Intracellular Fate and Impact on Gene Expression of Doxorubicin/Cyclodextrin-Graphene Nanomaterials at Sub-Toxic Concentration
Abstract
:1. Introduction
2. Results
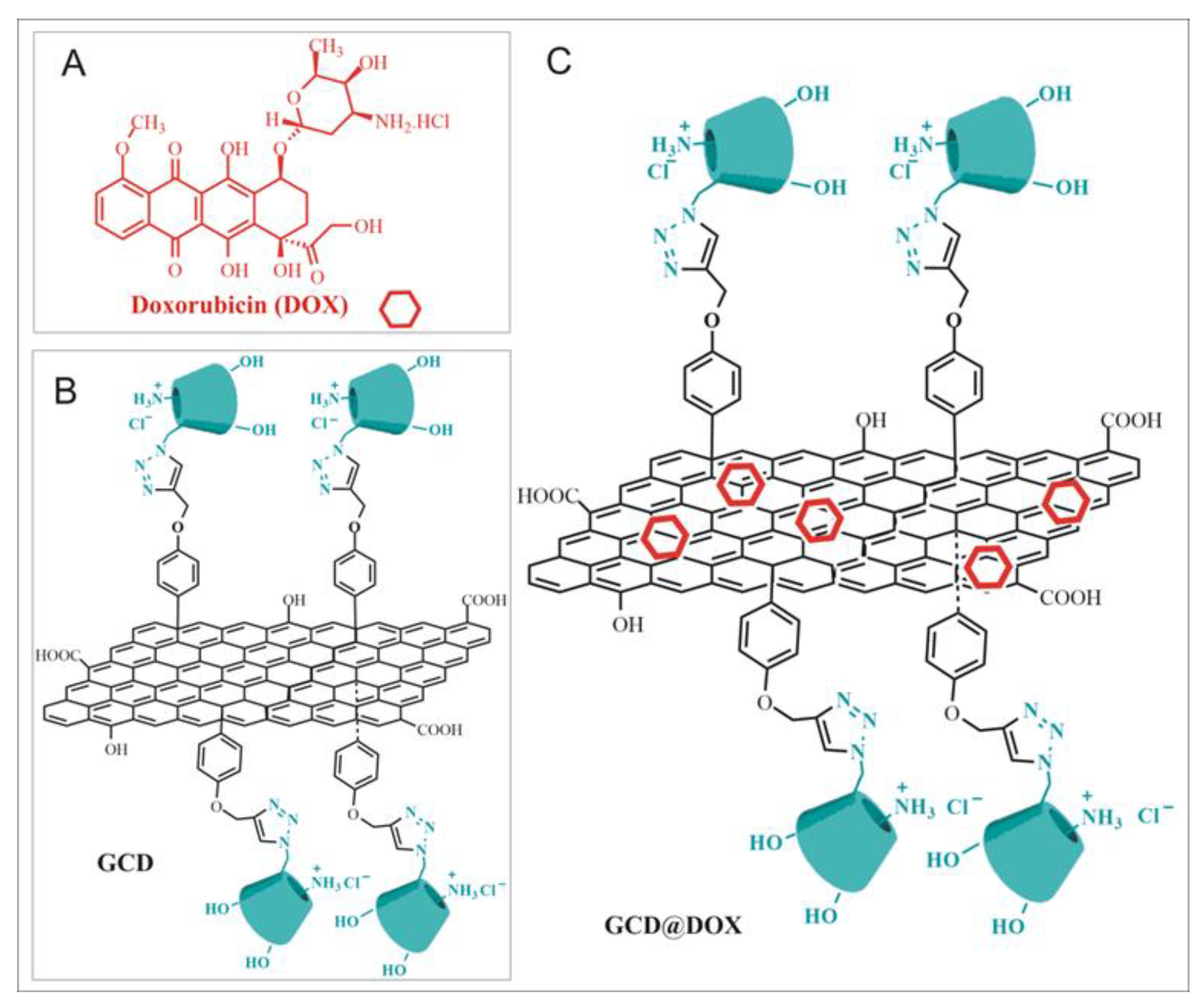
2.1. Preparation and Characterization of GCD@DOX Complex
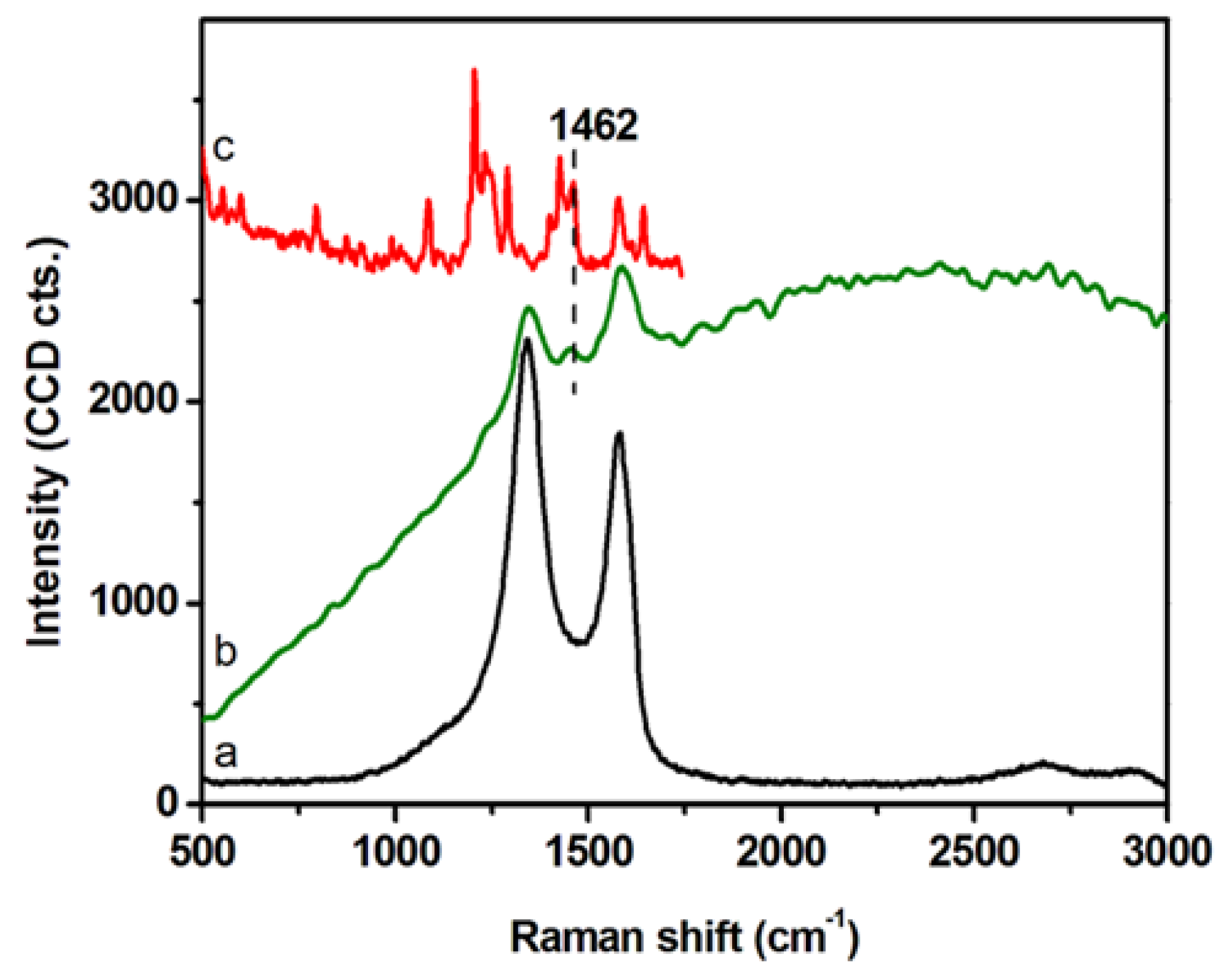
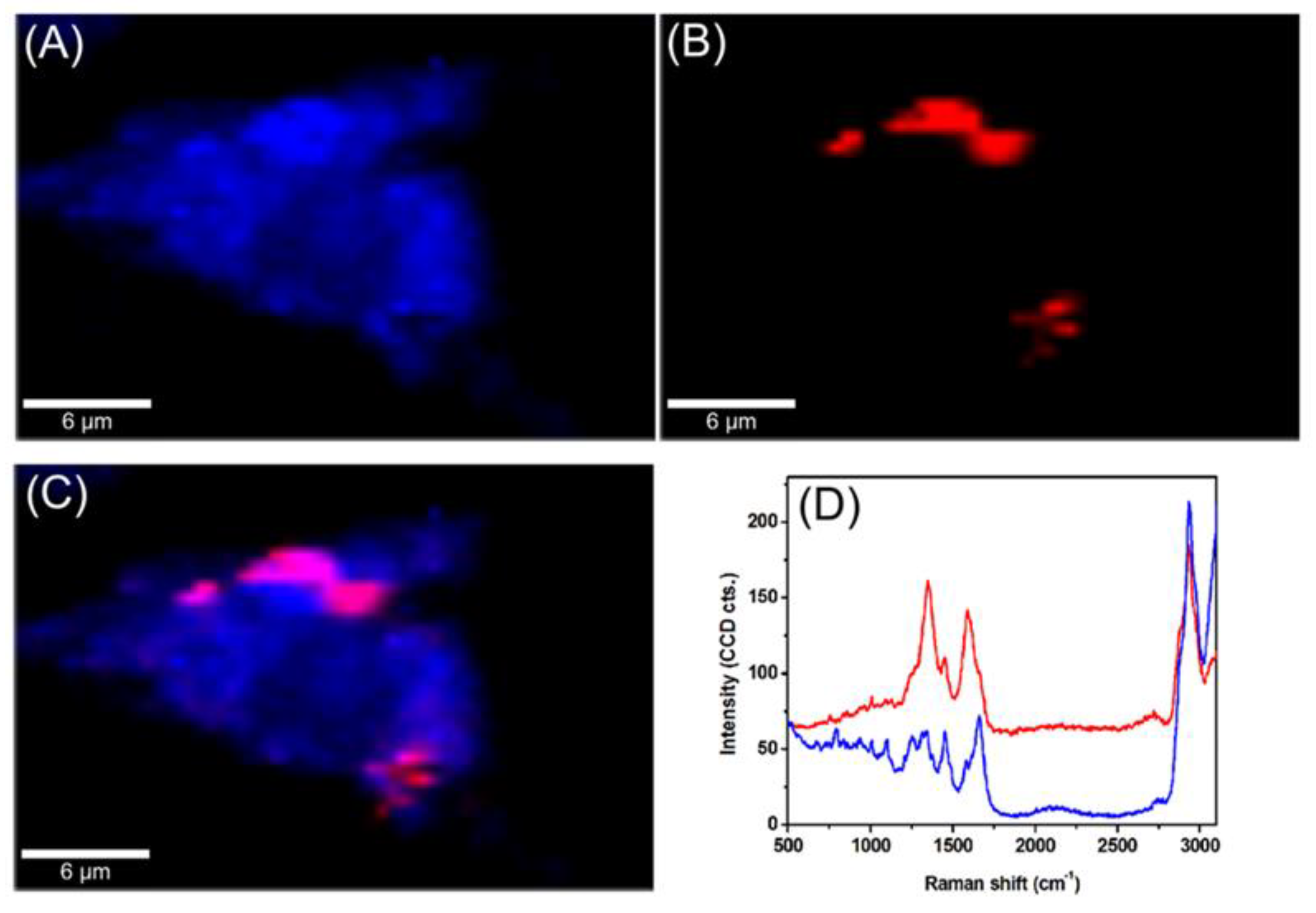
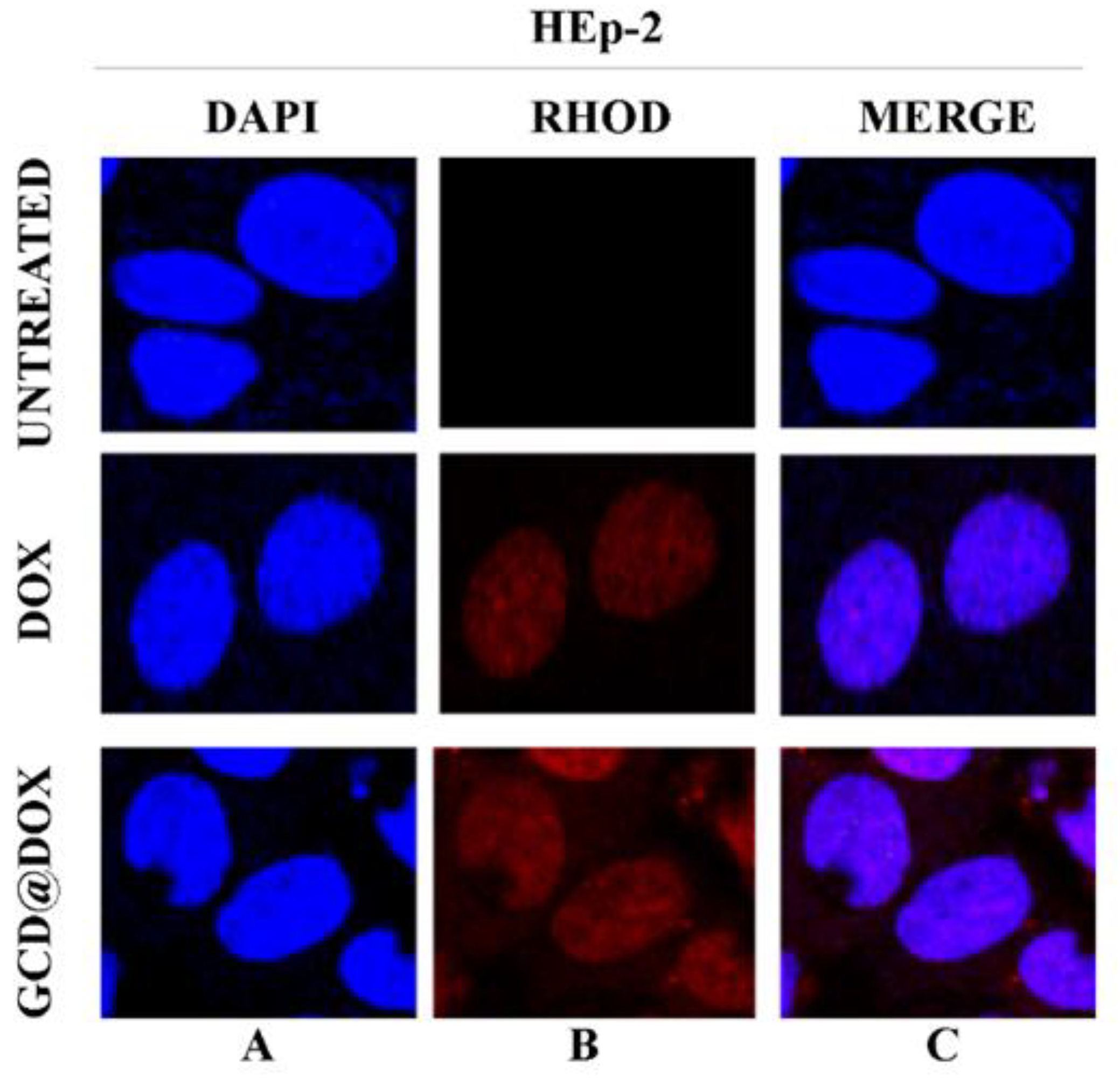
2.2. Intracellular Trafficking of GCD@DOX by FLIM and Raman Mapping and Fluorescent Microscopy
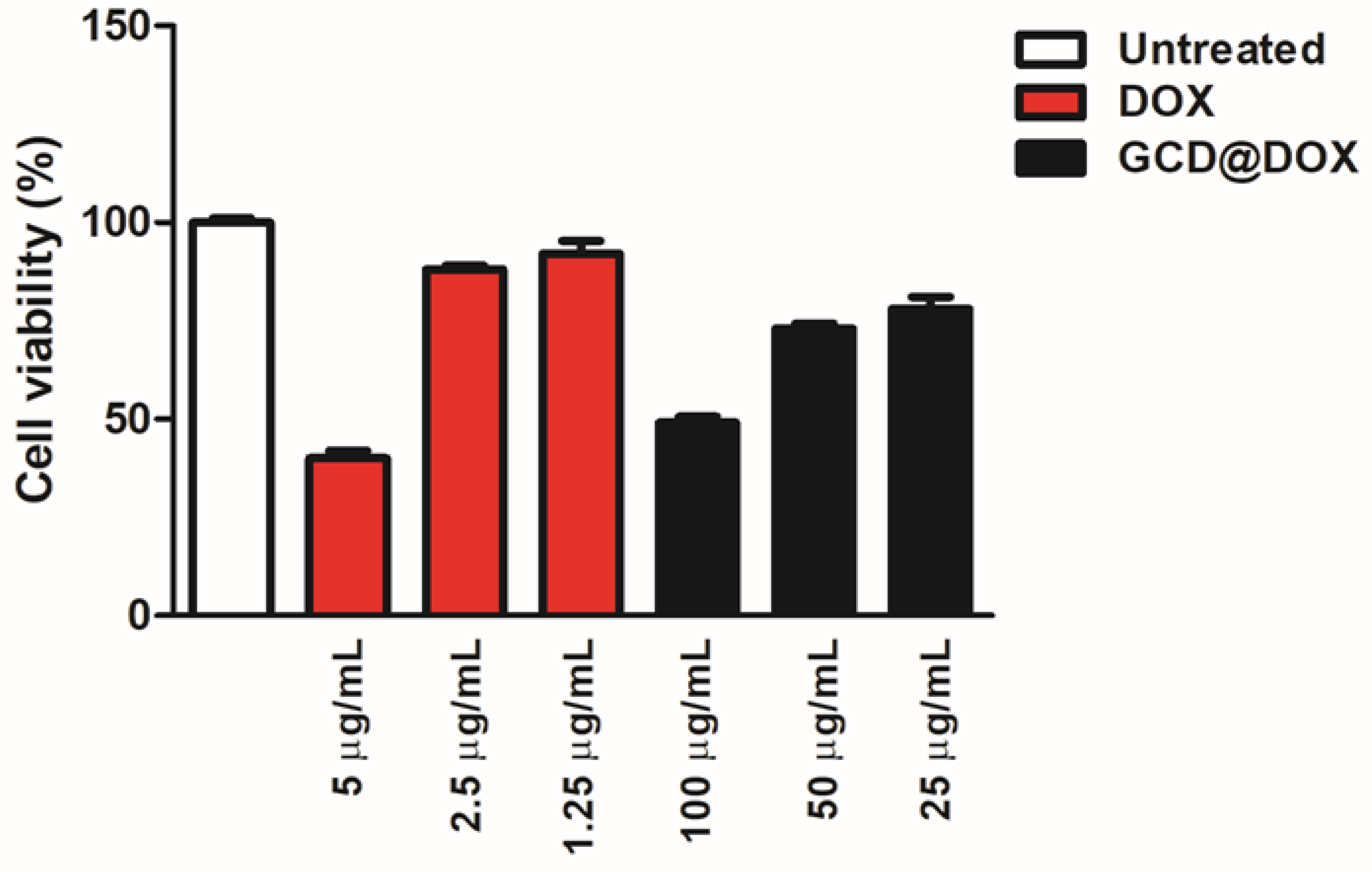
2.3. Cytotoxicity Study
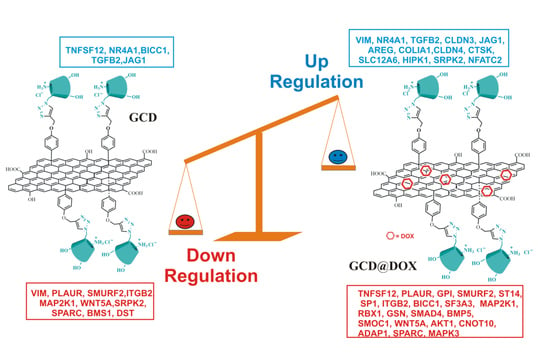
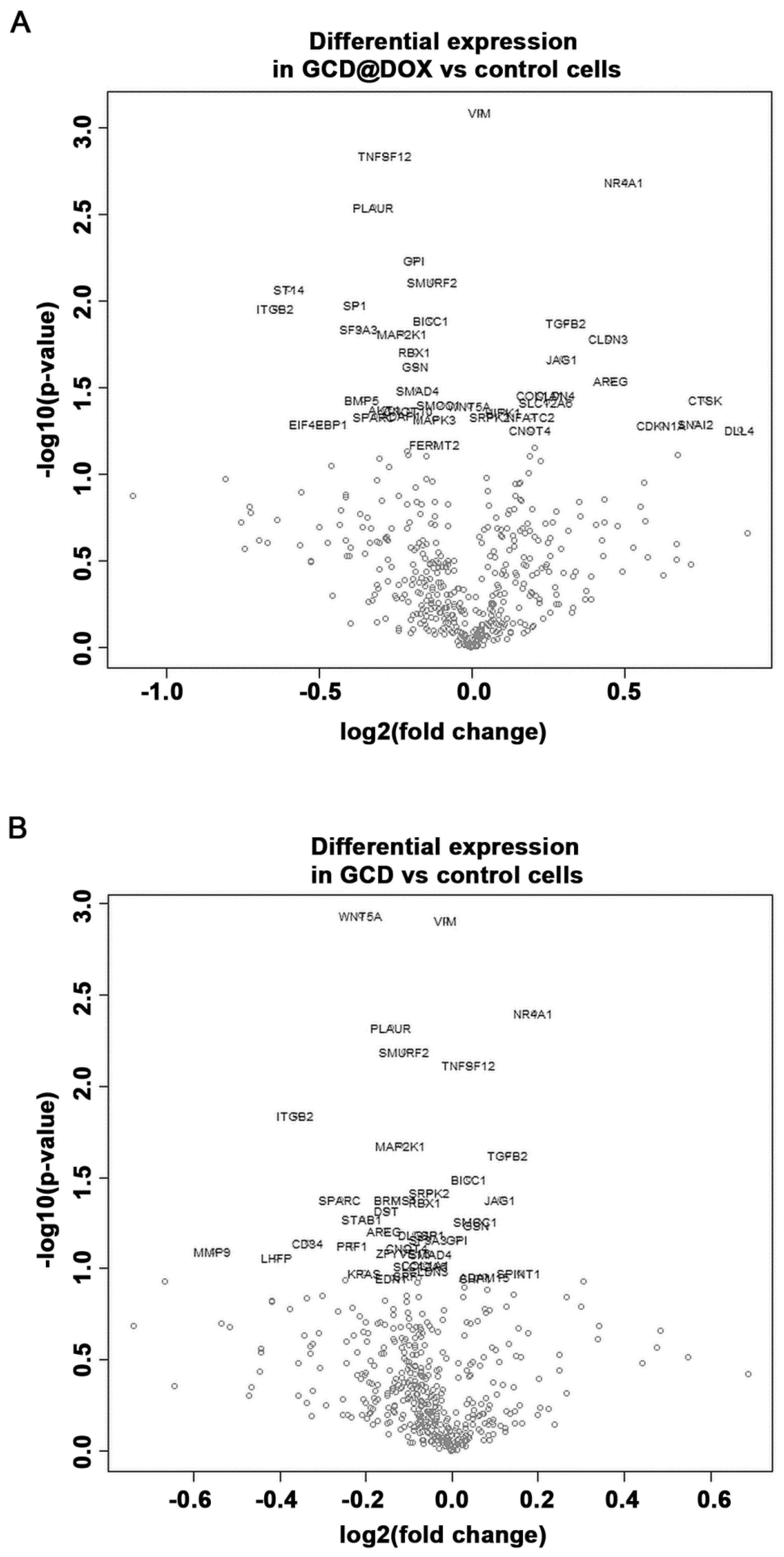
2.4. Gene Expression Modulation in HEp-2 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Viability Assay
4.3. Intracellular Uptake of GCD@DOX
4.4. Characterization Techniques
4.5. Preparation of GCD@DOX
4.6. RNA Isolation and NanoString® Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| G | Graphene |
| GCD | Graphene modified with cationic cyclodextrins |
| DOX | Doxorubicin |
| GCD@DOX | Graphene cationic cyclodextrin platform entrapping doxorubicin |
| DLS | Dynamic light scattering |
| FLIM | Fluorescence lifetime imaging microscopy |
| TEM | Transmission electron microscope |
| HEp-2 cells | Human epithelial type 2 cells |
| ECM | Extracellular matrix |
| ST14 | Matriptase |
| ITGB2 | Integrin-beta2 |
| JAG1 | Jagged 1 |
| AREG | Amphiregulin |
| CTSK | Cathepsin K |
| SPARC | Osteonectin |
| GSN | Gelsolin |
| BMP5 | Bone morphogenetic protein 5 |
| AKT1 | Serine/Threonine Kinase 1 |
| CLDN3 | Claudin-3 |
| NR4A1 | Nuclear receptor subfamily 4 group A member 1 |
| TGFβ2 | Transforming growth factor β |
| VIM | Vimentin |
| TNFSF12 | Tumor Necrosis Factor (Ligand) Superfamily Member 12 |
| BICC1 | BicC RNA Binding Protein 1 |
| SRPK2 | Serine/Arginine-Rich Protein-Specific Kinase 2 |
References
- Orecchioni, M.; Cabizza, R.; Bianco, A.; Delogu, L.G. Graphene as cancer theranostic tool: Progress and future challenges. Theranostics 2015, 5, 710–723. [Google Scholar] [CrossRef] [PubMed]
- Barreca, D.; Neri, G.; Scala, A.; Fazio, E.; Gentile, D.; Rescifina, A.; Piperno, A. Covalently immobilized catalase on functionalized graphene: Effect on the activity, immobilization efficiency, and tetramer stability. Biomater. Sci. 2018, 6, 3231–3240. [Google Scholar] [CrossRef] [PubMed]
- Cordaro, A.; Neri, G.; Sciortino, M.T.; Scala, A.; Piperno, A. Graphene-based strategies in liquid biopsy and in viral diseases diagnosis. Nanomaterials 2020, 10, 1014. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liang, X.; Zhang, J.; Yin, Y.; Zuo, T.; Wang, Y.; Yang, X.; Shen, Q. Inhibiting pulmonary metastasis of breast cancer based on dual-targeting graphene oxide with high stability and drug loading capacity. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1237–1248. [Google Scholar] [CrossRef]
- Xu, X.; Wang, J.; Wang, Y.; Zhao, L.; Li, Y.; Liu, C. Formation of graphene oxide-hybridized nanogels for combinative anticancer therapy. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 2387–2395. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, M.; Mignani, S.; Rodrigues, J.; Tomás, H. A glance over doxorubicin based-nanotherapeutics: From proof-of-concept studies to solutions in the market. J. Control. Release 2020, 317, 347–374. [Google Scholar] [CrossRef]
- Zhao, H.; Ding, R.; Zhao, X.; Li, Y.; Qu, L.; Pei, H.; Yildirimer, L.; Wu, Z.; Zhang, W. Graphene-based nanomaterials for drug and/or gene delivery, bioimaging, and tissue engineering. Drug Discov. Today 2017, 22, 1302–1317. [Google Scholar] [CrossRef]
- Li, Y.; Gao, X.; Yu, Z.; Liu, B.; Pan, W.; Li, N.; Tang, B. Reversing multidrug resistance by multiplexed gene silencing for enhanced breast cancer chemotherapy. ACS Appl. Mater. Interfaces 2018, 10, 15461–15466. [Google Scholar] [CrossRef]
- Piperno, A.; Scala, A.; Mazzaglia, A.; Neri, G.; Pennisi, R.; Sciortino, M.T.; Grassi, G. Cellular signaling pathways activated by functional graphene nanomaterials. Int. J. Mol. Sci. 2018, 19, 3365. [Google Scholar] [CrossRef] [Green Version]
- Martín, C.; Kostarelos, K.; Prato, M.; Bianco, A. Biocompatibility and biodegradability of 2d materials: Graphene and beyond. Chem. Commun. 2019, 55, 5540–5546. [Google Scholar] [CrossRef]
- Loeian, M.S.; Mehdi Aghaei, S.; Farhadi, F.; Rai, V.; Yang, H.W.; Johnson, M.D.; Aqil, F.; Mandadi, M.; Rai, S.N.; Panchapakesan, B. Liquid biopsy using the nanotube-ctc-chip: Capture of invasive ctcs with high purity using preferential adherence in breast cancer patients. Lab Chip 2019, 19, 1899–1915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neri, G.; Scala, A.; Fazio, E.; Mineo, P.G.; Rescifina, A.; Piperno, A.; Grassi, G. Repurposing of oxazolone chemistry: Gaining access to functionalized graphene nanosheets in a top-down approach from graphite. Chem. Sci. 2015, 6, 6961–6970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, E.; Li, B.L.; Ariga, K.; Lim, C.-T.; Garaj, S.; Leong, D.T. Toxicity of two-dimensional layered materials and their heterostructures. Bioconj. Chem. 2019, 30, 2287–2299. [Google Scholar] [CrossRef] [PubMed]
- Neri, G.; Scala, A.; Barreca, F.; Fazio, E.; Mineo, P.G.; Mazzaglia, A.; Grassi, G.; Piperno, A. Engineering of carbon based nanomaterials by ring-opening reactions of a reactive azlactone graphene platform. Chem. Commun. 2015, 51, 4846–4849. [Google Scholar] [CrossRef] [PubMed]
- Neri, G.; Micale, N.; Scala, A.; Fazio, E.; Mazzaglia, A.; Mineo, P.G.; Montesi, M.; Panseri, S.; Tampieri, A.; Grassi, G.; et al. Silibinin-conjugated graphene nanoplatform: Synthesis, characterization and biological evaluation. FlatChem 2017, 1, 34–41. [Google Scholar] [CrossRef]
- Piperno, A.; Mazzaglia, A.; Scala, A.; Pennisi, R.; Zagami, R.; Neri, G.; Torcasio, S.M.; Rosmini, C.; Mineo, P.G.; Potara, M.; et al. Casting light on intracellular tracking of a new functional graphene-based microrna delivery system by flim and raman imaging. ACS Appl. Mater. Interfaces 2019, 11, 46101–46111. [Google Scholar] [CrossRef]
- Yazdimamaghani, M.; Moos, P.J.; Ghandehari, H. Time- and dose-dependent gene expression analysis of macrophage response as a function of porosity of silica nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2019, 21, 102041. [Google Scholar] [CrossRef]
- Orecchioni, M.; Jasim, D.A.; Pescatori, M.; Manetti, R.; Fozza, C.; Sgarrella, F.; Bedognetti, D.; Bianco, A.; Kostarelos, K.; Delogu, L.G. Molecular and genomic impact of large and small lateral dimension graphene oxide sheets on human immune cells from healthy donors. Adv. Healthc. Mater. 2016, 5, 276–287. [Google Scholar] [CrossRef]
- Orecchioni, M.; Bedognetti, D.; Newman, L.; Fuoco, C.; Spada, F.; Hendrickx, W.; Marincola, F.M.; Sgarrella, F.; Rodrigues, A.F.; Ménard-Moyon, C.; et al. Single-cell mass cytometry and transcriptome profiling reveal the impact of graphene on human immune cells. Nat. Commun. 2017, 8, 1109. [Google Scholar] [CrossRef] [Green Version]
- de Lázaro, I.; Kostarelos, K. Exposure to graphene oxide sheets alters the expression of reference genes used for real-time rt-qpcr normalization. Sci. Rep. 2019, 9, 12520. [Google Scholar] [CrossRef] [Green Version]
- Farahani, M.; Rezaei-Tavirani, M.; Zali, H.; Hatamie, S.; Ghasemi, N. Systems toxicology assessment revealed the impact of graphene-based materials on cell cycle regulators. J. Biomed. Mater. Res. Part A 2020, 108, 1520–1533. [Google Scholar] [CrossRef] [PubMed]
- Kasprzak, A.; Poplawska, M. Recent developments in the synthesis and applications of graphene-family materials functionalized with cyclodextrins. Chem. Comm. 2018, 54, 8547–8562. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, P.; Parandhaman, T.; Ramalingam, B.; Duraipandy, N.; Kiran, M.S.; Das, S.K. Fabrication of nontoxic reduced graphene oxide protein nanoframework as sustained antimicrobial coating for biomedical application. ACS Appl. Mater. Interfaces 2017, 9, 38255–38269. [Google Scholar] [CrossRef] [PubMed]
- Neri, G.; Fazio, E.; Mineo, P.G.; Scala, A.; Piperno, A. Sers sensing properties of new graphene/gold nanocomposite. Nanomaterials 2019, 9, 1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farhane, Z.; Bonnier, F.; Casey, A.; Byrne, H.J. Raman micro spectroscopy for in vitro drug screening: Subcellular localisation and interactions of doxorubicin. Analyst 2015, 140, 4212–4223. [Google Scholar] [CrossRef] [Green Version]
- Schneckenburger, H.; Wagner, M.; Weber, P.; Strauss, W.S.L.; Sailer, R. Autofluorescence lifetime imaging of cultivated cells using a uv picosecond laser diode. J. Fluoresc. 2004, 14, 649–654. [Google Scholar] [CrossRef]
- Yang, F.; Teves, S.S.; Kemp, C.J.; Henikoff, S. Doxorubicin, DNA torsion, and chromatin dynamics. Biochim. Biophys. Acta BBA Rev. Cancer 2014, 1845, 84–89. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Xu, M.; Gao, M.; Zhang, Z.; Xu, Y.; Xia, T.; Liu, S. Graphene oxide induced perturbation to plasma membrane and cytoskeletal meshwork sensitize cancer cells to chemotherapeutic agents. ACS Nano 2017, 11, 2637–2651. [Google Scholar] [CrossRef]
- Imani, R.; Prakash, S.; Vali, H.; Faghihi, S. Polyethylene glycol and octa-arginine dual-functionalized nanographene oxide: An optimization for efficient nucleic acid delivery. Biomater. Sci. 2018, 6, 1636–1650. [Google Scholar] [CrossRef]
- Sun, Q.; Wang, X.; Cui, C.; Li, J.; Wang, Y. Doxorubicin and anti-vegf sirna co-delivery via nano-graphene oxide for enhanced cancer therapy in vitro and in vivo. Int. J. Nanomed. 2018, 13, 3713–3728. [Google Scholar] [CrossRef] [Green Version]
- Ashjaran, M.; Babazadeh, M.; Akbarzadeh, A.; Davaran, S.; Salehi, R. Stimuli-responsive polyvinylpyrrolidone-nippam-lysine graphene oxide nano-hybrid as an anticancer drug delivery on mcf7 cell line. Artif. Cell. Nanomed. B. 2019, 47, 443–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmanouilidi, A.; Paladin, D.; Greening, D.W.; Falasca, M. Oncogenic and non-malignant pancreatic exosome cargo reveal distinct expression of oncogenic and prognostic factors involved in tumor invasion and metastasis. Proteomics 2019, 19, 1800158. [Google Scholar] [CrossRef] [PubMed]
- Mazar, A.P. Urokinase plasminogen activator receptor choreographs multiple ligand interactions: Implications for tumor progression and therapy. Clin. Cancer Res. 2008, 14, 5649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wang, Y.; Kong, F.; Han, R.; Song, W.; Chen, D.; Bu, L.; Wang, S.; Yue, J.; Ma, L. Identification of a six-gene prognostic signature for oral squamous cell carcinoma. J. Cell. Physiol. 2020, 235, 3056–3068. [Google Scholar] [CrossRef]
- Safe, S.; Abbruzzese, J.; Abdelrahim, M.; Hedrick, E. Specificity protein transcription factors and cancer: Opportunities for drug development. Cancer Prev. Res. 2018, 11, 371. [Google Scholar] [CrossRef]
- Siebring-van Olst, E.; Blijlevens, M.; de Menezes, R.X.; van der Meulen-Muileman, I.H.; Smit, E.F.; van Beusechem, V.W. A genome-wide sirna screen for regulators of tumor suppressor p53 activity in human non-small cell lung cancer cells identifies components of the rna splicing machinery as targets for anticancer treatment. Mol. Oncol. 2017, 11, 534–551. [Google Scholar] [CrossRef] [Green Version]
- Deng, R.; Hao, J.; Han, W.; Ni, Y.; Huang, X.; Hu, Q. Gelsolin regulates proliferation, apoptosis, migration and invasion in human oral carcinoma cells. Oncol. Lett. 2015, 9, 2129–2134. [Google Scholar] [CrossRef]
- Chen, Z.; Li, K.; Yin, X.; Li, H.; Li, Y.; Zhang, Q.; Wang, H.; Qiu, Y. Lower expression of gelsolin in colon cancer and its diagnostic value in colon cancer patients. J. Cancer 2019, 10, 1288–1296. [Google Scholar] [CrossRef]
- Zhang, J.; Furuta, T.; Sabit, H.; Tamai, S.; Jiapaer, S.; Dong, Y.; Kinoshita, M.; Uchida, Y.; Ohtsuki, S.; Terasaki, T.; et al. Gelsolin inhibits malignant phenotype of glioblastoma and is regulated by mir-654-5p and mir-450b-5p. Cancer Sci. 2020. [Google Scholar] [CrossRef]
- Feldt, J.; Schicht, M.; Garreis, F.; Welss, J.; Schneider, U.W.; Paulsen, F. Structure, regulation and related diseases of the actin-binding protein gelsolin. Expert Rev. Mol. Med. 2019, 20, e7. [Google Scholar] [CrossRef]
- Rao, G.; Pierobon, M.; Kim, I.K.; Hsu, W.H.; Deng, J.; Moon, Y.W.; Petricoin, E.F.; Zhang, Y.W.; Wang, Y.; Giaccone, G. Inhibition of akt1 signaling promotes invasion and metastasis of non-small cell lung cancer cells with k-ras or egfr mutations. Sci. Rep. 2017, 7, 7066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoeli-Lerner, M.; Yiu, G.K.; Rabinovitz, I.; Erhardt, P.; Jauliac, S.; Toker, A. Akt blocks breast cancer cell motility and invasion through the transcription factor nfat. Mol. Cell 2005, 20, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.-L.; Tucker, D.F.; Bae, S.S.; Bhatheja, K.; Birnbaum, M.J.; Field, J. Opposing roles for akt1 and akt2 in rac/pak signaling and cell migration. J. Biol. Chem. 2006, 281, 36443–36453. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Shang, X.; Manorek, G.; Howell, S.B. Regulation of the epithelial-mesenchymal transition by claudin-3 and claudin-4. PLoS ONE 2013, 8, e67496. [Google Scholar] [CrossRef] [Green Version]
- Hewitt, K.J.; Agarwal, R.; Morin, P.J. The claudin gene family: Expression in normal and neoplastic tissues. BMC Cancer 2006, 6, 186. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, R.; Souza, T.; Morin, P.J. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res. 2005, 65, 7378–7385. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wang, Y.; Zhang, B.; Zhang, H.; Zhou, M.; Wei, M.; Dong, Q.; Xu, Y.; Wang, Z.; Gao, L.; et al. Claudin-3 expression increases the malignant potential of lung adenocarcinoma cells: Role of epidermal growth factor receptor activation. Oncotarget 2017, 8, 23033–23047. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Yang, Y.D.; Fu, L.; Xu, W.; Liu, D.; Liang, Q.; Zhang, X.; Xu, L.; Guan, X.Y.; Wu, B.; et al. Cldn3 inhibits cancer aggressiveness via wnt-emt signaling and is a potential prognostic biomarker for hepatocellular carcinoma. Oncotarget 2014, 5, 7663–7676. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.; Yang, F.; He, H.; Li, Q.; Zhang, W.; Xing, J.; Zhu, Z.; Jiang, J.; Wang, H.; Zhao, X.; et al. Alteration of tumor suppressor bmp5 in sporadic colorectal cancer: A genomic and transcriptomic profiling based study. Mol. Cancer 2018, 17, 176. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Dai, X.; Cao, X.; Yan, H.; Ji, X.; Zhang, H.; Shen, S.; Si, Y.; Zhang, H.; Chen, J.; et al. Prdm4 mediates yap-induced cell invasion by activating leukocyte-specific integrin β2 expression. EMBO Rep. 2018, 19, e45180. [Google Scholar] [CrossRef]
- Hu, G.; Zeng, W.; Xia, Y. Tweak/fn14 signaling in tumors. Tumor Biol. 2017, 39, 1010428317714624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.O.; Li, X.; Khan, S.; Safe, S. Targeting nr4a1 (tr3) in cancer cells and tumors. Expert Opin. Ther. Targets 2011, 15, 195–206. [Google Scholar] [CrossRef]
- Zhu, B.; Yang, J.R.; Jia, Y.; Zhang, P.; Shen, L.; Li, X.L.; Li, J.; Wang, B. Overexpression of nr4a1 is associated with tumor recurrence and poor survival in non-small-cell lung carcinoma. Oncotarget 2017, 8, 113977–113986. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, E.; Lee, S.O.; Doddapaneni, R.; Singh, M.; Safe, S. Nuclear receptor 4a1 as a drug target for breast cancer chemotherapy. Endocr. Relat. Cancer 2015, 22, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Bierie, B.; Moses, H.L. Tgfβ: The molecular jekyll and hyde of cancer. Nat. Rev. Cancer 2006, 6, 506–520. [Google Scholar] [CrossRef]
- Bierie, B.; Moses, H.L. Transforming growth factor beta (tgf-β) and inflammation in cancer. Cytokine Growth F. R. 2010, 21, 49–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Sun, J.; Liu, X.; Nan, J.; Qin, X.; Wang, X.; Guo, J.; Zhao, C.; Yang, J. Tgf-β2 antagonizes il-6-promoted cell survival. Mol. Cell. Biochem. 2019, 461, 119–126. [Google Scholar] [CrossRef]
- Herreño, A.M.; Ramírez, A.C.; Chaparro, V.P.; Fernandez, M.J.; Cañas, A.; Morantes, C.F.; Moreno, O.M.; Brugés, R.E.; Mejía, J.A.; Bustos, F.J.; et al. Role of runx2 transcription factor in epithelial mesenchymal transition in non-small cell lung cancer: Epigenetic control of the runx2 p1 promoter. Tumor Biol. 2019, 41, 1010428319851014. [Google Scholar] [CrossRef] [Green Version]
- Prattapong, P.; Ngernsombat, C.; Aimjongjun, S.; Janvilisri, T. Crispr/cas9-mediated double knockout of srpk1 and srpk2 in a nasopharyngeal carcinoma cell line. Cancer Rep. 2020, 3, e1224. [Google Scholar] [CrossRef]
- Hatcher, J.M.; Wu, G.; Zeng, C.; Zhu, J.; Meng, F.; Patel, S.; Wang, W.; Ficarro, S.B.; Leggett, A.L.; Powell, C.E.; et al. Srpkin-1: A covalent srpk1/2 inhibitor that potently converts vegf from pro-angiogenic to anti-angiogenic isoform. Cell Chem. Biol. 2018, 25, 460–470.e466. [Google Scholar] [CrossRef] [Green Version]

| Sample | DH (µm ± SD) 1,2 (%) 3 | PDI | ζ (mV ± SD) | Theoretical Loading (%) | 3 Actual Loading (%) | 4 EE (%) |
|---|---|---|---|---|---|---|
| GCD | >1 μm | ≥ 0.4 | −9.5 ± 0.3 | |||
| GCD@DOX | 1 μm ± 0.2 | ≥ 0.4 | −16.6 ± 4 | 2.6 | 2.5 | 95 |
| GCD@DOX vs. Ctrl 3 | GCD vs. Ctrl | |||
|---|---|---|---|---|
| Gene | Log2 Fold Change | p-Value | Log2 Fold Change | p-Value |
| VIM 1 | 0.0277 | 0.000813 | −0.0134 | 0.00124 |
| TNFSF12 1 | −0.281 | 0.00145 | 0.0395 | 0.00759 |
| NR4A1 | 0.5 | 0.00206 | 0.19 | 0.004 |
| PLAUR | −0.322 | 0.00286 | −0.142 | 0.00477 |
| GPI | −0.187 | 0.0058 | ||
| SMURF2 | −0.127 | 0.00774 | −0.112 | 0.00649 |
| ST142 | −0.597 | 0.00851 | ||
| SP1 | −0.379 | 0.0105 | ||
| ITGB22 | −0.641 | 0.011 | −0.362 | 0.0144 |
| BICC1 1 | −0.132 | 0.0128 | 0.0386 | 0.0323 |
| TGFβ2 | 0.309 | 0.0134 | 0.129 | 0.0238 |
| SF3A3 | −0.368 | 0.0144 | ||
| MAP2K1 | −0.225 | 0.0153 | −0.12 | 0.0212 |
| CLDN3 | 0.45 | 0.0164 | ||
| RBX1 | −0.185 | 0.0194 | −0.0617 | 0.0429 |
| JAG12 | 0.298 | 0.0214 | 0.113 | 0.0416 |
| GSN | −0.184 | 0.0238 | ||
| AREG2 | 0.456 | 0.0285 | ||
| SMAD4 | −0.174 | 0.0327 | ||
| COL1A1 | 0.226 | 0.0345 | ||
| CLDN4 | 0.278 | 0.0349 | ||
| CTSK2 | 0.766 | 0.037 | ||
| BMP5 | −0.358 | 0.0373 | ||
| SLC12A6 | 0.246 | 0.0385 | ||
| SMOC1 | −0.106 | 0.0394 | ||
| WNT5A | −0.00843 | 0.0398 | −0.211 | 0.00117 |
| AKT1 | −0.283 | 0.0423 | ||
| CNOT10 | −0.202 | 0.0428 | ||
| HIPK1 | 0.103 | 0.0438 | ||
| ADAP1 | −0.23 | 0.0455 | ||
| SRPK2 1 | 0.0604 | 0.0464 | −0.0541 | 0.0382 |
| SPARC2 | −0.319 | 0.0464 | −0.259 | 0.0421 |
| NFATC2 | −0.194 | 0.0466 | ||
| MAPK3 | −0.122 | 0.0481 | ||
| BRMS1 | −0.13 | 0.0416 | ||
| DST | −0.152 | 0.0481 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caccamo, D.; Currò, M.; Ientile, R.; Verderio, E.A.; Scala, A.; Mazzaglia, A.; Pennisi, R.; Musarra-Pizzo, M.; Zagami, R.; Neri, G.; et al. Intracellular Fate and Impact on Gene Expression of Doxorubicin/Cyclodextrin-Graphene Nanomaterials at Sub-Toxic Concentration. Int. J. Mol. Sci. 2020, 21, 4891. https://doi.org/10.3390/ijms21144891
Caccamo D, Currò M, Ientile R, Verderio EA, Scala A, Mazzaglia A, Pennisi R, Musarra-Pizzo M, Zagami R, Neri G, et al. Intracellular Fate and Impact on Gene Expression of Doxorubicin/Cyclodextrin-Graphene Nanomaterials at Sub-Toxic Concentration. International Journal of Molecular Sciences. 2020; 21(14):4891. https://doi.org/10.3390/ijms21144891
Chicago/Turabian StyleCaccamo, Daniela, Monica Currò, Riccardo Ientile, Elisabetta AM Verderio, Angela Scala, Antonino Mazzaglia, Rosamaria Pennisi, Maria Musarra-Pizzo, Roberto Zagami, Giulia Neri, and et al. 2020. "Intracellular Fate and Impact on Gene Expression of Doxorubicin/Cyclodextrin-Graphene Nanomaterials at Sub-Toxic Concentration" International Journal of Molecular Sciences 21, no. 14: 4891. https://doi.org/10.3390/ijms21144891
APA StyleCaccamo, D., Currò, M., Ientile, R., Verderio, E. A., Scala, A., Mazzaglia, A., Pennisi, R., Musarra-Pizzo, M., Zagami, R., Neri, G., Rosmini, C., Potara, M., Focsan, M., Astilean, S., Piperno, A., & Sciortino, M. T. (2020). Intracellular Fate and Impact on Gene Expression of Doxorubicin/Cyclodextrin-Graphene Nanomaterials at Sub-Toxic Concentration. International Journal of Molecular Sciences, 21(14), 4891. https://doi.org/10.3390/ijms21144891