From Cell Entry to Engraftment of Exogenous Mitochondria
Abstract
:1. Introduction
2. Cell Entry
2.1. Passage through the Plasma Membrane of Viruses
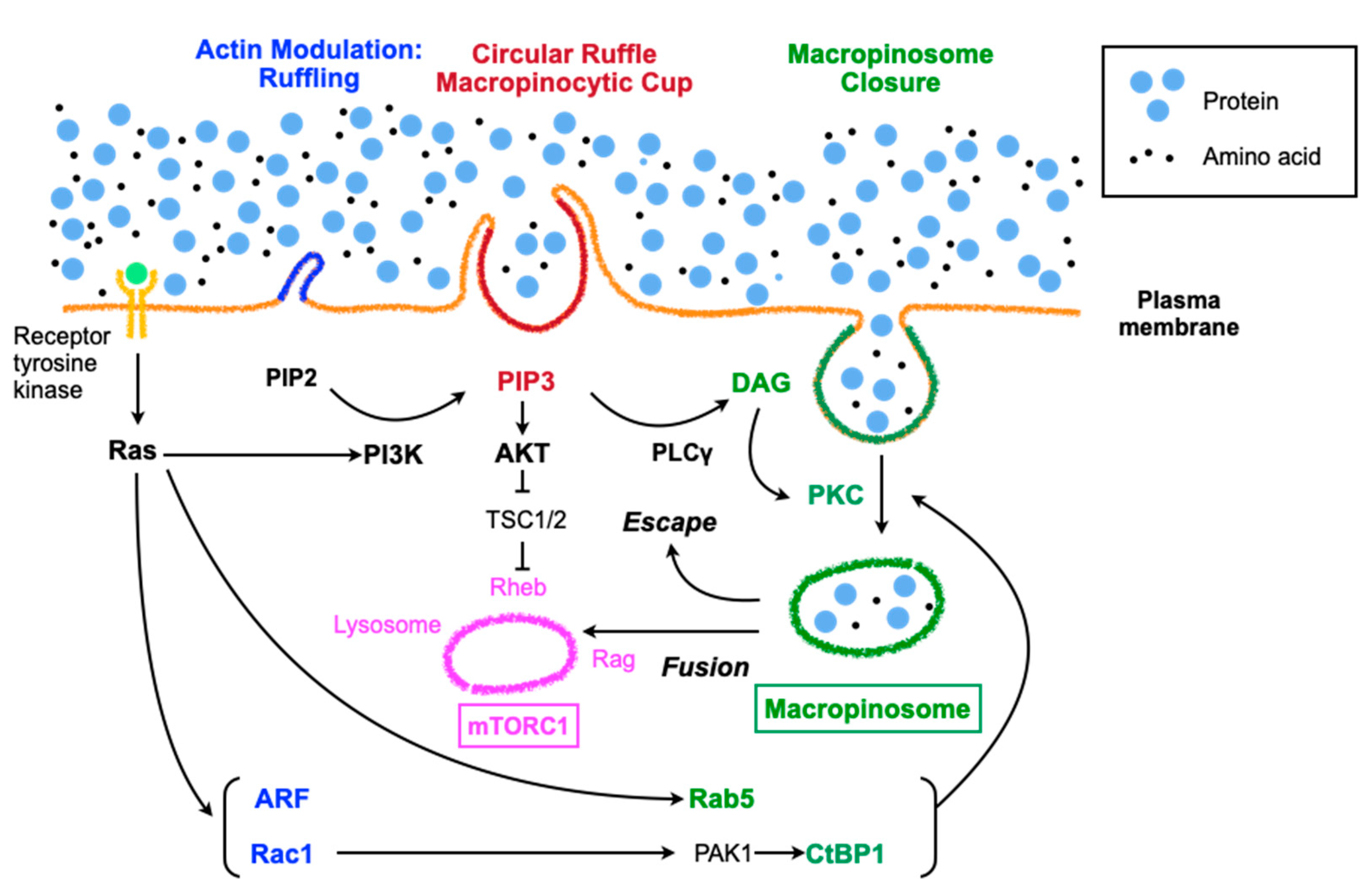
2.2. Molecular Aspects of Macropinocytosis
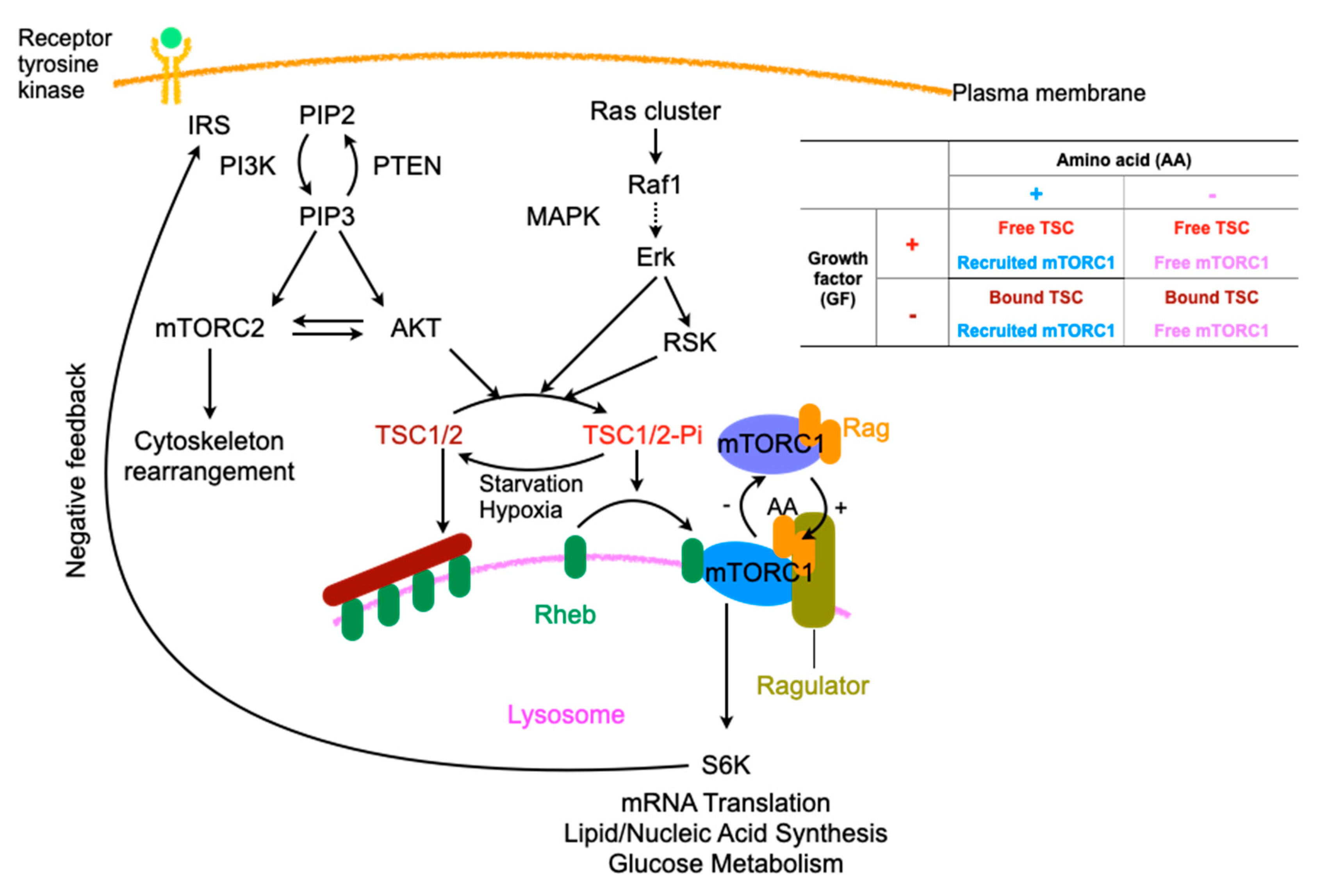
2.3. Regulation of Macropinocytosis
3. Relations with Internalized Mitochondria and Endosomes
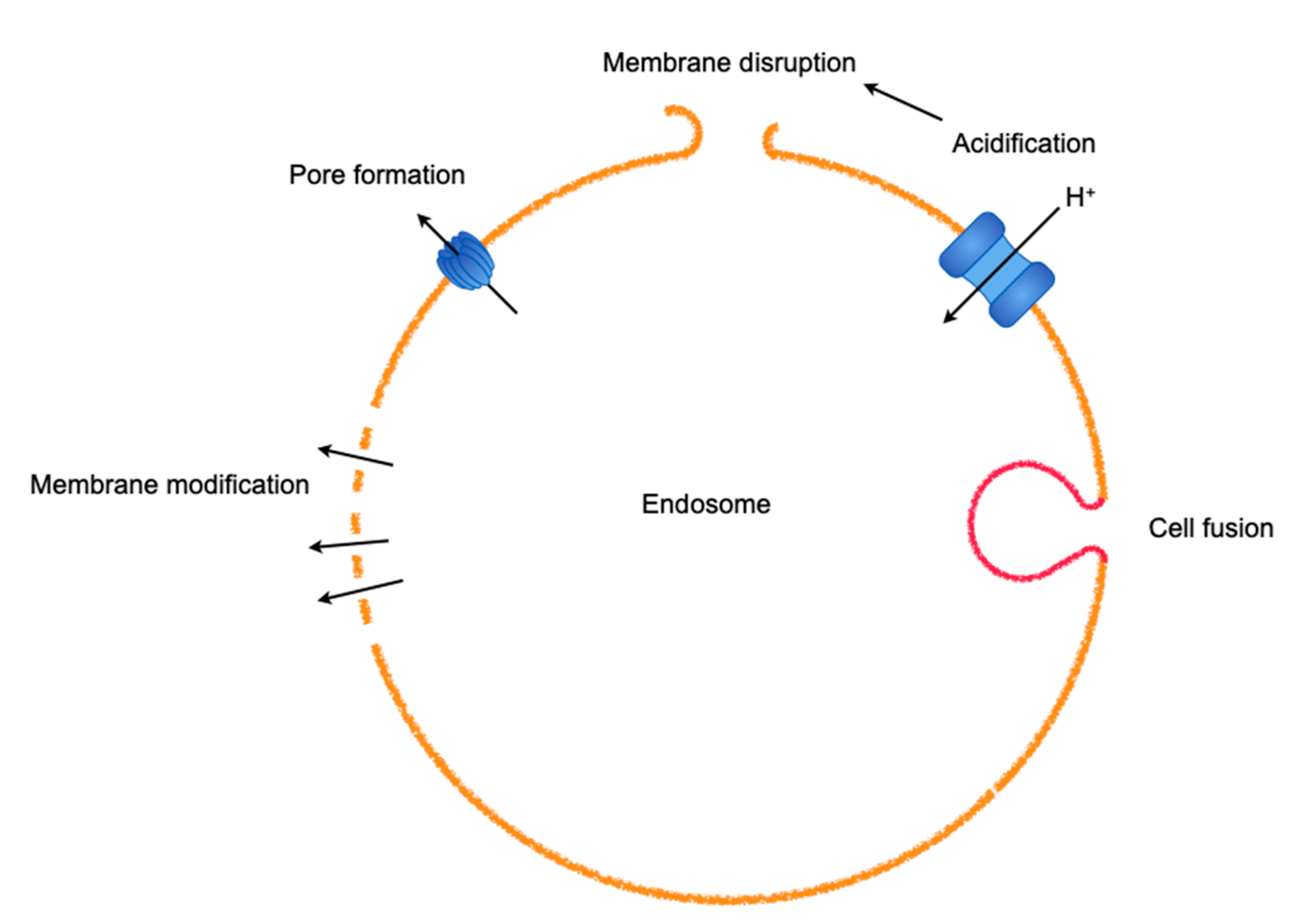
3.1. Endosomal Escapes for Viruses
3.2. Mitochondria–Endosome Interactions and Mitochondrial Exports
4. Artificial Mitochondrial Transfer
4.1. Cell Penetrating Peptide
4.2. Centrifugal Force
4.3. Magnetic Force Using Anti-Mitochondria Specific Protein with Magnetic Nanoparticles
4.4. Generation of Vapor Bubbles by a Photothermal Nanoblade
5. Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CAR | Coxsackievirus adenovirus receptor |
| VSAP | Vasodilator-stimulated phosphoprotein |
| PIP3 | Phosphatidylinositol (3,4,5)-triphosphate |
| RTKs | Receptor tyrosine kinases |
| M-CSF | Macrophage colony-stimulating factor |
| PH | Pleckstrin homology |
| PLCγ | Phospholipase C-γ |
| IP3 | Inositol 1,4,5-trisphosphate |
| DAG | Diacylglycerol |
| PKC | Protein kinase C |
| mTOR | Mechanistic target of rapamycin |
| MAP | Mitogen-activated protein |
| PIP2 | Phosphatidylinositol (4,5)-bisphosphate |
| TSC2 | Tuberous sclerosis complex 2 |
| Rheb | Ras homolog enriched in the brain |
| GAP | GTPase-activating protein |
| PTEN | Phosphatase and tensin homologue |
| IRS-1 | Insulin receptor substrate-1 |
| GM-CSF | Granulocyte/macrophage colony-stimulating factor |
| FLT3L | FMS-related tyrosine kinase 3 ligand |
| TLRs | Toll-like receptors |
| CRM1 | Chromosome region maintenance 1 |
| NPC | Nuclear pore complex |
| ER | Endoplasmic reticulum |
| MAM | Mitochondria-associated membrane |
| MDVs | Mitochondria-derived vesicles |
| PVM | Parasitophorous vacuole membrane |
| CPPs | Cell penetrating peptides |
| TAT | Trans-Activator of Transcription protein |
References
- Lane, N. Bioenergetic Constraints on the Evolution of Complex Life. Cold Spring Harb. Perspect. Boil. 2014, 6, a015982. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C.; Chalkia, D. Mitochondrial DNA Genetics and the Heteroplasmy Conundrum in Evolution and Disease. Cold Spring Harb. Perspect. Boil. 2013, 5, a021220. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, T.M.U.; Barrias, E.S.; De Souza, W. Macropinocytosis: a pathway to protozoan infection. Front. Physiol. 2015, 6, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitani, T.; Kami, D.; Matoba, S.; Gojo, S. Internalization of isolated functional mitochondria: involvement of macropinocytosis. J. Cell. Mol. Med. 2014, 18, 1694–1703. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Pacitto, R.; Inoki, K.; Swanson, J.A. Macropinocytosis, mTORC1 and cellular growth control. Cell. Mol. Life Sci. 2017, 75, 1227–1239. [Google Scholar] [CrossRef] [Green Version]
- Spees, J.L.; Olson, S.D.; Whitney, M.J.; Prockop, D.J. Mitochondrial transfer between cells can rescue aerobic respiration. Proc. Natl. Acad. Sci. USA 2006, 103, 1283–1288. [Google Scholar] [CrossRef] [Green Version]
- Maeda, H.; Kami, D.; Maeda, R.; Murata, Y.; Jo, J.-I.; Kitani, T.; Tabata, Y.; Matoba, S.; Gojo, S. TAT-dextran–mediated mitochondrial transfer enhances recovery from models of reperfusion injury in cultured cardiomyocytes. J. Cell. Mol. Med. 2020, 24, 5007–5020. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Hwang, J.W.; Yun, C.-K.; Lee, Y.; Choi, Y.-S. Delivery of exogenous mitochondria via centrifugation enhances cellular metabolic function. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Macheiner, T.; Fengler, V.H.I.; Agreiter, M.; Eisenberg, T.; Madeo, F.; Kolb, D.; Huppertz, B.; Ackbar, R.; Sargsyan, K. Magnetomitotransfer: An efficient way for direct mitochondria transfer into cultured human cells. Sci. Rep. 2016, 6, 35571. [Google Scholar] [CrossRef]
- Wu, T.-H.; Sagullo, E.; Case, D.; Zheng, X.; Li, Y.; Hong, J.S.; TeSlaa, T.; Patananan, A.N.; McCaffery, J.M.; Niazi, K.; et al. Mitochondrial Transfer by Photothermal Nanoblade Restores Metabolite Profile in Mammalian Cells. Cell Metab. 2016, 23, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Rustom, A.; Saffrich, R.; Walther, P.; Marković, I.; Gerdes, H.-H. Nanotubular Highways for Intercellular Organelle Transport. Science 2004, 303, 1007–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenfield, A.; Braude, P.; Flinter, F.; Lovell-Badge, R.; Ogilvie, C.; Perry, A.C.F. Assisted reproductive technologies to prevent human mitochondrial disease transmission. Nat. Biotechnol. 2017, 35, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Tan, A.S.; Baty, J.W.; Dong, L.; Bezawork-Geleta, A.; Endaya, B.; Goodwin, J.; Bajzikova, M.; Kovarova, J.; Peterka, M.; Yan, B.; et al. Mitochondrial Genome Acquisition Restores Respiratory Function and Tumorigenic Potential of Cancer Cells without Mitochondrial DNA. Cell Metab. 2015, 21, 81–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdinocci, D.; Simões, R.; Kovarova, J.; Cunha-Oliveira, T.; Neuzil, J.; Pountney, D. Intracellular and Intercellular Mitochondrial Dynamics in Parkinson’s Disease. Front. Mol. Neurosci. 2019, 13, 930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayakawa, K.; Esposito, E.; Wang, X.; Terasaki, Y.; Liu, Y.; Xing, C.; Ji, X.; Lo, E.H. Transfer of mitochondria from astrocytes to neurons after stroke. Nature 2016, 535, 551–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, P.-J.; Kuo, C.-C.; Lee, H.-C.; Shen, C.-I.; Cheng, F.-C.; Wu, S.-F.; Chang, J.C.; Pan, H.-C.; Lin, S.-Z.; Liu, C.-S.; et al. Transferring Xenogenic Mitochondria Provides Neural Protection against Ischemic Stress in Ischemic Rat Brains. Cell Transplant. 2016, 25, 913–927. [Google Scholar] [CrossRef] [Green Version]
- Gollihue, J.L.; Patel, S.P.; Eldahan, K.C.; Cox, D.H.; Donahue, R.; Taylor, B.; Sullivan, P.G.; Rabchevsky, A.G. Effects of Mitochondrial Transplantation on Bioenergetics, Cellular Incorporation, and Functional Recovery after Spinal Cord Injury. J. Neurotrauma 2018, 35, 1800–1818. [Google Scholar] [CrossRef]
- Masuzawa, A.; Black, K.M.; Pacak, C.A.; Ericsson, M.; Barnett, R.J.; Drumm, C.; Seth, P.; Bloch, D.B.; Levitsky, S.; Cowan, D.B.; et al. Transplantation of autologously derived mitochondria protects the heart from ischemia-reperfusion injury. Am. J. Physiol. Circ. Physiol. 2013, 304, H966–H982. [Google Scholar] [CrossRef]
- Lin, H.-C.; Liu, S.-Y.; Lai, H.-S.; Lai, I.-R. Isolated Mitochondria Infusion Mitigates Ischemia-Reperfusion Injury of the Liver in Rats. Shock 2013, 39, 304–310. [Google Scholar] [CrossRef]
- Moskowitzova, K.; Orfany, A.; Liu, K.; Ramirez-Barbieri, G.; Thedsanamoorthy, J.K.; Yao, R.; Guariento, A.; Doulamis, I.P.; Blitzer, D.; Shin, B.; et al. Mitochondrial transplantation enhances murine lung viability and recovery after ischemia-reperfusion injury. Am. J. Physiol. Cell. Mol. Physiol. 2019, 318, L78–L88. [Google Scholar] [CrossRef]
- Bloomfield, G.; Kay, R.R. Uses and abuses of macropinocytosis. J. Cell Sci. 2016, 129, 2697–2705. [Google Scholar] [CrossRef] [Green Version]
- Mercer, J.; Helenius, A. Virus entry by macropinocytosis. Nature 2009, 11, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Helmuth, J.A.; Burckhardt, C.J.; Koumoutsakos, P.; Greber, U.F.; Sbalzarini, I. A novel supervised trajectory segmentation algorithm identifies distinct types of human adenovirus motion in host cells. J. Struct. Boil. 2007, 159, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Greber, U.F.; Flatt, J.W. Adenovirus Entry: From Infection to Immunity. Annu. Rev. Virol. 2019, 6, 177–197. [Google Scholar] [CrossRef] [PubMed]
- Luisoni, S.; Suomalainen, M.; Boucke, K.; Tanner, L.B.; Wenk, M.R.; Guan, X.L.; Grzybek, M.; Coskun, Ü.; Greber, U.F. Co-option of Membrane Wounding Enables Virus Penetration into Cells. Cell Host Microbe 2015, 18, 75–85. [Google Scholar] [CrossRef] [Green Version]
- Burckhardt, C.J.; Greber, U.F. Virus Movements on the Plasma Membrane Support Infection and Transmission between Cells. PLoS Pathog. 2009, 5, e1000621. [Google Scholar] [CrossRef]
- Pache, L.; Venkataraman, S.; Reddy, V.S.; Nemerow, G. Structural Variations in Species B Adenovirus Fibers Impact CD46 Association. J. Virol. 2008, 82, 7923–7931. [Google Scholar] [CrossRef] [Green Version]
- Amstutz, B.; Gastaldelli, M.; Kälin, S.; Imelli, N.; Boucke, K.; Wandeler, E.; Mercer, J.; Hemmi, S.; Greber, U.F. Subversion of CtBP1-controlled macropinocytosis by human adenovirus serotype 3. EMBO J. 2008, 27, 956–969. [Google Scholar] [CrossRef] [Green Version]
- Wolfrum, N.; Greber, U.F. Adenovirus signalling in entry. Cell. Microbiol. 2012, 15, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Kälin, S.; Amstutz, B.; Gastaldelli, M.; Wolfrum, N.; Boucke, K.; Havenga, M.; Digennaro, F.; Liska, N.; Hemmi, S.; Greber, U.F. Macropinocytotic Uptake and Infection of Human Epithelial Cells with Species B2 Adenovirus Type 35. J. Virol. 2010, 84, 5336–5350. [Google Scholar] [CrossRef] [Green Version]
- Liberali, P.; Kakkonen, E.; Turacchio, G.; Valente, C.; Spaar, A.; Perinetti, G.; Bockmann, R.A.; Corda, D.; Colanzi, A.; Marjomaki, V.; et al. The closure of Pak1-dependent macropinosomes requires the phosphorylation of CtBP1/BARS. EMBO J. 2008, 27, 970–981. [Google Scholar] [CrossRef] [Green Version]
- Mercer, J.; Helenius, A. Vaccinia Virus Uses Macropinocytosis and Apoptotic Mimicry to Enter Host Cells. Science 2008, 320, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Niebuhr, K.; Giuriato, S.; Pedron, T.; Philpott, D.J.; Gaits-Iacovoni, F.; Sable, J.; Sheetz, M.P.; Parsot, C.; Sansonetti, P.J.; Payrastre, B. Conversion of PtdIns(4,5)P2 into PtdIns(5)P by the S.flexneri effector IpgD reorganizes host cell morphology. EMBO J. 2002, 21, 5069–5078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moser, T.S.; Jones, R.G.; Thompson, C.B.; Coyne, C.B.; Cherry, S. A Kinome RNAi Screen Identified AMPK as Promoting Poxvirus Entry through the Control of Actin Dynamics. PLoS Pathog. 2010, 6, e1000954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondratowicz, A.S.; Hunt, C.L.; Davey, R.A.; Cherry, S.; Maury, W. AMP-Activated Protein Kinase Is Required for the Macropinocytic Internalization of Ebolavirus. J. Virol. 2012, 87, 746–755. [Google Scholar] [CrossRef] [Green Version]
- Albert, M.L. Death-defying immunity: do apoptotic cells influence antigen processing and presentation? Nat. Rev. Immunol. 2004, 4, 223–231. [Google Scholar] [CrossRef]
- González, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017, 36, 397–408. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, S.; Hoppe, A.D.; Araki, N.; Swanson, J.A. Sequential signaling in plasma-membrane domains during macropinosome formation in macrophages. J. Cell Sci. 2009, 122, 3250–3261. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-L.; Wang, Y.; Sesaki, H.; Iijima, M. Myosin I Links PIP3 Signaling to Remodeling of the Actin Cytoskeleton in Chemotaxis. Sci. Signal. 2012, 5, ra10. [Google Scholar] [CrossRef] [Green Version]
- Vanhaesebroeck, B.; Stephens, L.R.; Hawkins, P. PI3K signalling: the path to discovery and understanding. Nat. Rev. Mol. Cell Boil. 2012, 13, 195–203. [Google Scholar] [CrossRef]
- Azzi, A.; Boscoboinik, D.; Hensey, C. The protein kinase C family. JBIC J. Boil. Inorg. Chem. 1992, 208, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Apgar, J.R. Activation of protein kinase C in rat basophilic leukemia cells stimulates increased production of phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: correlation with actin polymerization. Mol. Boil. Cell 1995, 6, 97–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nussinov, R.; Tsai, C.-J.; Jang, H. Is Nanoclustering essential for all oncogenic KRas pathways? Can it explain why wild-type KRas can inhibit its oncogenic variant? Semin. Cancer Boil. 2019, 54, 114–120. [Google Scholar] [CrossRef]
- Nussinov, R.; Tsai, C.-J.; Jang, H. Does Ras Activate Raf and PI3K Allosterically? Front. Oncol. 2019, 9, 9. [Google Scholar] [CrossRef]
- Yang, G.; Murashige, D.S.; Humphrey, S.J.; James, D.E. A Positive Feedback Loop between Akt and mTORC2 via SIN1 Phosphorylation. Cell Rep. 2015, 12, 937–943. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Chen, Z.; Erdjument-Bromage, H.; Tempst, P.; Pandolfi, P.P. Phosphorylation and Functional Inactivation of TSC2 by Erk. Cell 2005, 121, 179–193. [Google Scholar] [CrossRef] [Green Version]
- Inoki, K.; Li, Y.; Zhu, T.; Wu, J.; Guan, K.-L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature 2002, 4, 648–657. [Google Scholar] [CrossRef]
- Dibble, C.; Elis, W.; Menon, S.; Qin, W.; Klekota, J.; Asara, J.M.; Finan, P.M.; Kwiatkowski, D.J.; Murphy, L.O.; Manning, B.D. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell 2012, 47, 535–546. [Google Scholar] [CrossRef] [Green Version]
- Betz, C.; Hall, M.N. Where is mTOR and what is it doing there? J. Cell Boil. 2013, 203, 563–574. [Google Scholar] [CrossRef] [Green Version]
- Menon, S.; Dibble, C.; Talbott, G.; Hoxhaj, G.; Valvezan, A.J.; Takahashi, H.; Cantley, L.C.; Manning, B.D. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 2014, 156, 771–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bar-Peled, L.; Schweitzer, L.D.; Zoncu, R.; Sabatini, D.M. Ragulator Is a GEF for the Rag GTPases that Signal Amino Acid Levels to mTORC1. Cell 2012, 150, 1196–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demetriades, C.; Doumpas, N.; A Teleman, A. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 2014, 156, 786–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrington, L.S.; Findlay, G.M.; Lamb, R.F. Restraining PI3K: mTOR signalling goes back to the membrane. Trends Biochem. Sci. 2005, 30, 35–42. [Google Scholar] [CrossRef]
- Maehama, T.; Dixon, J.E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Boil. Chem. 1998, 273, 13375–13378. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.J.; DeStefano, M.A.; Oh, W.J.; Wu, C.-C.; Vega-Cotto, N.M.; Finlan, M.; Liu, U.; Su, B.; Jacinto, E. mTOR complex 2 regulates proper turnover of insulin receptor substrate-1 via the ubiquitin ligase subunit Fbw8. Mol. Cell 2012, 48, 875–887. [Google Scholar] [CrossRef] [Green Version]
- Harrington, L.S.; Findlay, G.M.; Gray, A.; Tolkacheva, T.; Wigfield, S.; Rebholz, H.; Barnett, J.; Leslie, N.R.; Cheng, S.; Shepherd, P.R.; et al. The TSC1-2 tumor suppressor controls insulin–PI3K signaling via regulation of IRS proteins. J. Cell Boil. 2004, 166, 213–223. [Google Scholar] [CrossRef] [Green Version]
- Shah, O.J.; Wang, Z.; Hunter, T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 2004, 14, 1650–1656. [Google Scholar] [CrossRef] [Green Version]
- Hackstein, H.; Taner, T.; Logar, A.J.; Thomson, A.W. Rapamycin inhibits macropinocytosis and mannose receptor–mediated endocytosis by bone marrow–derived dendritic cells. Blood 2002, 100, 1084–1087. [Google Scholar] [CrossRef] [Green Version]
- Weichhart, T.; Hengstschläger, M.; Linke, M. Regulation of innate immune cell function by mTOR. Nat. Rev. Immunol. 2015, 15, 599–614. [Google Scholar] [CrossRef]
- Sung, S.; Choi, J.; Cheong, H. Catabolic pathways regulated by mTORC1 are pivotal for survival and growth of cancer cells expressing mutant Ras. Oncotarget 2015, 6, 40405–40417. [Google Scholar] [CrossRef] [Green Version]
- Palm, W.; Park, Y.; Wright, K.; Pavlova, N.; Tuveson, D.A.; Thompson, C.B. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell 2015, 162, 259–270. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.; Kovarova, J.; Bajzikova, M.; Bezawork-Geleta, A.; Svec, D.; Endaya, B.; Sachaphibulkij, K.; Coelho, A.; Sebkova, N.; Ruzickova, A.; et al. Horizontal transfer of whole mitochondria restores tumorigenic potential in mitochondrial DNA-deficient cancer cells. eLife 2017, 6, 529. [Google Scholar] [CrossRef] [Green Version]
- Phinney, D.; Di Giuseppe, M.; Njah, J.; Sala-Llinas, E.; Shiva, S.; Croix, C.M.S.; Stolz, N.B.; Watkins, S.C.; Di, Y.; Leikauf, G.D.; et al. Mesenchymal stem cells use extracellular vesicles to outsource mitophagy and shuttle microRNAs. Nat. Commun. 2015, 6, 8472. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.-H.O.; Kim, K.-Y.; Bushong, E.A.; Mills, E.A.; Boassa, D.; Shih, T.; Kinebuchi, M.; Phan, S.; Zhou, Y.; Bihlmeyer, N.; et al. Transcellular degradation of axonal mitochondria. Proc. Natl. Acad. Sci. USA 2014, 111, 9633–9638. [Google Scholar] [CrossRef] [Green Version]
- Seth, P.; Pastan, I.; Willingham, M.C. Adenovirus-dependent changes in cell membrane permeability: role of Na+, K+-ATPase. J. Virol. 1987, 61, 883–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, S.C. Viral membrane fusion. Virology 2015, 479, 498–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, N.; Taiz, L. The evolution of H+-ATPases. Trends Biochem. Sci. 1989, 14, 113–116. [Google Scholar] [CrossRef]
- Jae, L.T.; Raaben, M.; Riemersma, M.; Van Beusekom, E.; Blomen, V.A.; Velds, A.; Kerkhoven, R.M.; Carette, J.E.; Topaloglu, H.; Meinecke, P.; et al. Deciphering the Glycosylome of Dystroglycanopathies Using Haploid Screens for Lassa Virus Entry. Science 2013, 340, 479–483. [Google Scholar] [CrossRef] [Green Version]
- Brecher, M.; Schornberg, K.L.; Delos, S.E.; Fusco, M.L.; Saphire, E.O.; White, J.M. Cathepsin Cleavage Potentiates the Ebola Virus Glycoprotein To Undergo a Subsequent Fusion-Relevant Conformational Change. J. Virol. 2011, 86, 364–372. [Google Scholar] [CrossRef] [Green Version]
- Staring, J.; Raaben, M.; Brummelkamp, T.R. Viral escape from endosomes and host detection at a glance. J. Cell Sci. 2018, 131, jcs216259. [Google Scholar] [CrossRef] [Green Version]
- Cady, S.D.; Luo, W.; Hu, F.; Hong, M. Structure and Function of the Influenza A M2 Proton Channel. Biochemistry 2009, 48, 7356–7364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leopold, P.L.; Kreitzer, G.; Miyazawa, N.; Rempel, S.; Pfister, K.K.; Rodriguez-Boulan, E.; Crystal, R.G. Dynein- and Microtubule-Mediated Translocation of Adenovirus Serotype 5 Occurs after Endosomal Lysis. Hum. Gene Ther. 2000, 11, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Dodding, M.P.; Way, M. Coupling viruses to dynein and kinesin-1. EMBO J. 2011, 30, 3527–3539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gazzola, M.; Burckhardt, C.J.; Bayati, B.; Engelke, M.F.; Greber, U.F.; Koumoutsakos, P. A Stochastic Model for Microtubule Motors Describes the In Vivo Cytoplasmic Transport of Human Adenovirus. PLoS Comput. Boil. 2009, 5, e1000623. [Google Scholar] [CrossRef] [Green Version]
- Kural, C.; Kim, H.; Syed, S.; Goshima, G.; Gelfand, V.I.; Selvin, P.R. Kinesin and Dynein Move a Peroxisome in Vivo: A Tug-of-War or Coordinated Movement? Science 2005, 308, 1469–1472. [Google Scholar] [CrossRef] [Green Version]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I. CRM1 Is an Export Receptor for Leucine-Rich Nuclear Export Signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, R.; Delphin, C.; Guan, T.; Gerace, L.; Melchior, F. A Small Ubiquitin-Related Polypeptide Involved in Targeting RanGAP1 to Nuclear Pore Complex Protein RanBP2. Cell 1997, 88, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Strunze, S.; Trotman, L.C.; Boucke, K.; Greber, U.F. Nuclear Targeting of Adenovirus Type 2 Requires CRM1-mediated Nuclear Export. Mol. Boil. Cell 2005, 16, 2999–3009. [Google Scholar] [CrossRef]
- Trotman, L.C.; Mosberger, N.; Fornerod, M.; Stidwill, R.P.; Greber, U.F. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 2001, 3, 1092–1100. [Google Scholar] [CrossRef]
- Jahed, Z.; Soheilypour, M.; Peyro, M.; Mofrad, M.R.K. The LINC and NPC relationship - it’s complicated! J. Cell Sci. 2016, 129, 3219–3229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniele, T.; Schiaffino, M.V. Organelle biogenesis and interorganellar connections: Better in contact than in isolation. Commun. Integr. Boil. 2014, 7, e29587. [Google Scholar] [CrossRef] [PubMed]
- Szymański, J.; Janikiewicz, J.; Michalska, B.; Patalas-Krawczyk, P.; Perrone, M.; Ziółkowski, W.; Duszynski, J.; Pinton, P.; Dobrzyń, A.; Wieckowski, M.R. Interaction of Mitochondria with the Endoplasmic Reticulum and Plasma Membrane in Calcium Homeostasis, Lipid Trafficking and Mitochondrial Structure. Int. J. Mol. Sci. 2017, 18, 1576. [Google Scholar] [CrossRef] [PubMed]
- Rieusset, J. The role of endoplasmic reticulum-mitochondria contact sites in the control of glucose homeostasis: an update. Cell Death Dis. 2018, 9, 388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinvalet, D. The role of the mitochondria and the endoplasmic reticulum contact sites in the development of the immune responses. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sheftel, A.D.; Zhang, A.-S.; Brown, C.M.; Shirihai, O.S.; Ponka, P. Direct interorganellar transfer of iron from endosome to mitochondrion. Blood 2007, 110, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charman, M.; Kennedy, B.E.; Osborne, N.; Karten, B. MLN64 mediates egress of cholesterol from endosomes to mitochondria in the absence of functional Niemann-Pick Type C1 protein. J. Lipid Res. 2009, 51, 1023–1034. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.; Leonzino, M.; Hancock-Cerutti, W.; Horenkamp, F.A.; Li, P.; Lees, J.A.; Wheeler, H.; Reinisch, K.M.; De Camilli, P. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Boil. 2018, 217, 3625–3639. [Google Scholar] [CrossRef] [Green Version]
- Park, J.-S.; Thorsness, M.K.; Policastro, R.; McGoldrick, L.L.; Hollingsworth, N.M.; Thorsness, P.E.; Neiman, A.M. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Boil. Cell 2016, 27, 2435–2449. [Google Scholar] [CrossRef] [Green Version]
- Daniele, T.; Hurbain, I.; Vago, R.; Casari, G.; Raposo, G.; Tacchetti, C.; Schiaffino, M.V. Mitochondria and Melanosomes Establish Physical Contacts Modulated by Mfn2 and Involved in Organelle Biogenesis. Curr. Boil. 2014, 24, 393–403. [Google Scholar] [CrossRef] [Green Version]
- Naon, D.P.; Zaninello, M.; Giacomello, M.; Varanita, T.; Grespi, F.; Lakshminaranayan, S.; Serafini, A.; Semenzato, M.; Herkenne, S.; Hernandez-Alvarez, M.I.; et al. Critical reappraisal confirms that Mitofusin 2 is an endoplasmic reticulum-mitochondria tether. Proc. Natl. Acad. Sci. USA 2016, 113, 11249–11254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abuaita, B.H.; Schultz, T.L.; O’Riordan, M. Mitochondria-Derived Vesicles Deliver Antimicrobial Reactive Oxygen Species to Control Phagosome-Localized Staphylococcus aureus. Cell Host Microbe 2018, 24, 625–636.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pernas, L.; Bean, C.; Boothroyd, J.C.; Scorrano, L. Mitochondria Restrict Growth of the Intracellular Parasite Toxoplasma gondii by Limiting Its Uptake of Fatty Acids. Cell Metab. 2018, 27, 886–897.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todkar, K.; Chikhi, L.; Germain, M. Mitochondrial interaction with the endosomal compartment in endocytosis and mitochondrial transfer. Mitochondrion 2019, 49, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Chernyak, B.V. Mitochondrial Transplantation: A Critical Analysis. Biochemistry (Mosc.) 2020, 85, 636–641. [Google Scholar] [CrossRef]
- Bernardi, P.; Rasola, A.; Forte, M.; Lippe, G. The Mitochondrial Permeability Transition Pore: Channel Formation by F-ATP Synthase, Integration in Signal Transduction, and Role in Pathophysiology. Physiol. Rev. 2015, 95, 1111–1155. [Google Scholar] [CrossRef]
- Marlein, C.R.; Zaitseva, L.; Piddock, R.E.; Robinson, S.D.; Edwards, D.R.; Shafat, M.S.; Zhou, Z.; Lawes, M.; Bowles, K.M.; Rushworth, S.A. NADPH oxidase-2 derived superoxide drives mitochondrial transfer from bone marrow stromal cells to leukemic blasts. Blood 2017, 130, 1649–1660. [Google Scholar] [CrossRef]
- Pour, P.A.; Kenney, M.C.; Kheradvar, A. Bioenergetics Consequences of Mitochondrial Transplantation in Cardiomyocytes. J. Am. Hear. Assoc. 2020, 9, e014501. [Google Scholar] [CrossRef]
- Sinha, P.; Islam, M.N.; Bhattacharya, S.; Bhattacharya, J. Intercellular mitochondrial transfer: bioenergetic crosstalk between cells. Curr. Opin. Genet. Dev. 2016, 38, 97–101. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-Y.; Liang, M.-Z.; Chen, L. Current progress of mitochondrial transplantation that promotes neuronal regeneration. Transl. Neurodegener. 2019, 8, 17. [Google Scholar] [CrossRef] [Green Version]
- Emani, S.M.; McCully, J.D. Mitochondrial transplantation: applications for pediatric patients with congenital heart disease. Transl. Pediatr. 2018, 7, 169–175. [Google Scholar] [CrossRef] [Green Version]
- Commisso, C.; Davidson, S.M.; Soydaner-Azeloglu, R.G.; Parker, S.J.; Kamphorst, J.J.; Hackett, S.; Grabocka, E.; Nofal, M.; Drebin, J.A.; Thompson, C.B.; et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 2013, 497, 633–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von der Haar, T.; Josse, L.; Wright, P.; Zenthon, J.; Tuite, M.F. Development of a novel yeast cell-based system for studying the aggregation of Alzheimer’s disease-associated Abeta peptides in vivo. Neurodegener Dis. 2007, 4, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Kruth, H.S.; Jones, N.L.; Huang, W.; Zhao, B.; Ishii, I.; Chang, J.; Combs, C.A.; Malide, D.; Zhang, W.-Y. Macropinocytosis Is the Endocytic Pathway That Mediates Macrophage Foam Cell Formation with Native Low 105 Density Lipoprotein. J. Boil. Chem. 2004, 280, 2352–2360. [Google Scholar] [CrossRef] [Green Version]
- West, A.P.; Shadel, G.S.; Ghosh, S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011, 11, 389–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Target | Diseases | Method | Outcome | Animal | Ref./Year |
|---|---|---|---|---|---|
| Brain | Ischemia reperfusion injury | Direct injection | Enhanced survival-related gene expression | Mice | [15]/2016 |
| Brain | Ischemia reperfusion injury | Intra-arterial injection | Functional recovery | Rats | [16]/2016 |
| Spine | Spinal cord injury | Direct injection | Improved mitochondrial respiration | Rats | [17]/2018 |
| Heart | Ischemia reperfusion injury | Direct injection | Enhanced post-infarct cardiac function | Rabbits | [18]/2013 |
| Liver | Ischemia reperfusion injury | Direct injection into the spleen | Suppressed necrosis and apoptosis | Rats | [19]/2013 |
| Lung | Ischemia reperfusion injury | Intra-arterial or trans-tracheal injection | Increased compliance and inspiratory capacity | Mice | [20]/2020 |
| Method | Recipient Cell | Donor Mitochondria | Mitochondria Marker | Ref./Year | ||
|---|---|---|---|---|---|---|
| TAT-dextran | Human | Uterine endometrial gland-derived mesenchymal cells | Rat | C2C12, an immortalized myoblast cell line | MitoDsRed | [7]/2020 |
| Rat | Neonate cardiomyocytes, primary | Rat | C2C12, an immortalized myoblast cell line | MitoDsRed | ||
| Centrifugation | Rat | L6, muscle | Human | Umbilical cord-derived mesenchymal stem cells | MitoTracker | [8]/2018 |
| Magnetomitotransfer | Human | MRC-5, fibroblasts | Human | MRC-5, a diploid human cell culture line composed of fibroblasts, originally developed from research deriving lung tissue of a 14 week old aborted Caucasian male fetus | MitoTracker | [9]/2016 |
| Photothermal nanoblade | Human | 143BTK-, osteosarcoma | Human | MDA-MB-453, breast carcinoma | MitoDsRed | [10]/2016 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kami, D.; Gojo, S. From Cell Entry to Engraftment of Exogenous Mitochondria. Int. J. Mol. Sci. 2020, 21, 4995. https://doi.org/10.3390/ijms21144995
Kami D, Gojo S. From Cell Entry to Engraftment of Exogenous Mitochondria. International Journal of Molecular Sciences. 2020; 21(14):4995. https://doi.org/10.3390/ijms21144995
Chicago/Turabian StyleKami, Daisuke, and Satoshi Gojo. 2020. "From Cell Entry to Engraftment of Exogenous Mitochondria" International Journal of Molecular Sciences 21, no. 14: 4995. https://doi.org/10.3390/ijms21144995
APA StyleKami, D., & Gojo, S. (2020). From Cell Entry to Engraftment of Exogenous Mitochondria. International Journal of Molecular Sciences, 21(14), 4995. https://doi.org/10.3390/ijms21144995




