Label-Free Multiple Reaction Monitoring, a Promising Method for Quantification Analyses of Specific Proteins in Bacteria
Abstract
1. Introduction
2. Results
2.1. Selection of Specific Peptides for Detection of AprBp and GseBp Proteins
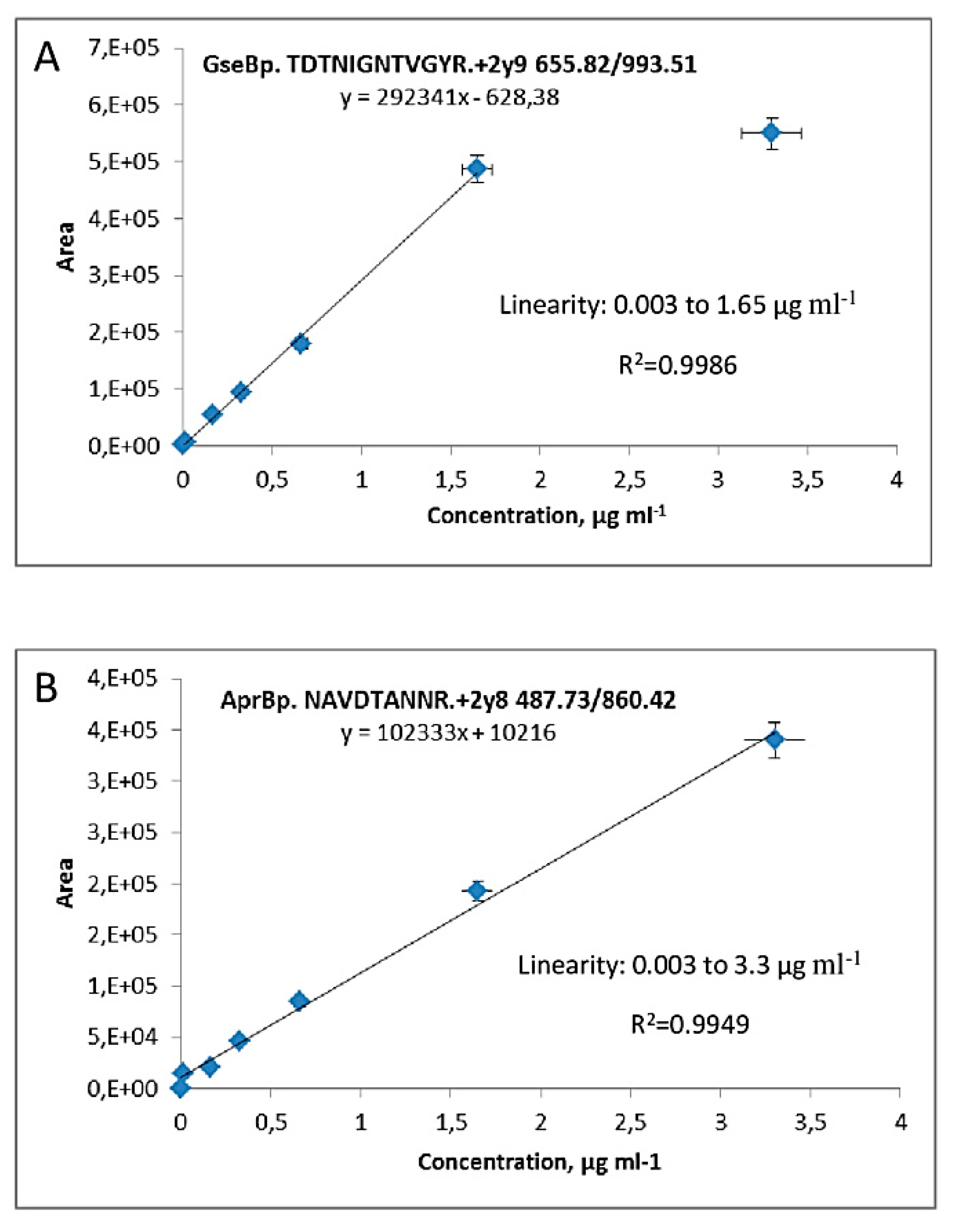
2.2. Calibration Plots for Quantification of Serine Proteases AprBp and GseBp
2.3. Quantification of AprBp and GseBp Serine Proteases by the Selected Peptides
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Bacterial Strains and Growth Conditions
4.3. Sample Preparation
4.4. Assessment of Specific Proteolytic Activity
4.5. SDS-PAGE
4.6. In-Gel Tryptic Digestion and Peptide Extraction
4.7. In Silico Peptide Selection and Skyline Settings
4.8. Quantitative LC–MRM–MS Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AprBp | Subtilisin-like protease |
| DTT | Dithiothreitol |
| ELISA | Enzyme-linked immunosorbent assay |
| GseBp | Glutamyl endopeptidase |
| GRAS | Generally recognized as safe |
| IAA | Iodoacetamide |
| IPTG | Isopropyl β-galactopyranoside |
| iTRAQ | Isobaric tags for relative and absolute quantitation |
| LB | Lysogeny broth |
| LC–MS | Liquid chromatography-tandem mass spectrometry |
| LIKE | LIa-kontrollierte expression |
| LPS | Lipopolysaccharide |
| MRM | Multiple reaction monitoring |
| OD | Optical density |
| QQQ | Triple-quadrupole |
| Q1 | First quadrupole |
| Q2 | Second quadrupole |
| Q3 | Third quadrupole |
| rcf | Relative centrifugal field |
| rpm | Revolutions per minute |
| SILAC | Stable isotope labeling by amino acids in cell culture |
| SN | Supernatant |
| TCA | Trichloroacetic acid |
References
- Wolfe, M.S. Intramembrane Proteolysis. Chem. Rev. 2009, 109, 1599–1612. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, R.T.; Badgujar, S.B. Biological aspects of proteolytic enzymes: A Review. J. Pharm. Res. 2010, 3, 2048–2068. [Google Scholar]
- Li, Q.; Yi, L.; Marek, P.; Iverson, B.L. Commercial proteases: Present and future. FEBS Lett. 2013, 587, 1155–1163. [Google Scholar] [CrossRef] [PubMed]
- Bongers, R.S.; Veening, J.W.; Wieringen, V.M.; Kuipers, O.P.; Kleerebezem, M. Development and characterization of a subtilin-regulated expression system in Bacillus subtilis: Strict control of gene expression by addition of subtilin. Appl. Environ. Microbiol. 2005, 71, 8818–8824. [Google Scholar] [CrossRef]
- Vavrova, L.; Muchova, K.; Barak, I. Comparison of different Bacillus subtilis expression systems. Res. Microbiol. 2010, 161, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Zukowski, M.M.; Miller, L. Hyperproduction of an intracellular heterologous protein in a sacUh mutant of Bacillus subtilis. Gene 1986, 46, 247–255. [Google Scholar] [CrossRef]
- Conrad, B.; Savchenko, R.S.; Breves, R.; Hofemeister, J. A T7 promoter-specific, inducible protein expression system for Bacillus subtilis. Mol. Gen. Genet. 1996, 250, 230–236. [Google Scholar]
- Bhavsar, A.P.; Zhao, X.; Brown, E.D. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: Conditional complementation of a teichoic acid mutant. Appl. Environ. Microbiol. 2001, 67, 403–410. [Google Scholar] [CrossRef]
- Fukushima, T.; Yamamoto, H.; Atrih, A.; Foster, S.J.; Sekiguchi, J. A polysaccharide deacetylase gene (pdaA) is required for germination and for production of muramic delta-lactam residues in the spore cortex of Bacillus subtilis. J. Bacteriol. 2002, 184, 6007–6015. [Google Scholar] [CrossRef]
- Ho, K.M.; Lim, B.L. Co-expression of a prophage system and a plasmid system in Bacillus subtilis. Protein. Expr. Purif. 2003, 32, 293–301. [Google Scholar] [CrossRef]
- Phan, T.T.P.; Schumann, W. Development of glycine-inducible expression system for Bacillus subtilis. J. Biotechnol. 2007, 128, 486–499. [Google Scholar] [CrossRef] [PubMed]
- Toymentseva, A.A.; Schrecke, K.; Sharipova, M.R.; Mascher, T. The LIKE system, a novel protein expression toolbox for based on the liI promoter. Microb. Cell Fact. 2012, 11, 143. [Google Scholar] [CrossRef] [PubMed]
- Vidova, V.; Spacil, Z. A review on mass spectrometry-based quantitative proteomics: Targeted and data independent acquisition. Anal. Chim. Acta. 2017, 964, 7–23. [Google Scholar] [CrossRef]
- Resta, D.; Brambilla, F.; Arnoldi, A. HPLC-Chip-Multiple Reaction Monitoring (MRM) method for the label-free absolute quantification of ɣ-conglutin in lupin: Proteotypic peptides and standard addition method. Food Chem. 2012, 131, 126–133. [Google Scholar] [CrossRef]
- Blankley, R.T.; Fisher, C.; Westwood, M.; North, R.; Baker, P.N.; Walker, M.J.; Williamson, A.; Whetton, A.D.; Lin, W.; McCowan, L.; et al. A label-free selected reaction monitoring workflow identifies a subset of pregnancy specific glycoproteins as potential predictive markers of early-onset pre-eclampsia. Mol. Cell Proteomics. 2013, 12, 3148–3159. [Google Scholar] [CrossRef]
- Al Ali, A.; Touboul, D.; Le Caër, J.P.; Schmitz-Afonso, I.; Flinois, J.P.; Marchetti, C.; De Waziers, I.; Brunelle, A.; Laprévote, O.; Beaune, P. Optimization and validation of a label-free MRM method for the quantification of cytochrome P450 isoforms in biological samples. Anal. Bioanal. Chem. 2014, 406, 4861–4874. [Google Scholar] [CrossRef] [PubMed]
- Sharipova, M.R. Late stages of protein secretion in bacilli. Biochemistry 2002, 67, 1207–1216. [Google Scholar]
- Sharipova, M.; Balaban, N.; Kayumov, A. The expression of the serine proteinase gene of B. intermedius in B. subtilis. Microbiol. Res. 2008, 163, 39–50. [Google Scholar] [CrossRef]
- Lange, V.; Picotti, P.; Domon, B.; Aebersold, R. Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2008, 4, 222. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010, 26, 966–968. [Google Scholar] [CrossRef]
- Sharipova, M.R.; Shagimardanova, E.I.; Chastukhina, I.B.; Shamsutdinov, T.R.; Balaban, N.P.; Mardanova, A.M.; Rudenskaya, G.N.; Demidyuk, I.V.; Kostrov, S.V. The expression of Bacillus intermedius glutamyl endopeptidase gene in Bacillus subtilis recombinant strains. Mol. Biol. Rep. 2007, 34, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Mikhailova, E.O.; Mardanova, A.M.; Balaban, N.P.; Rudenskaya, G.N.; Ilyinskaya, O.N.; Sharipova, M.R. Biochemical properties of Bacillus intermedius subtilisin-like proteinase secreted by a Bacillus subtilis recombinant strain in its stationary phase of growth. Biochemistry 2009, 74, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Tikhonova, A.O.; Toymentseva, A.A.; Sharipova, M.R. Screening of heterologous signal peptides for optimization of the LIKE-expression system. BioNanoScience 2017, 7, 408–414. [Google Scholar] [CrossRef]
- Kuzyk, M.A.; Smith, D.; Yang, J.; Cross, T.J.; Jackson, A.M.; Hardie, D.B.; Anderson, N.L.; Borchers, G.H. CH Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell Proteomics. 2009, 8, 1860–1877. [Google Scholar] [CrossRef]
- Rezeli, M.; Végvári, Á.; Silajdžić, E.; Björkqvist, M.; Tabrizi, J.S.; Laurell, T.; Marko-Varga, G. Inflammatory markers in Huntington’s disease plasma—A robust nanoLC–MRM-MS assay development. EuPA Open Proteom. 2014, 3, 68–75. [Google Scholar] [CrossRef][Green Version]
- Park, S.L.; Carmella, S.G.; Chen, M.; Patel, Y.; Stram, D.O.; Haiman, C.A.; Le Marchand, L.; Hecht, S.S. Mercapturic acids derived from the toxicants acrolein and crotonaldehyde in the urine of cigarette smokers from five ethnic groups with differing risks for lung cancer. PLoS ONE 2015, 10, e0124841. [Google Scholar] [CrossRef] [PubMed]
- Romanova, Y.; Laikov, A.; Markelova, M.; Khadiullna, R.; Makseev, A.; Hasanova, M.; Rizvanov, A.; Khaiboullina, S.; Salafutdinov, I. Proteomic analysis of human serum from patients with chronic kidney disease. Biomolecules 2020, 10, 257. [Google Scholar] [CrossRef]
- Schaafsma, A.; Limay-Rios, V.; Baute, T.; Smith, J.; Xue, Y. Neonicotinoid insecticide residues in surface water and soil associated with commercial maize (corn) fields in southwestern Ontario. PLoS ONE 2015, 10, e0118139. [Google Scholar] [CrossRef]
- Kumar, A.; Gangadharan, B.; Zitzmann, N. Multiple reaction monitoring and multiple reaction monitoring cubed based assays for the quantitation of apolipoprotein F. J. Chromatogr B Analyt. Technol. Biomed. Life Sci. 2016, 1033, 278–286. [Google Scholar] [CrossRef]
- Zheng, J.; Mandal, R.; Wishart, D.S. A sensitive, high-throughput LC-MS/MS method for measuring catecholamines in low volume serum. Anal. Chim. Acta. 2018, 1037, 159–167. [Google Scholar] [CrossRef]
- Chenau, J.; Fenaille, F.; Ezan, E.; Morel, N.; Lamourette, P.; Goossens, P.L.; Becher, F. Sensitive detection of Bacillus anthracis spores by immunocapture and liquid chromatography-tandem mass spectrometry. Anal. Chem. 2011, 83, 8675–8682. [Google Scholar] [CrossRef] [PubMed]
- Mullins, E.A.; Rubinson, E.H.; Pereira, K.N.; Calcutt, M.W.; Christov, P.P.; Eichman, B.F. An HPLC-tandem mass spectrometry method for simultaneous detection of alkylated base excision repair products. Methods 2013, 64, 59–66. [Google Scholar] [CrossRef][Green Version]
- Domanski, D.; Percy, A.J.; Yang, J.; Chambers, A.G.; Hill, J.S.; Freue, G.V.; Borchers, G.H. MRM-based multiplexed quantitation of 67 putative cardiovascular disease biomarkers in human plasma. Proteomics 2012, 12, 1222–1243. [Google Scholar] [CrossRef]
- Keshishian, H.; Addona, T.; Burgess, M.; Kuhn, E.; Carr, S.A. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell Proteomics 2007, 6, 2212–2229. [Google Scholar] [CrossRef] [PubMed]
- Onisko, B.; Dynin, I.; Requena, J.R.; Silva, C.J.; Erickson, M.; Carter, J.M. Mass spectrometric detection of attomole amounts of the prion protein by nanoLC/MS/MS. J. Am. Soc. Mass Spectr. 2007, 18, 1070–1079. [Google Scholar] [CrossRef] [PubMed]
- Gillette, M.A.; Carr, S.A. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat. Methods. 2013, 10, 28–34. [Google Scholar] [CrossRef]
- Stammen, S.; Müller, K.; Korneli, C.; Biedendieck, R.; Franco-Lara, E.; Jahn, D. High-yield intra- and extracellular protein production using Bacillus megaterium. Appl. Environ. Microbiol. 2010, 76, 4037–4046. [Google Scholar] [CrossRef]
- Speicher, K.; Kolbas, O.; Harper, S.; Speicher, D. Systematic analysis of peptide recoveries from in-gel digestions for protein identifications in proteome studies. J. Biomol. Tech. 2000, 11, 74–86. [Google Scholar]
- Yang, J.K.; Shihb, I.L.; Tzengc, Y.M.; Wang, S.L. Production and purification of protease from a Bacillus subtilis that can deproteinize crustacean wastes. Enzyme Microb. Technol. 2000, 26, 406–413. [Google Scholar] [CrossRef]
- Lyublinskaya, L.; Belyaev, S.; Strongin, A.; Matyash, L.; Levin, E.; Stepanov, V. A new chromogenic substrate for subtilisin. Anal. Biochem. 1974, 62, 371–376. [Google Scholar] [CrossRef]
- Lyublinskaya, L.; Voyushina, T.; Stepanov, V. p-Nitroanilides of pyroglutamyl peptides as chromogenic substrates of serine proteinases. Bioorg. Khim. 1974, 13, 748–753. [Google Scholar]
- Mikhailova, E.O.; Balaban, N.P.; Mardanova, A.M.; Rudakova, N.L.; Ilyinskaya, O.N.; Rudenskaya, G.N.; Rizvanov, A.A.; Sharipova, M.R. Purification of a subtilisin-like serine proteinase from recombinant Bacillus subtilis during different phases of growth. Ann. Microbiol. 2009, 59, 1–8. [Google Scholar] [CrossRef]
- Balaban, N.P.; Mardanova, A.M.; Sharipova, M.R.; Gabdrakhmanova, L.A.; Sokolova, E.A.; Garusov, A.V.; Milgotina, E.I.; Rudenskaya, G.N.; Leshchinskaya, I.B. Isolation and characterization of glutamyl endopeptidase from Bacillus intermedius 3-19. Biochemistry 2003, 68, 1514–1521. [Google Scholar] [CrossRef] [PubMed]
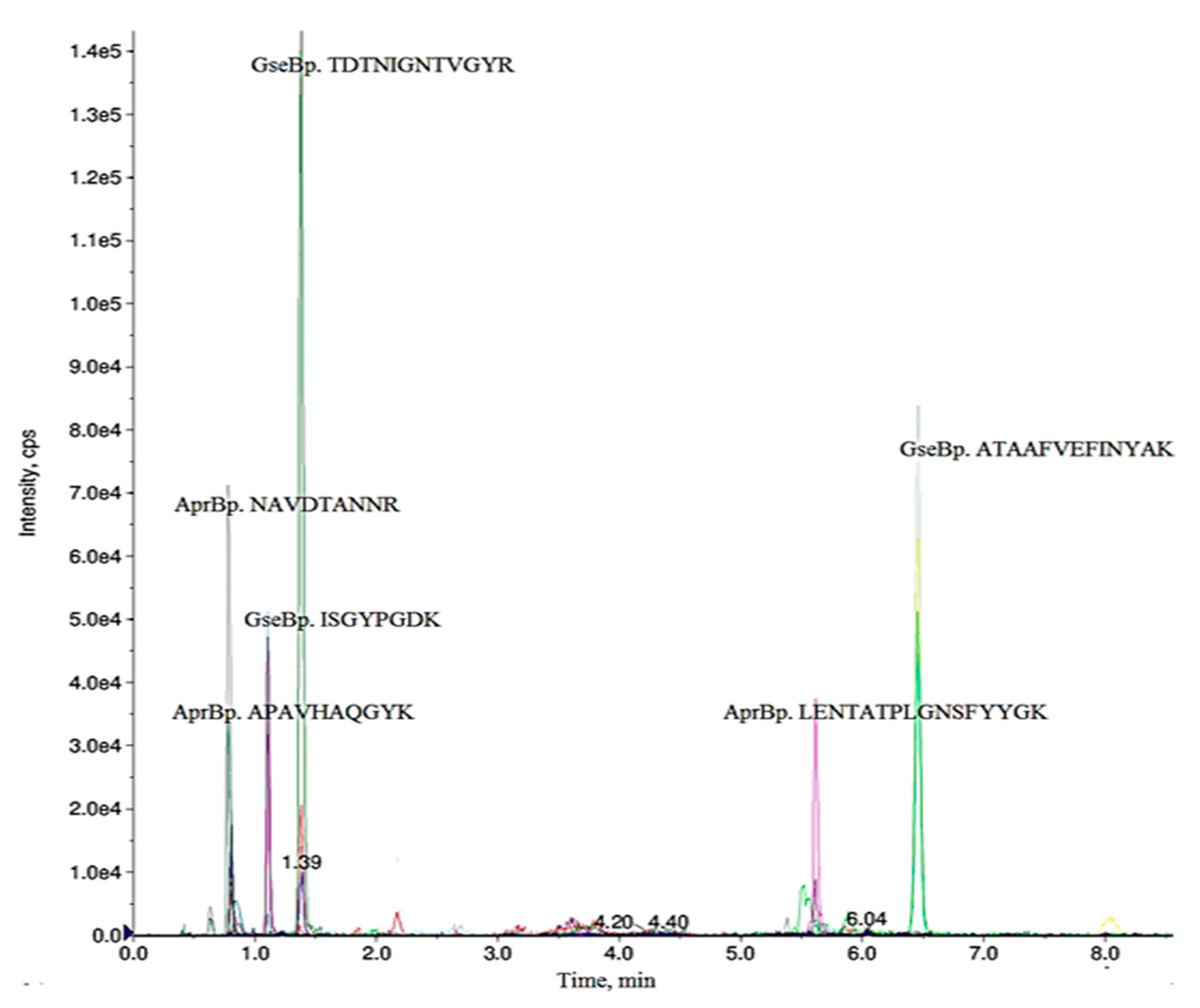
| Precursor Ion Q1, m/z | Ion Charge | Product Ion Q3, m/z | Peptide | Declustering Potential (V) | Collision Energy (V) | Retention Time (min) |
|---|---|---|---|---|---|---|
| AprBp | ||||||
| 521.27 | 802.42 | APAVHAQGYK | 69.1 | 27.6 | 1.03 | |
| +2 | 703.35 | |||||
| 566.29 | ||||||
| 487.73 | 860.42 * | NAVDTANNR | 66.7 | 26.4 | 0.79 | |
| +2 | 690.31 | |||||
| 575.28 | ||||||
| 887.93 | 1145.56 | LENTATPLGNSFYYGK | 95.8 | 40.8 | 5.62 | |
| +2 | 1048.50 | |||||
| 935.42 | ||||||
| GseBp | ||||||
| 655.82 | 993.51 * | TDTNIGNTVGYR | 78.9 | 32.5 | 1.39 | |
| +2 | 766.38 | |||||
| 709.36 | ||||||
| 418.71 | 723.33 | ISGYPGDK | 61.6 | 23.9 | 1.11 | |
| +2 | 636.29 | |||||
| 416.21 | ||||||
| 722.87 | 1130.58 | ATAAFVEFINYAK | 83.8 | 34.9 | 6.46 | |
| +2 | 983.51 | |||||
| 884.45 | ||||||
| Strain | Total Protein Concentration (μg mL−1) in SN, Inducer "−" | Total Protein Concentration (μg mL−1) in SN, Inducer "+" | Target Protein Concentration (μg mL−1) in the Vial, Inducer "−"/"+" | Target Protein Concentration (μg mL−1) in SN, Inducer "−"/"+" |
|---|---|---|---|---|
| Subtilisin-like protease (AprBp) | ||||
| B. pumilus 3–19 | 4.5 | – | 6 | 1.63 |
| B. subtilis AT1 | 0.32 | 0.33 | 0 | 0 |
| B. subtilis MRB044 (SPAprBp) | 2 | 5 | 0.2/5 | 0.024/1.5 |
| B. subtilis MRB046 (SPYngk) | 1.3 | 1.8 | 0.45/0.25 | 0.005/0.03 |
| Glutamyl endopeptidase (GseBp) | ||||
| B. pumilus 3–19 | 4.5 | – | 5 | 1.3 |
| B. subtilis AT1 | 0.32 | 0.33 | 0/0 | 0/0 |
| B. subtilis MRB047 (SPGseBp) | 1.2 | 1.65 | 0.005/0.05 | 0.00036/0.005 |
| B. subtilis MRB049 (SPYngk) | 2.06 | 3.2 | 0.05/0.3 | 0.006/0.06 |
| Strain | Relevant Genotype | Source |
|---|---|---|
| B. pumilus 3–19 | StrR | Laboratory of Biosynthesis and Bioengineering of Enzymes, KFU |
| B. subtilis: | ||
| BG2036 | ΔaprE-684, ΔnprE522 | (Yang et al [39]) |
| AT1 | ΔaprE-684, ΔnprE522, pLIKE-rep | (Tikhonova et al [23]) |
| MRB044 | pLIKE-rep + SPAprBp + AprBp | (Tikhonova et al [23]) |
| MRB046 | pLIKE-rep + SPYngk + AprBp | (Tikhonova et al [23]) |
| MRB047 | pLIKE-rep + SPGseBp + GseBp | (Tikhonova et al [23]) |
| MRB049 | pLIKE-rep + SPYngk + GseBp | (Tikhonova et al [23]) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toymentseva, A.A.; Koryagina, A.O.; Laikov, A.V.; Sharipova, M.R. Label-Free Multiple Reaction Monitoring, a Promising Method for Quantification Analyses of Specific Proteins in Bacteria. Int. J. Mol. Sci. 2020, 21, 4924. https://doi.org/10.3390/ijms21144924
Toymentseva AA, Koryagina AO, Laikov AV, Sharipova MR. Label-Free Multiple Reaction Monitoring, a Promising Method for Quantification Analyses of Specific Proteins in Bacteria. International Journal of Molecular Sciences. 2020; 21(14):4924. https://doi.org/10.3390/ijms21144924
Chicago/Turabian StyleToymentseva, Anna A., Anastasia O. Koryagina, Alexander V. Laikov, and Margarita R. Sharipova. 2020. "Label-Free Multiple Reaction Monitoring, a Promising Method for Quantification Analyses of Specific Proteins in Bacteria" International Journal of Molecular Sciences 21, no. 14: 4924. https://doi.org/10.3390/ijms21144924
APA StyleToymentseva, A. A., Koryagina, A. O., Laikov, A. V., & Sharipova, M. R. (2020). Label-Free Multiple Reaction Monitoring, a Promising Method for Quantification Analyses of Specific Proteins in Bacteria. International Journal of Molecular Sciences, 21(14), 4924. https://doi.org/10.3390/ijms21144924





