Gram-Negative Bacterial Endotoxin LPS Induces NeuGc Loss through Ets1-Dependent Downregulation of Intestine-Specific pcmah Transcript in Porcine Intestinal Cells
Abstract
1. Introduction
2. Results
2.1. LPS Exposure Leads to Loss of NeuGc Biosynthesis in Pig Small Intestinal IPI-2I Cells
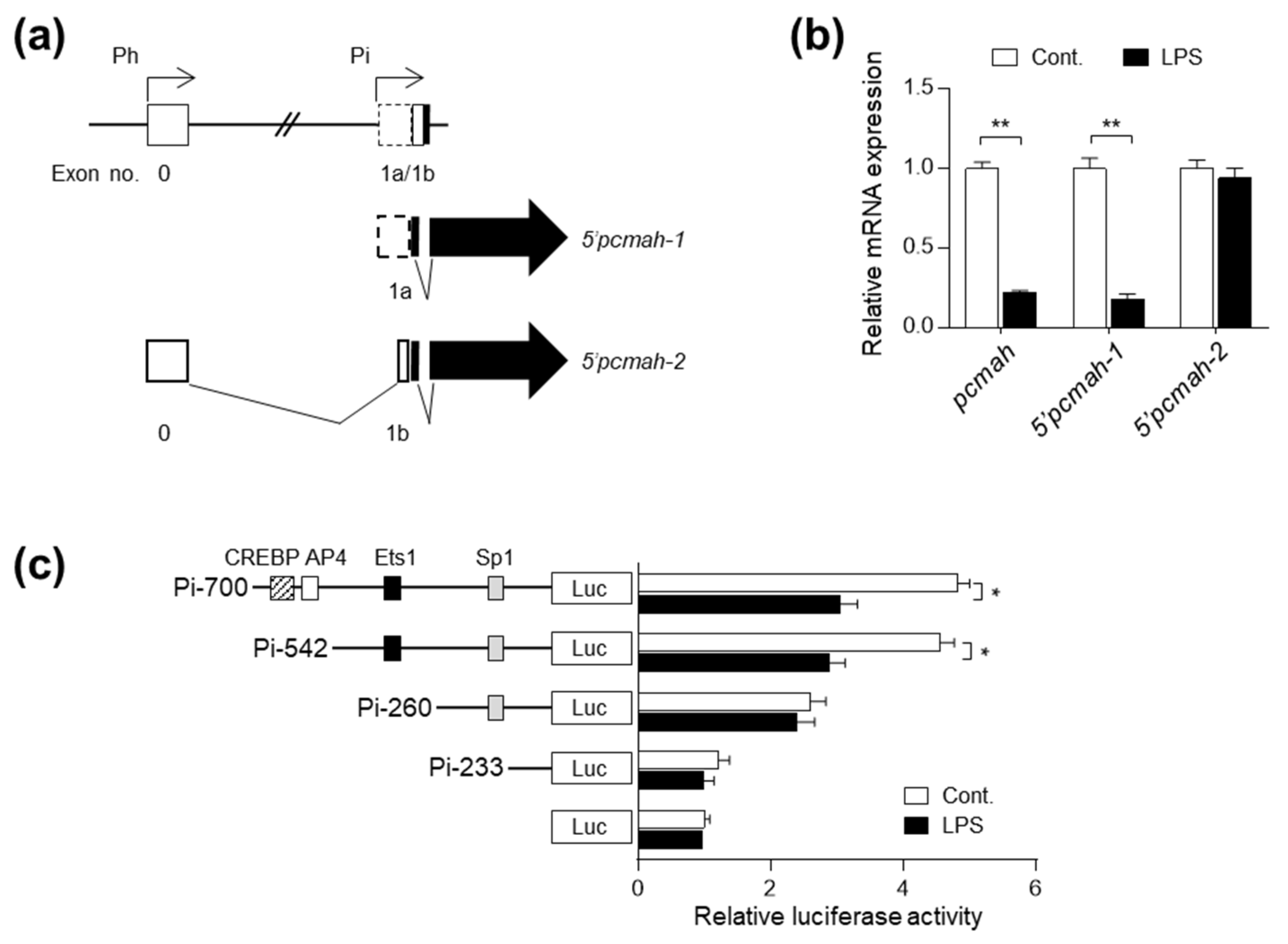
2.2. LPS-Induced pcmah Loss Is Mediated by the Intestine-Specific 5′pcmah-1 Transcript
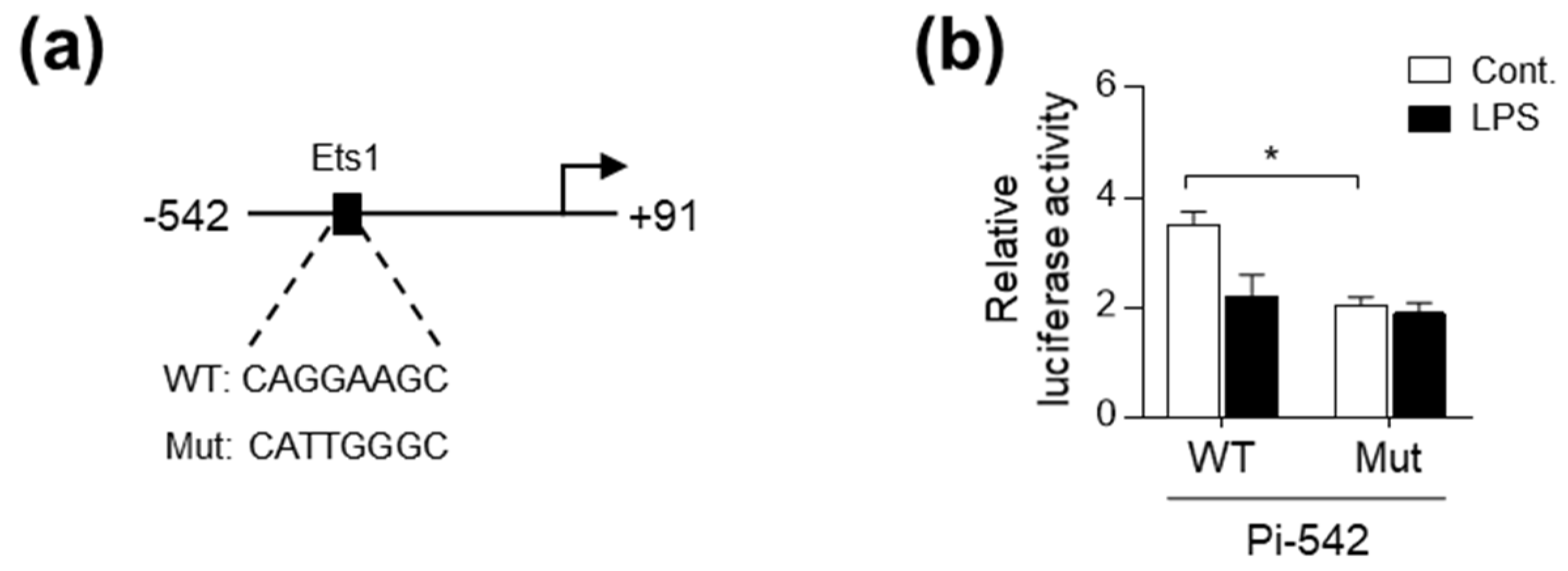
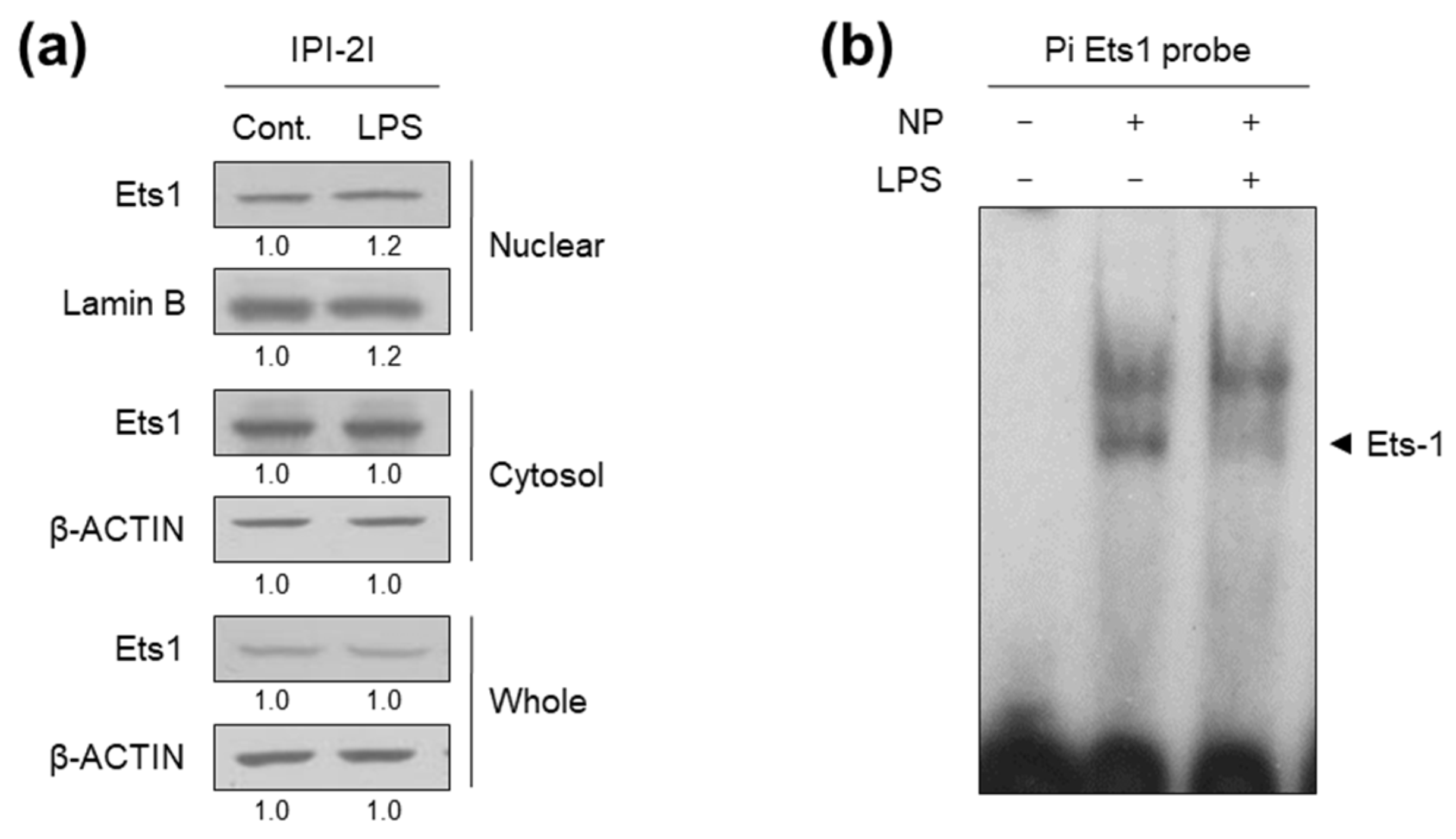
2.3. LPS Interferes with Ets1 Binding to the Pi Promoter Region of pcmah
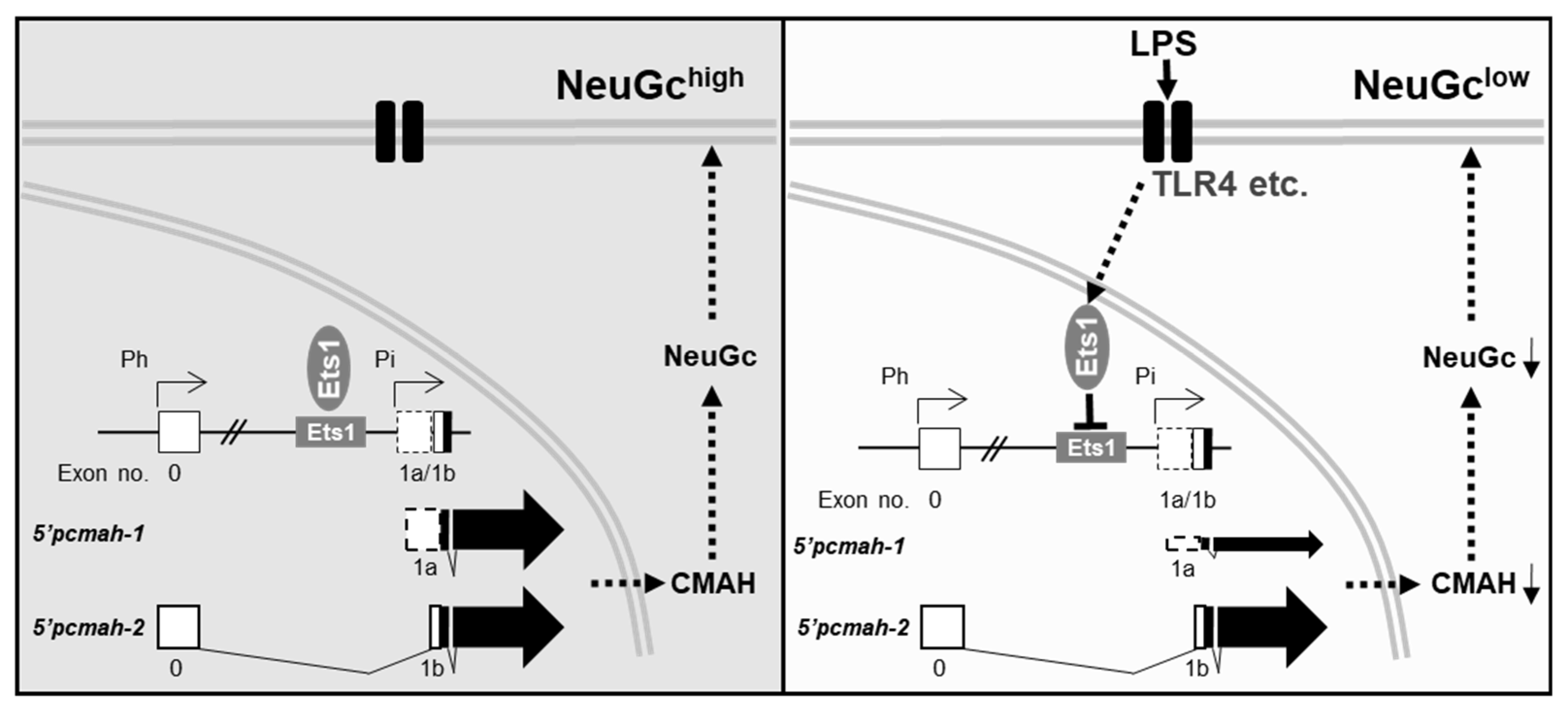
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Enzyme-Linked Immunosorbent Assay (ELISA) for NeuGc
4.3. Western Blot Analysis
4.4. Reverse Transcription-Polymerase Chain Reaction and Real-Time Quantitative PCR
4.5. DNA Constructs and Site-Directed Mutagenesis
4.6. Luciferase Reporter Assay
4.7. Electromobility Shift Assay (EMSA)
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| NeuGc | N-glycolylneuraminic acid |
| CMAH | Cytidine-5′-monophospho-N-acetylneuraminic acid hydroxylase |
| LPS | Lipopolysaccharide |
| ELISA | Enzyme-linked immunosorbent assay |
| RT-PCR | Reverse transcription-polymerase chain reaction |
| EMSA | Electromobility shift assay |
| Pi | Intestine specific promoter |
References
- Angata, T.; Varki, A. Chemical diversity in the sialic acids and related α-keto acids: An evolutionary perspective. Chem. Rev. 2002, 102, 439–470. [Google Scholar] [CrossRef] [PubMed]
- Kelm, S.; Schauer, R. Sialic acids in molecular and cellular interactions. In International Review of Cytology; Elsevier: Amsterdam, The Netherlands, 1997; Volume 175, pp. 137–240. [Google Scholar]
- Chou, H.-H.; Takematsu, H.; Diaz, S.; Iber, J.; Nickerson, E.; Wright, K.L.; Muchmore, E.A.; Nelson, D.L.; Warren, S.T.; Varki, A. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc. Natl. Acad. Sci. USA 1998, 95, 11751–11756. [Google Scholar] [CrossRef] [PubMed]
- Teneberg, S.; Willemsen, P.; de Graaf, F.K.; Karlsson, K.-A. Receptor-active glycolipids of epithelial cells of the small intestine of young and adult pigs in relation to susceptibility to infection with Escherichia coli K99. FEBS Lett. 1990, 263, 10–14. [Google Scholar] [CrossRef]
- Karlsson, N.G.; Olson, F.J.; Jovall, P.-Å.; Andersch, Y.; Enerbäck, L.; Hansson, G.C. Identification of transient glycosylation alterations of sialylated mucin oligosaccharides during infection by the rat intestinal parasite Nippostrongylus brasiliensis. Biochem. J. 2000, 350, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Rolsma, M.D.; Kuhlenschmidt, T.B.; Gelberg, H.B.; Kuhlenschmidt, M.S. Structure and function of a ganglioside receptor for porcine rotavirus. J. Virol. 1998, 72, 9079–9091. [Google Scholar] [CrossRef] [PubMed]
- Byres, E.; Paton, A.W.; Paton, J.C.; Löfling, J.C.; Smith, D.F.; Wilce, M.C.; Talbot, U.M.; Chong, D.C.; Yu, H.; Huang, S. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature 2008, 456, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Willemsen, P.; De Graaf, F. Multivalent binding of K99 fimbriae to the N-glycolyl-GM3 ganglioside receptor. Infect. Immun. 1993, 61, 4518–4522. [Google Scholar] [CrossRef]
- Malykh, Y.N.; King, T.P.; Logan, E.; Kelly, D.; Schauer, R.; Shaw, L. Regulation of N-glycolylneuraminic acid biosynthesis in developing pig small intestine. Biochem. J. 2003, 370, 601–607. [Google Scholar] [CrossRef]
- Kawano, T.; Koyama, S.; Takematsu, H.; Kozutsumi, Y.; Kawasaki, H.; Kawashima, S.; Kawasaki, T.; Suzuki, A. Molecular cloning of cytidine monophospho-N-acetylneuraminic acid hydroxylase. Regulation of species-and tissue-specific expression of N-glycolylneuraminic acid. J. Biol. Chem. 1995, 270, 16458–16463. [Google Scholar] [CrossRef]
- Muchmore, E.; Milewski, M.; Varki, A.; Diaz, S. Biosynthesis of N-glycolyneuraminic acid. The primary site of hydroxylation of N-acetylneuraminic acid is the cytosolic sugar nucleotide pool J. Biol. Chem. 1989, 264, 20216–20223. [Google Scholar]
- Schlenzka, W.; Shaw, L.; Kelm, S.; Schmidt, C.L.; Bill, E.; Trautwein, A.X.; Lottspeich, F.; Schauer, R. CMP-N-acetylneuraminic acid hydroxylase: The first cytosolic Rieske iron-sulphur protein to be described in Eukarya. FEBS Lett. 1996, 385, 197–200. [Google Scholar] [CrossRef]
- Naito, Y.; Takematsu, H.; Koyama, S.; Miyake, S.; Yamamoto, H.; Fujinawa, R.; Sugai, M.; Okuno, Y.; Tsujimoto, G.; Yamaji, T. Germinal center marker GL7 probes activation-dependent repression of N-glycolylneuraminic acid, a sialic acid species involved in the negative modulation of B-cell activation. Mol. Cell. Biol. 2007, 27, 3008–3022. [Google Scholar] [CrossRef] [PubMed]
- Song, K.-H.; Kwak, C.-H.; Jin, U.-H.; Ha, S.-H.; Park, J.-Y.; Abekura, F.; Chang, Y.-C.; Cho, S.-H.; Lee, K.; Chung, T.-W. Housekeeping promoter 5′pcmah-2 of pig CMP-N-acetylneuraminic acid hydroxylase gene for NeuGc expression. Glycoconj. J. 2016, 33, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Song, K.-H.; Kang, Y.-J.; Jin, U.-H.; Park, Y.-I.; Kim, S.-M.; Seong, H.-H.; Hwang, S.; Yang, B.-S.; Im, G.-S.; Min, K.-S. Cloning and functional characterization of pig CMP-N-acetylneuraminic acid hydroxylase for the synthesis of N-glycolylneuraminic acid as the xenoantigenic determinant in pig–human xenotransplantation. Biochem. J. 2010, 427, 179–188. [Google Scholar] [CrossRef]
- Song, K.-H.; Kwak, C.-H.; Chung, T.-W.; Ha, S.-H.; Park, J.-Y.; Ha, K.-T.; Cho, S.-H.; Lee, Y.-C.; Kim, C.-H. Intestine specific regulation of pig cytidine-5′-monophospho-N-acetylneuraminic acid hydroxylase gene for N-glycolylneuraminic acid biosynthesis. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Metzler-Zebeli, B.U.; Mann, E.; Schmitz-Esser, S.; Wagner, M.; Ritzmann, M.; Zebeli, Q. Changing dietary calcium-phosphorus level and cereal source selectively alters abundance of bacteria and metabolites in the upper gastrointestinal tracts of weaned pigs. Appl. Environ. Microbiol. 2013, 79, 7264–7272. [Google Scholar] [CrossRef]
- Seki, E.; Schnabl, B. Role of innate immunity and the microbiota in liver fibrosis: Crosstalk between the liver and gut. J. Physiol. 2012, 590, 447–458. [Google Scholar] [CrossRef]
- Guo, S.; Nighot, M.; Al-Sadi, R.; Alhmoud, T.; Nighot, P.; Ma, T.Y. Lipopolysaccharide regulation of intestinal tight junction permeability is mediated by TLR4 signal transduction pathway activation of FAK and MyD88. J. Immunol. 2015, 195, 4999–5010. [Google Scholar] [CrossRef]
- Dittmer, J. The biology of the Ets1 proto-oncogene. Mol. Cancer 2003, 2, 29. [Google Scholar] [CrossRef]
- Benoit, R.; Rowe, S.; Watkins, S.C.; Boyle, P.; Garrett, M.; Alber, S.; Wiener, J.; Rowe, M.I.; Ford, H.R. Pure endotoxin does not pass across the intestinal epithelium in vitro. Shock 1998, 10, 43–48. [Google Scholar] [CrossRef]
- Yagi, S.; Takaki, A.; Hori, T.; Sugimachi, K. Enteric lipopolysaccharide raises plasma IL-6 levels in the hepatoportal vein during non-inflammatory stress in the rat. Fukuoka Igaku Zasshi 2002, 93, 38–51. [Google Scholar] [PubMed]
- Guo, S.; Al-Sadi, R.; Said, H.M.; Ma, T.Y. Lipopolysaccharide causes an increase in intestinal tight junction permeability in vitro and in vivo by inducing enterocyte membrane expression and localization of TLR-4 and CD14. Am. J. Pathol. 2013, 182, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.C. Endotoxemia: Methods of detection and clinical correlates. Clin. Microbiol. Rev. 1995, 8, 268–292. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.R.; Kushwah, R.; Hu, J. Multiple roles of the epithelium-specific ETS transcription factor, ESE-1, in development and disease. Lab. Invest. 2012, 92, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Rabault, B.; Ghysdael, J. Calcium-induced phosphorylation of ETS1 inhibits its specific DNA binding activity. J. Biol. Chem. 1994, 269, 28143–28151. [Google Scholar]
- Cowley, D.O.; Graves, B.J. Phosphorylation represses Ets-1 DNA binding by reinforcing autoinhibition. Genes Dev. 2000, 14, 366–376. [Google Scholar]
- Suriano, A.R.; Sanford, A.N.; Kim, N.; Oh, M.; Kennedy, S.; Henderson, M.J.; Dietzmann, K.; Sullivan, K.E. GCF2/LRRFIP1 represses tumor necrosis factor alpha expression. Mol. Cell. Biol. 2005, 25, 9073–9081. [Google Scholar] [CrossRef]
- West, A.P.; Koblansky, A.A.; Ghosh, S. Recognition and signaling by toll-like receptors. Annu. Rev. Cell Dev. Biol. 2006, 22, 409–437. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwak, C.-H.; Song, K.-H.; Kim, C.-H. Gram-Negative Bacterial Endotoxin LPS Induces NeuGc Loss through Ets1-Dependent Downregulation of Intestine-Specific pcmah Transcript in Porcine Intestinal Cells. Int. J. Mol. Sci. 2020, 21, 4892. https://doi.org/10.3390/ijms21144892
Kwak C-H, Song K-H, Kim C-H. Gram-Negative Bacterial Endotoxin LPS Induces NeuGc Loss through Ets1-Dependent Downregulation of Intestine-Specific pcmah Transcript in Porcine Intestinal Cells. International Journal of Molecular Sciences. 2020; 21(14):4892. https://doi.org/10.3390/ijms21144892
Chicago/Turabian StyleKwak, Choong-Hwan, Kwon-Ho Song, and Cheorl-Ho Kim. 2020. "Gram-Negative Bacterial Endotoxin LPS Induces NeuGc Loss through Ets1-Dependent Downregulation of Intestine-Specific pcmah Transcript in Porcine Intestinal Cells" International Journal of Molecular Sciences 21, no. 14: 4892. https://doi.org/10.3390/ijms21144892
APA StyleKwak, C.-H., Song, K.-H., & Kim, C.-H. (2020). Gram-Negative Bacterial Endotoxin LPS Induces NeuGc Loss through Ets1-Dependent Downregulation of Intestine-Specific pcmah Transcript in Porcine Intestinal Cells. International Journal of Molecular Sciences, 21(14), 4892. https://doi.org/10.3390/ijms21144892




