression and tumor grade. Acta Neuropathol. Commun. 2017, 5, 4. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, J.; Zhang, L.; Zhang, Z.; He, L.; Mou, Y.; Deng, Y.; Cao, Y.; Yang, P.; Su, Y.; et al. Role of C/EBP homologous protein and endoplasmic reticulum stress in asthma exacerbation by regulating the IL-4/signal transducer and activator of transcription 6/transcription factor EC/IL-4 receptor α positive feedback loop in M2 macrophages. J. Allergy Clin. Immunol. 2017, 140, 1550–1561.e8. [Google Scholar] [CrossRef]
- Rehli, M.; Sulzbacher, S.; Pape, S.; Ravasi, T.; Wells, C.A.; Heinz, S.; Söllner, L.; Chartouni, C.E.; Krause, S.W.; Steingrimsson, E.; et al. Transcription Factor Tfec Contributes to the IL-4-Inducible Expression of a Small Group of Genes in Mouse Macrophages Including the Granulocyte Colony-Stimulating Factor Receptor. J. Immunol. 2005, 174, 7111–7122. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, T.; Sumiyoshi, H.; Higashi, K.; Nakao, S.; Higashiyama, R.; Fukumitsu, H.; Minakawa, K.; Chiba, Y.; Suzuki, Y.; Sumida, K.; et al. Identification of a Novel Bone Marrow Cell-Derived Accelerator of Fibrotic Liver Regeneration Through Mobilization of Hepatic Progenitor Cells in Mice. STEM CELLS 2019, 37, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, H.; Mao, Y.; Wang, X.; Zhang, X.; Yu, X.; Tian, J.; Lei, Z.; Li, C.; Han, Q.; et al. Apoptotic SKOV3 cells stimulate M0 macrophages to differentiate into M2 macrophages and promote the proliferation and migration of ovarian cancer cells by activating the ERK signaling pathway. Int. J. Mol. Med. 2019. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, H.; Shuman, L.; Warrick, J.I.; Raman, J.D.; Degraff, D.J. Androgen represses opioid growth factor receptor (OGFR) in human prostate cancer LNCaP cells and OGFR expression in human prostate cancer tissue. Am. J. Clin. Exp. Urol. 2018, 6, 164–171. [Google Scholar]
- Tannenbaum, J.; Bennett, B.T. Russell and Burch’s 3Rs then and now: The need for clarity in definition and purpose. J. Am. Assoc. Lab. Anim. Sci. JAALAS 2015, 54, 120–132. [Google Scholar]
- Alhadlaq, H.A.; Akhtar, M.J.; Ahamed, M. Zinc ferrite nanoparticle-induced cytotoxicity and oxidative stress in different human cells. Cell Biosci. 2015, 5. [Google Scholar] [CrossRef]
- Oberdörster, G.; Oberdörster, E.; Oberdörster, J. Nanotoxicology: An Emerging Discipline Evolving from Studies of Ultrafine Particles. Environmental Health Perspectives 2005, 113, 823–839. [Google Scholar] [CrossRef]
- Geiser, M. Update on Macrophage Clearance of Inhaled Micro- and Nanoparticles. J. Aerosol Med. Pulm. Drug Deliv. 2010, 23, 207–217. [Google Scholar] [CrossRef]
- Lehnert, B.E. Pulmonary and thoracic macrophage subpopulations and clearance of particles from the lung. Environ. Health Perspect. 1992, 97, 17–46. [Google Scholar] [CrossRef]
- Semmler-Behnke, M.; Takenaka, S.; Fertsch, S.; Wenk, A.; Seitz, J.; Mayer, P.; Oberdörster, G.; Kreyling, W.G. Efficient Elimination of Inhaled Nanoparticles from the Alveolar Region: Evidence for Interstitial Uptake and Subsequent Reentrainment onto Airways Epithelium. Environ. Health Perspect. 2007, 115, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Eidi, H.; Joubert, O.; Attik, G.; Duval, R.E.; Bottin, M.C.; Hamouia, A.; Maincent, P.; Rihn, B.H. Cytotoxicity assessment of heparin nanoparticles in NR8383 macrophages. Int. J. Pharm. 2010, 396, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Paul, G.; Ong, H.X.; Young, P.M.; Gu, Y.T.; Saha, S.C. A Review of Respiratory Anatomical Development, Air Flow Characterization and Particle Deposition. IJERPH 2020, 17, 380. [Google Scholar] [CrossRef] [PubMed]
- Heyder, J. Deposition of Inhaled Particles in the Human Respiratory Tract and Consequences for Regional Targeting in Respiratory Drug Delivery. Proc. Am. Thorac. Soc. 2004, 1, 315–320. [Google Scholar] [CrossRef]
- Gerloff, K.; Fenoglio, I.; Carella, E.; Kolling, J.; Albrecht, C.; Boots, A.W.; Förster, I.; Schins, R.P.F. Distinctive Toxicity of TiO2 Rutile/Anatase Mixed Phase Nanoparticles on Caco-2 Cells. Chem. Res. Toxicol. 2012, 25, 646–655. [Google Scholar] [CrossRef]
- Uboldi, C.; Urbán, P.; Gilliland, D.; Bajak, E.; Valsami-Jones, E.; Ponti, J.; Rossi, F. Role of the crystalline form of titanium dioxide nanoparticles: Rutile, and not anatase, induces toxic effects in Balb/3T3 mouse fibroblasts. Toxicol. In Vitro 2016, 31, 137–145. [Google Scholar] [CrossRef]
- Huang, Y.-W.; Wu, C.; Aronstam, R.S. Toxicity of Transition Metal Oxide Nanoparticles: Recent Insights from in vitro Studies. Materials 2010, 3, 4842–4859. [Google Scholar] [CrossRef]
- Haase, A.; Dommershausen, N.; Schulz, M.; Landsiedel, R.; Reichardt, P.; Krause, B.-C.; Tentschert, J.; Luch, A. Genotoxicity testing of different surface-functionalized SiO2, ZrO2 and silver nanomaterials in 3D human bronchial models. Arch. Toxicol. 2017, 91, 3991–4007. [Google Scholar] [CrossRef]
- Wiemann, M.; Vennemann, A.; Sauer, U.G.; Wiench, K.; Ma-Hock, L.; Landsiedel, R. An in vitro alveolar macrophage assay for predicting the short-term inhalation toxicity of nanomaterials. J. Nanobiotechnol. 2016, 14, 16. [Google Scholar] [CrossRef]
- Karkossa, I.; Bannuscher, A.; Hellack, B.; Bahl, A.; Buhs, S.; Nollau, P.; Luch, A.; Schubert, K.; von Bergen, M.; Haase, A. An in-depth multi-omics analysis in RLE-6TN rat alveolar epithelial cells allows for nanomaterial categorization. Part. Fibre Toxicol. 2019, 16, 38. [Google Scholar] [CrossRef]
- Cohen, J.M.; Teeguarden, J.G.; Demokritou, P. An integrated approach for the in vitro dosimetry of engineered nanomaterials. Part. Fibre Toxicol. 2014, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Moore, T.L.; Urban, D.A.; Rodriguez-Lorenzo, L.; Milosevic, A.; Crippa, F.; Spuch-Calvar, M.; Balog, S.; Rothen-Rutishauser, B.; Lattuada, M.; Petri-Fink, A. Nanoparticle administration method in cell culture alters particle-cell interaction. Sci. Rep. 2019, 9, 900. [Google Scholar] [CrossRef]
- Thomas, D.G.; Smith, J.N.; Thrall, B.D.; Baer, D.R.; Jolley, H.; Munusamy, P.; Kodali, V.; Demokritou, P.; Cohen, J.; Teeguarden, J.G. ISD3: A particokinetic model for predicting the combined effects of particle sedimentation, diffusion and dissolution on cellular dosimetry for in vitro systems. Part. Fibre Toxicol. 2018, 15, 6. [Google Scholar] [CrossRef] [PubMed]
- Doumandji, Z.; Safar, R.; Lovera-Leroux, M.; Nahle, S.; Cassidy, H.; Matallanas, D.; Rihn, B.H.; Ferrari, L.; Joubert, O. Protein and lipid homeostasis altered in rat macrophages after exposure to metallic oxide nanoparticles. Cell Biol. Toxicol. 2019. [Google Scholar] [CrossRef]
- Hans, C.P.; Sharma, N.; Sen, S.; Zeng, S.; Dev, R.; Jiang, Y.; Mahajan, A.; Joshi, T. Transcriptomics Analysis Reveals New Insights into the Roles of Notch1 Signaling on Macrophage Polarization. Sci. Rep. 2019, 9, 7999. [Google Scholar] [CrossRef] [PubMed]
- Nahle, S.; Cassidy, H.; Leroux, M.M.; Mercier, R.; Ghanbaja, J.; Doumandji, Z.; Matallanas, D.; Rihn, B.H.; Joubert, O.; Ferrari, L. Genes expression profiling of alveolar macrophages exposed to non-functionalized, anionic and cationic multi-walled carbon nanotubes shows three different mechanisms of toxicity. J. Nanobiotechnol. 2020, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- Sano, H.; Kuroki, Y. The lung collectins, SP-A and SP-D, modulate pulmonary innate immunity. Mol. Immunol. 2005, 42, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Stringer, B.; Kobzik, L. Alveolar macrophage uptake of the environmental particulate titanium dioxide: Role of surfactant components. Am. J. Respir. Cell Mol. Biol. 1996, 14, 155–160. [Google Scholar] [CrossRef]
- Tofte, R.W.; Peterson, P.K.; Kim, Y.; Quie, P.G. Influence of serum concentration on opsonization by the classical and alternative complement pathways. Infect. Immun. 1980, 27, 693–696. [Google Scholar] [CrossRef]
- Che Hak, C.R.; Fatanah, D.N.E.; Abdullah, Y.; Meor Sulaiman, M.Y. The Effect of Surfactants on the Stability of TiO2 Aqueous Suspension. Int. J. Curr. Res. Sci. Eng. Technol. 2018, 1, 172. [Google Scholar] [CrossRef]
- Sato, T.; Kohnosu, S. Effect of surfactant concentration on the stability of aqueous titanium dioxide suspensions. J. Colloid Interface Sci. 1991, 143, 434–439. [Google Scholar] [CrossRef]
- Heijman, S.G.J.; Stein, H.N. Electrostatic and Sterical Stabilization of TiO2 Dispersions. Langmuir 1995, 11, 422–427. [Google Scholar] [CrossRef][Green Version]
- Tkachenko, N.H.; Yaremko, Z.M.; Bellmann, C.; Soltys, M.M. The influence of ionic and nonionic surfactants on aggregative stability and electrical surface properties of aqueous suspensions of titanium dioxide. J. Colloid Interface Sci. 2006, 299, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Zhang, X.; Ding, B.; Wang, L.; Lu, N. Synergistic action of TiO 2 particles and surfactants on the foamability and stabilization of aqueous foams. RSC Adv. 2017, 7, 44972–44978. [Google Scholar] [CrossRef]
- Gehr, P.; Schürch, S.; Berthiaume, Y.; Hof, V.I.; Geiser, M. Particle Retention in Airways by Surfactant. J. Aerosol Med. 1990, 3, 27–43. [Google Scholar] [CrossRef]
- Imae, T.; Hasegawa, E.; Iwamoto, T. Dispersion Stability of TiO2 Particles in Aqueous Surfactant Solutions. J. Jpn. Oil Chem. Soc. 1993, 42, 501–506. [Google Scholar] [CrossRef][Green Version]
- Godinez, I.G.; Darnault, C.J.G.; Khodadoust, A.P.; Bogdan, D. Deposition and release kinetics of nano-TiO2 in saturated porous media: Effects of solution ionic strength and surfactants. Environ. Pollut. 2013, 174, 106–113. [Google Scholar] [CrossRef]
- Zhang, C.; Lohwacharin, J.; Takizawa, S. Properties of residual titanium dioxide nanoparticles after extended periods of mixing and settling in synthetic and natural waters. Sci. Rep. 2017, 7, 9943. [Google Scholar] [CrossRef]
- Vranic, S.; Garcia-Verdugo, I.; Darnis, C.; Sallenave, J.-M.; Boggetto, N.; Marano, F.; Boland, S.; Baeza-Squiban, A. Internalization of SiO2 nanoparticles by alveolar macrophages and lung epithelial cells and its modulation by the lung surfactant substitute Curosurf®. Environ. Sci. Pollut. Res. 2013, 20, 2761–2770. [Google Scholar] [CrossRef]
- Chézeau, L.; Kohlstaedt, L.A.; Le Faou, A.; Cosnier, F.; Rihn, B.; Gaté, L. Proteomic analysis of bronchoalveolar lavage fluid in rat exposed to TiO2 nanostructured aerosol by inhalation. J. Proteomics 2019, 207, 103451. [Google Scholar] [CrossRef]
- NIOSH Occupational exposure to titanium dioxide current intelligence. Natl. Inst. Occup. Saf. Health 2011.
- Gaté, L.; Disdier, C.; Cosnier, F.; Gagnaire, F.; Devoy, J.; Saba, W.; Brun, E.; Chalansonnet, M.; Mabondzo, A. Biopersistence and translocation to extrapulmonary organs of titanium dioxide nanoparticles after subacute inhalation exposure to aerosol in adult and elderly rats. Toxicol. Lett. 2017, 265, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, E.F.; Lindesmith, L.C.; Lobue, A.D.; Baric, R.S. Norovirus pathogenesis: Mechanisms of persistence and immune evasion in human populations. Immunol. Rev. 2008, 225, 190–211. [Google Scholar] [CrossRef] [PubMed]
- Barin, J.G.; Baldeviano, G.C.; Talor, M.V.; Wu, L.; Ong, S.; Quader, F.; Chen, P.; Zheng, D.; Caturegli, P.; Rose, N.R.; et al. Macrophages participate in IL-17-mediated inflammation. Eur. J. Immunol. 2012, 42, 726–736. [Google Scholar] [CrossRef] [PubMed]
- Nakai, K.; He, Y.-Y.; Nishiyama, F.; Naruse, F.; Haba, R.; Kushida, Y.; Katsuki, N.; Moriue, T.; Yoneda, K.; Kubota, Y. IL-17A induces heterogeneous macrophages, and it does not alter the effects of lipopolysaccharides on macrophage activation in the skin of mice. Sci. Rep. 2017, 7, 12473. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Chen, L.; Wang, S.; Wang, Y.; Zhang, W.; Ma, K.; Hu, C.; Zhu, H.; Liang, S.; Liu, M.; Xu, N. IL-6 influences the polarization of macrophages and the formation and growth of colorectal tumor. Oncotarget 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Han, Z.C. STAT3: A critical transcription activator in angiogenesis. Med. Res. Rev. 2008, 28, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. c-Myc Target Genes Involved in Cell Growth, Apoptosis, and Metabolism. Mol. Cell. Biol. 1999, 19, 1–11. [Google Scholar] [CrossRef]
- Biroccio, A.; Amodei, S.; Antonelli, A.; Benassi, B.; Zupi, G. Inhibition of c-Myc Oncoprotein Limits the Growth of Human Melanoma Cells by Inducing Cellular Crisis. J. Biol. Chem. 2003, 278, 35693–35701. [Google Scholar] [CrossRef] [PubMed]
- Cano-Ramos, E.; Lavin, B.; Pello, O.M. Inhibition of MYC in macrophages: Tumor vs inflammation-related diseases. OncoImmunology 2014, 3, e956013. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pello, O.M.; Chèvre, R.; Laoui, D.; De Juan, A.; Lolo, F.; Andrés-Manzano, M.J.; Serrano, M.; Van Ginderachter, J.A.; Andrés, V. In vivo Inhibition of c-MYC in Myeloid Cells Impairs Tumor-Associated Macrophage Maturation and Pro-Tumoral Activities. PLoS ONE 2012, 7, e45399. [Google Scholar] [CrossRef] [PubMed]
- Pello, O.M.; De Pizzol, M.; Mirolo, M.; Soucek, L.; Zammataro, L.; Amabile, A.; Doni, A.; Nebuloni, M.; Swigart, L.B.; Evan, G.I.; et al. Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood 2012, 119, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Demeterco, C.; Itkin-Ansari, P.; Tyrberg, B.; Ford, L.P.; Jarvis, R.A.; Levine, F. c-Myc Controls Proliferation Versus Differentiation in Human Pancreatic Endocrine Cells. J. Clin. Endocrinol. Metab. 2002, 87, 3475–3485. [Google Scholar] [CrossRef]
- Okuyama, H.; Endo, H.; Akashika, T.; Kato, K.; Inoue, M. Downregulation of c-MYC Protein Levels Contributes to Cancer Cell Survival under Dual Deficiency of Oxygen and Glucose. Cancer Res. 2010, 70, 10213–10223. [Google Scholar] [CrossRef]
- Zhang, P.; Li, H.; Wu, M.-L.; Chen, X.-Y.; Kong, Q.-Y.; Wang, X.-W.; Sun, Y.; Wen, S.; Liu, J. c-Myc downregulation: A critical molecular event in resveratrol-induced cell cycle arrest and apoptosis of human medulloblastoma cells. J. Neurooncol. 2006, 80, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.; Hung, M.E.; Cangelose, B.K.; Leonard, J.N. Regulation of the IL-10-driven macrophage phenotype under incoherent stimuli. Innate Immun. 2016, 22, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Staitieh, B.S.; Egea, E.E.; Fan, X.; Azih, N.; Neveu, W.; Guidot, D.M. Activation of Alveolar Macrophages with Interferon-gamma Promotes Antioxidant Defenses via the Nrf2-ARE Pathway. J. Clin. Cell. Immunol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Toda, M.; Mizuguchi, S.; Minamiyama, Y.; Yamamoto-Oka, H.; Aota, T.; Kubo, S.; Nishiyama, N.; Shibata, T.; Takemura, S. Pirfenidone suppresses polarization to M2 phenotype macrophages and the fibrogenic activity of rat lung fibroblasts. J. Clin. Biochem. Nutr. 2018, 63, 58–65. [Google Scholar] [CrossRef]
- Scrima, M.; De Marco, C.; De Vita, F.; Fabiani, F.; Franco, R.; Pirozzi, G.; Rocco, G.; Malanga, D.; Viglietto, G. The Nonreceptor-Type Tyrosine Phosphatase PTPN13 Is a Tumor Suppressor Gene in Non–Small Cell Lung Cancer. Am. J. Pathol. 2012, 180, 1202–1214. [Google Scholar] [CrossRef]
- Takahashi, K.; Sivina, M.; Hoellenriegel, J.; Oki, Y.; Hagemeister, F.B.; Fayad, L.; Romaguera, J.E.; Fowler, N.; Fanale, M.A.; Kwak, L.W.; et al. CCL3 and CCL4 are biomarkers for B cell receptor pathway activation and prognostic serum markers in diffuse large B cell lymphoma. Br. J. Haematol. 2015, 171, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cai, Y.; Liu, L.; Wu, Y.; Xiong, X. Crucial biological functions of CCL7 in cancer. PeerJ 2018, 6, e4928. [Google Scholar] [CrossRef] [PubMed]
- Parikh, N.; Shuck, R.L.; Gagea, M.; Shen, L.; Donehower, L.A. Enhanced inflammation and attenuated tumor suppressor pathways are associated with oncogene-induced lung tumors in aged mice. Aging Cell 2018, 17, e12691. [Google Scholar] [CrossRef]
- Cho, Y.B.; Lee, W.Y.; Choi, S.-J.; Kim, J.; Hong, H.K.; Kim, S.-H.; Choi, Y.-L.; Kim, H.C.; Yun, S.H.; Chun, H.-K.; et al. CC chemokine ligand 7 expression in liver metastasis of colorectal cancer. Oncol. Rep. 2012, 28, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Kim, S.-Y.; Song, S.J.; Hong, H.K.; Lee, Y.; Oh, B.Y.; Lee, W.Y.; Cho, Y.B. Crosstalk between CCL7 and CCR3 promotes metastasis of colon cancer cells via ERK-JNK signaling pathways. Oncotarget 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Wyler, L.; Napoli, C.U.; Ingold, B.; Sulser, T.; Heikenwälder, M.; Schraml, P.; Moch, H. Brain metastasis in renal cancer patients: Metastatic pattern, tumour-associated macrophages and chemokine/chemoreceptor expression. Br. J. Cancer 2014, 110, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Saio Tumor-associated macrophage/microglia infiltration in human gliomas is correlated with MCP-3, but not MCP-1. Int. J. Oncol. 2009, 34.
- Pradeep, A.R.; Thorat Manojkumar, S.; Garima, G.; Raju, A. Serum levels of oncostatin M (a gp 130 cytokine): An inflammatory biomarker in periodontal disease. Biomarkers 2010, 15, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Yan, X.; Sun, X.; Chen, T.; Liu, Y.; Cao, J. Oncostatin M Is a Prognostic Biomarker and Inflammatory Mediator for Sepsis. J. Infect. Dis. 2020, jiaa009. [Google Scholar] [CrossRef]
- Oxford IBD Cohort Investigators; West, N.R.; Hegazy, A.N.; Owens, B.M.J.; Bullers, S.J.; Linggi, B.; Buonocore, S.; Coccia, M.; Görtz, D.; This, S.; et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor–neutralizing therapy in patients with inflammatory bowel disease. Nat. Med. 2017, 23, 579–589. [Google Scholar] [CrossRef]
- Verstockt, S.; Verstockt, B.; Vermeire, S. Oncostatin M as a new diagnostic, prognostic and therapeutic target in inflammatory bowel disease (IBD). Expert Opin. Ther. Targets 2019, 23, 943–954. [Google Scholar] [CrossRef]
- Junk, D.J.; Bryson, B.L.; Smigiel, J.M.; Parameswaran, N.; Bartel, C.A.; Jackson, M.W. Oncostatin M promotes cancer cell plasticity through cooperative STAT3-SMAD3 signaling. Oncogene 2017, 36, 4001–4013. [Google Scholar] [CrossRef] [PubMed]
- Ayaub, E.A.; Dubey, A.; Imani, J.; Botelho, F.; Kolb, M.R.J.; Richards, C.D.; Ask, K. Overexpression of OSM and IL-6 impacts the polarization of pro-fibrotic macrophages and the development of bleomycin-induced lung fibrosis. Sci. Rep. 2017, 7, 13281. [Google Scholar] [CrossRef] [PubMed]
- Wulczyn, F.G.; Naumann, M.; Scheidereit, C. Candidate proto-oncogene bcl-3 encodes a subunit-specific inhibitor of transcription factor NF-κB. Nature 1992, 358, 597–599. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Didonato, J.A.; Karin, M.; McKeithan, T.W. BCL3 encodes a nuclear protein which can alter the subcellular location of NF-kappa B proteins. Mol. Cell. Biol. 1994, 14, 3915–3926. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-P.; Vancurova, I. Bcl3 regulates pro-survival and pro-inflammatory gene expression in cutaneous T-cell lymphoma. Biochim. Biophys. Acta BBA Mol. Cell Res. 2014, 1843, 2620–2630. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, V.; Melendez-Zajgla, J. Role of Bcl-3 in solid tumors. Mol. Cancer 2011, 10, 152. [Google Scholar] [CrossRef]
- Guan, Y.; Yao, H.; Zheng, Z.; Qiu, G.; Sun, K. MiR-125b targets BCL3 and suppresses ovarian cancer proliferation: miR-125b inhibits ovarian cancer cell proliferation. Int. J. Cancer 2011, 128, 2274–2283. [Google Scholar] [CrossRef]
- Maldonado, V.; Espinosa, M.; Pruefer, F.; Patiño, N.; Ceballos-Canciono, G.; Urzua, U.; Juretic, N.; Melendez-Zajgla, J. Gene regulation by BCL3 in a cervical cancer cell line. Folia Biol. (Praha) 2010, 56, 183–193. [Google Scholar]
- Zhao, H.; Wang, W.; Zhao, Q.; Hu, G.; Deng, K.; Liu, Y. BCL3 exerts an oncogenic function by regulating STAT3 in human cervical cancer. OncoTargets Ther. 2016, 9, 6619–6629. [Google Scholar] [CrossRef]
- Wakefield, A.; Soukupova, J.; Montagne, A.; Ranger, J.; French, R.; Muller, W.J.; Clarkson, R.W.E. Bcl3 Selectively Promotes Metastasis of ERBB2-Driven Mammary Tumors. Cancer Res. 2013, 73, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Lee, J.M.; Kim, H.; Nam, H.J.; Shin, H.-J.R.; Kim, D.; Ko, E.; Noh, D.-Y.; Kim, K.I.; Kim, J.H.; et al. Bcl3-dependent stabilization of CtBP1 is crucial for the inhibition of apoptosis and tumor progression in breast cancer. Biochem. Biophys. Res. Commun. 2010, 400, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Phuyal, S.; Kasem, M.; Rubio, L.; Karlsson, H.L.; Marcos, R.; Skaug, V.; Zienolddiny, S. Effects on Human Bronchial Epithelial Cells Following Low-Dose Chronic Exposure to Nanomaterials: A 6-Month Transformation Study. Toxicol. In Vitro 2017, 44, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Sze, A.; Erickson, D.; Ren, L.; Li, D. Zeta-potential measurement using the Smoluchowski equation and the slope of the current–time relationship in electroosmotic flow. J. Colloid Interface Sci. 2003, 261, 402–410. [Google Scholar] [CrossRef]
- Leroux, M.M.; Hocquel, R.; Bourge, K.; Joubert, O. Optimization of an innovative air-liquid interface exposure system for transcriptomic assays with semi-adherent cells. 2020; Submitted. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Pirooznia, M.; Nagarajan, V.; Deng, Y. GeneVenn - A Web Application for Comparing Gene Lists Using Venn Diagrams. Bioinformation 2007, 1, 420–422. [Google Scholar] [CrossRef]
- Abu-Jamous, B.; Kelly, S. Clust: Automatic Extraction of Optimal Co-Expressed Gene Clusters from Gene Expression Data. Genome Biol. 2018, 19, 172. [Google Scholar] [CrossRef]



| Nanoparticle | Primary Size (nm) | Secondary Size (nm) | Zeta Potential (mV) | Specific Surface Aera (m2/g) | Provider |
|---|---|---|---|---|---|
| TiO2 (NM-105) | 21.5 ± 7.2 | 170 ± 1.5 | 11.1 ± 0.7 | 51 | Joint Research Center |
| Groups | FC 1.3 | FC 1.5 | FC3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Total | Up | Down | Total | Up | Down | Total | |
| in vitro submerged | 4612 | 5224 | 9836 | 3939 | 3956 | 7895 | 345 | 1376 | 1721 |
| in vitro ALI | 439 | 412 | 851 | 108 | 94 | 202 | 0 | 1 | 1 |
| lung | 898 | 579 | 1477 | 559 | 221 | 780 | 64 | 5 | 69 |
| Gene Set | p-Value in vivo Lung | p-Value in vitro ALI | p-Value in vitro Submerged |
|---|---|---|---|
| Common gene sets between the three exposures | |||
| IL6_JAK_STAT3_SIGNALING | 1.25 × 10−7 | 7.42 × 10−2 | 3.82 × 10−2 |
| MYC_TARGETS_V2 | 4.30 × 10−6 | 7.42 × 10−2 | 3.65 × 10−2 |
| Common gene sets between in vivo and ALI exposures | |||
| E2F_TARGETS | 1.04 × 10−14 | 8.40 × 10−7 | – |
| G2M_CHECKPOINT | 7.96 × 10−11 | 7.00 × 10−3 | – |
| MYC_TARGETS_V1 | 1.08 × 10−4 | 7.00 × 10−3 | – |
| EPITHELIAL_MESENCHYMAL_TRANSITION | 9.03 × 10−2 | 3.84 × 10−2 | – |
| Common gene sets between in vivo and in vitro submerged exposures | |||
| UV_RESPONSE_DN | 6.21 × 10−3 | – | 6.73 × 10−2 |
| TNFA_SIGNALING_VIA_NFKB | 9.43 × 10−3 | – | 1.46 × 10−3 |
| Non common gene sets | |||
| ALLOGRAFT_REJECTION | 1.25 × 10−7 | – | – |
| MTORC1_SIGNALING | 5.73 × 10−7 | – | – |
| INFLAMMATORY_RESPONSE | 1.48 × 10−6 | – | – |
| MYOGENESIS | 4.30 × 10−6 | – | – |
| INTERFERON_GAMMA_RESPONSE | 8.61 × 10−4 | – | – |
| COMPLEMENT | 5.05 × 10−3 | – | – |
| ANGIOGENESIS | 5.82 × 10−3 | – | – |
| IL2_STAT5_SIGNALING | 2.32 × 10−2 | – | – |
| CHOLESTEROL_HOMEOSTASIS | 3.89 × 10−2 | – | – |
| APICAL_JUNCTION | 4.49 × 10−2 | – | – |
| GLYCOLYSIS | 6.27 × 10−2 | – | – |
| HEDGEHOG_SIGNALING | 6.27 × 10−2 | – | – |
| WNT_BETA_CATENIN_SIGNALING | 8.13 × 10−2 | – | – |
| COAGULATION | 8.89 × 10−2 | – | – |
| TGF_BETA_SIGNALING | 9.03 × 10−2 | – | – |
| OXIDATIVE_PHOSPHORYLATION | 9.03 × 10−2 | – | – |
| UNFOLDED_PROTEIN_RESPONSE | 9.55 × 10−2 | – | – |
| UV_RESPONSE_UP | – | 7.42 × 10−2 | – |
| MITOTIC_SPINDLE | – | – | 9.38 × 10−3 |
| DNA_REPAIR | – | – | 3.65 × 10−2 |
| PROTEIN_SECRETION | – | – | 4.11 × 10−2 |
| Name Gene Protein | FC | Protein Function | Function Group *KEGG Pathway | Pathologies Associated | ||
|---|---|---|---|---|---|---|
| Vivo | Vitro | ALI | ||||
| Cenpf Centromere protein F | 1.97 | −8.43 | −1.33 | The CENPF protein is a part of the corona of kinetochore complex which interacts with microtubules and participate to a precise and rapid chromosome segregation. [23] | Mitosis kinetochore–centromere complex | |
| Nuf2 kinetochore protein Nuf2 | 2.66 | −4.05 | −1.31 | Component of the essential kinetochore-associated NDC80 complex, required for chromosome segregation and spindle checkpoint activity, required for kinetochore integrity. [24] | Mitosis kinetochore–centromere complex | |
| Kif15 kinesin-like protein KIF15 | 2.11 | −3.35 | −1.38 | Plus-end directed kinesin-like motor enzyme involved in mitotic spindle assembly [25] | Mitosis kinetochore–centromere complex | |
| Kif20b kinesin family member 20B | 2.50 | −8.91 | −1.39 | Belongs to the TRAFAC class myosin–kinesin ATPase superfamily. kinesin family (String DB) | Mitosis kinetochore–centromere complex | |
| Plk4 serine/threonine protein kinase | 1.67 | −4.64 | −1.36 | serine/threonine–protein kinase that plays a central role in centriole duplication; (UniProt) | Mitosis kinetochore–centromere complex | |
| Depdc1 DEP domain containing 1 | 3.31 | −4.45 | −1.33 | DEP domain containing 1 (DEPDC1) is a highly conserved protein among many species. DEPDC1 was overexpressed in different types of cancers. [26] | Mitosis regulation [26] | Cancer [26] |
| Hmgb2 high mobility group box 2 | 1.46 | −3.59 | −1.44 | Multifunctional protein with various roles in different cellular compartments. May act in a redox sensitive manner. In the nucleus is an abundant chromatin-associated non-histone protein involved in transcription, chromatin remodeling and V(D)J recombination. HMGBs act as architectural facilitators in the assembly of nucleoprotein complexes. [27] | Mitosis chromatin remodeling T cells differentiation | Cancer [28] [29] |
| P2ry12 P2Y purinoceptor 12 | 1.43 | −3.54 | −1.58 | Receptor for ADP and ATP coupled to G-proteins. Required for normal platelet aggregation and blood coagulation. [30] | Inflammation: chemotaxis receptor Cell differentiation Macrophages M1/M2 [31] | Cancer [31] |
| Ccl4 C–C motif chemokine 4 | 1.71 | 3.72 | 2.15 | Monokine with inflammatory and chemokinetic properties; (UniProt) | Inflammation: chemotaxis *Cytokine–cytokine receptor interaction | Inflammation diseases |
| Ccl7 C–C motif chemokine 7 | 5.61 | 3.53 | 1.47 | Chemotactic factor attracts monocytes and eosinophils, but not neutrophils. (String DB) | Inflammation: chemotaxis *Cytokine–cytokine receptor interaction | Inflammation diseases Cancer |
| Osm oncostatin-M | 1.38 | 6.84 | 1.41 | Growth regulator. It regulates cytokine production, including IL-6, G-CSF and GM-CSF; (UniProt) | Inflammation: regulation *Cytokine–cytokine receptor interaction | Cancer |
| Tfec transcription factor EC | 1.35 | −5.86 | −1.32 | transcriptional regulator that acts as a repressor or an activator; (UniProt) | Cell differentiation Macrophages M2 activation [32] Inflammation [33] | Cancer |
| Ogfrl1 opioid growth factor receptor-like 1 | 1.47 | −7.00 | −1.32 | Mobilization and differentiation of bone marrow (BM)-derived cells [34] | Cell differentiation Upregulated in M2 macrophages [35] | Cancer [36] |
| Bcl3 B-cell CLL/lymphoma 3 | 1.69 | 4.60 | 1.41 | BCL3 (BCL3 transcription Coactivator) is a proto-oncogene candidate. Its related pathways are Apoptosis-related network and Common cytokine receptor gamma-chain family signaling pathways. Contributes to the regulation of cell proliferation and to the regulation of transcriptional activation of NF-kappa-B target genes. (GeneCards) | Cell proliferation Apoptosis regulation | Cancer |
| Osgin1 oxidative stress induced growth inhibitor 1 | −1.38 | 7.10 | 1.74 | This gene encodes an oxidative stress response protein that regulates cell death. Expression regulated by p53 and induced by DNA damage. The protein regulates apoptosis by inducing cytochrome c release from mitochondria. Key regulator of both inflammatory and anti-inflammatory molecules. The loss of this protein correlates with uncontrolled cell growth and tumor formation. (GeneCards) | Inflammation regulation Apoptosis regulation | Cancer |
| Ptpn13 protein tyrosine phosphatase non-receptor type 13 | −1.41 | 3.01 | −1.57 | Member of the protein tyrosine phosphatase (PTP) family. PTPs are signaling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle and oncogenic transformation. Regulates negatively FAS-induced apoptosis and NGFR-mediated pro-apoptotic signaling. (GeneCards) | Cell proliferation and differentiation Apoptosis regulation Mitosis Oncogenic transformation | Amyloidosis Cancer |
| Ppp2r5b serine/threonine protein phosphatase regulatory subunit beta isoform | −1.57 | 3.32 | 1.48 | The product of this gene belongs to the phosphatase 2A (PP2A) regulatory subunit B family. PP2A it is implicated in the negative control of cell growth and division. The phosphorylated form mediates the interaction between PP2A and AKT1. (GeneCards) | Cell proliferation | Cancer |
| Bcap29 B-cell receptor-associated protein 29 | 1.54 | −2.90 | −1.38 | Among its related pathways are B Cell Receptor Signaling Pathway and AKT Signaling Pathway. May play a role in transport of membrane proteins. May be involved in CASP8-mediated apoptosis. (GeneCards) | Transport of membrane protein Apoptosis | |
| (A) GO-term | Description | Count in Gene Set | False Discovery Rate |
| GO:0071346 | cellular response to interferon-gamma | 2 of 39 | 0.0265 |
| GO:0070098 | chemokine-mediated signaling pathway | 2 of 30 | 0.0265 |
| GO:0050921 | positive regulation of chemotaxis | 2 of 68 | 0.0265 |
| GO:0048522 | positive regulation of cellular process | 7 of 2201 | 0.0265 |
| GO:0048247 | lymphocyte chemotaxis | 2 of 20 | 0.0265 |
| GO:0045087 | innate immune response | 3 of 217 | 0.0265 |
| GO:0044089 | positive regulation of cellular component biogenesis | 3 of 220 | 0.0265 |
| GO:0040011 | locomotion | 4 of 404 | 0.0265 |
| GO:0030593 | neutrophil chemotaxis | 2 of 23 | 0.0265 |
| GO:0016477 | cell migration | 3 of 293 | 0.0265 |
| GO:0010469 | regulation of signaling receptor activity | 4 of 325 | 0.0265 |
| GO:0009967 | positive regulation of signal transduction | 4 of 638 | 0.0265 |
| GO:0006955 | immune response | 4 of 386 | 0.0265 |
| GO:0006954 | inflammatory response | 3 of 250 | 0.0265 |
| GO:0006935 | chemotaxis | 3 of 172 | 0.0265 |
| GO:0006928 | movement of cell or subcellular component | 4 of 486 | 0.0265 |
| GO:0002687 | positive regulation of leukocyte migration | 2 of 63 | 0.0265 |
| GO:0002548 | monocyte chemotaxis | 2 of 12 | 0.0265 |
| GO:0071347 | cellular response to interleukin-1 | 2 of 75 | 0.0267 |
| GO:0051173 | positive regulation of nitrogen compound metabolic process | 5 of 1184 | 0.0267 |
| (B) KEGG Pathways | Pathway Description | Count in Gene Set | False Discovery Rate |
| rno04060 | Cytokine–cytokine receptor interaction | 3 of 217 | 0.0130 |
| (C) Reactome Pathways | Pathway Description | Count in Gene Set | False Discovery Rate |
| RNO-68877 | Mitotic prometaphase | 4 of 168 | 0.00063 |
| RNO-5663220 | RHO GTPases activate formins | 3 of 116 | 0.0013 |
| RNO-2500257 | Resolution of sister chromatid cohesion | 3 of 100 | 0.0013 |
| RNO-141444 | Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal | 3 of 80 | 0.0013 |
| RNO-2467813 | Separation of sister chromatids | 3 of 149 | 0.0019 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leroux, M.M.; Doumandji, Z.; Chézeau, L.; Gaté, L.; Nahle, S.; Hocquel, R.; Zhernovkov, V.; Migot, S.; Ghanbaja, J.; Bonnet, C.; et al. Toxicity of TiO2 Nanoparticles: Validation of Alternative Models. Int. J. Mol. Sci. 2020, 21, 4855. https://doi.org/10.3390/ijms21144855
Leroux MM, Doumandji Z, Chézeau L, Gaté L, Nahle S, Hocquel R, Zhernovkov V, Migot S, Ghanbaja J, Bonnet C, et al. Toxicity of TiO2 Nanoparticles: Validation of Alternative Models. International Journal of Molecular Sciences. 2020; 21(14):4855. https://doi.org/10.3390/ijms21144855
Chicago/Turabian StyleLeroux, Mélanie M., Zahra Doumandji, Laetitia Chézeau, Laurent Gaté, Sara Nahle, Romain Hocquel, Vadim Zhernovkov, Sylvie Migot, Jafar Ghanbaja, Céline Bonnet, and et al. 2020. "Toxicity of TiO2 Nanoparticles: Validation of Alternative Models" International Journal of Molecular Sciences 21, no. 14: 4855. https://doi.org/10.3390/ijms21144855
APA StyleLeroux, M. M., Doumandji, Z., Chézeau, L., Gaté, L., Nahle, S., Hocquel, R., Zhernovkov, V., Migot, S., Ghanbaja, J., Bonnet, C., Schneider, R., Rihn, B. H., Ferrari, L., & Joubert, O. (2020). Toxicity of TiO2 Nanoparticles: Validation of Alternative Models. International Journal of Molecular Sciences, 21(14), 4855. https://doi.org/10.3390/ijms21144855







