Mid-Gestation lethality of Atxn2l-Ablated Mice
Abstract
1. Introduction
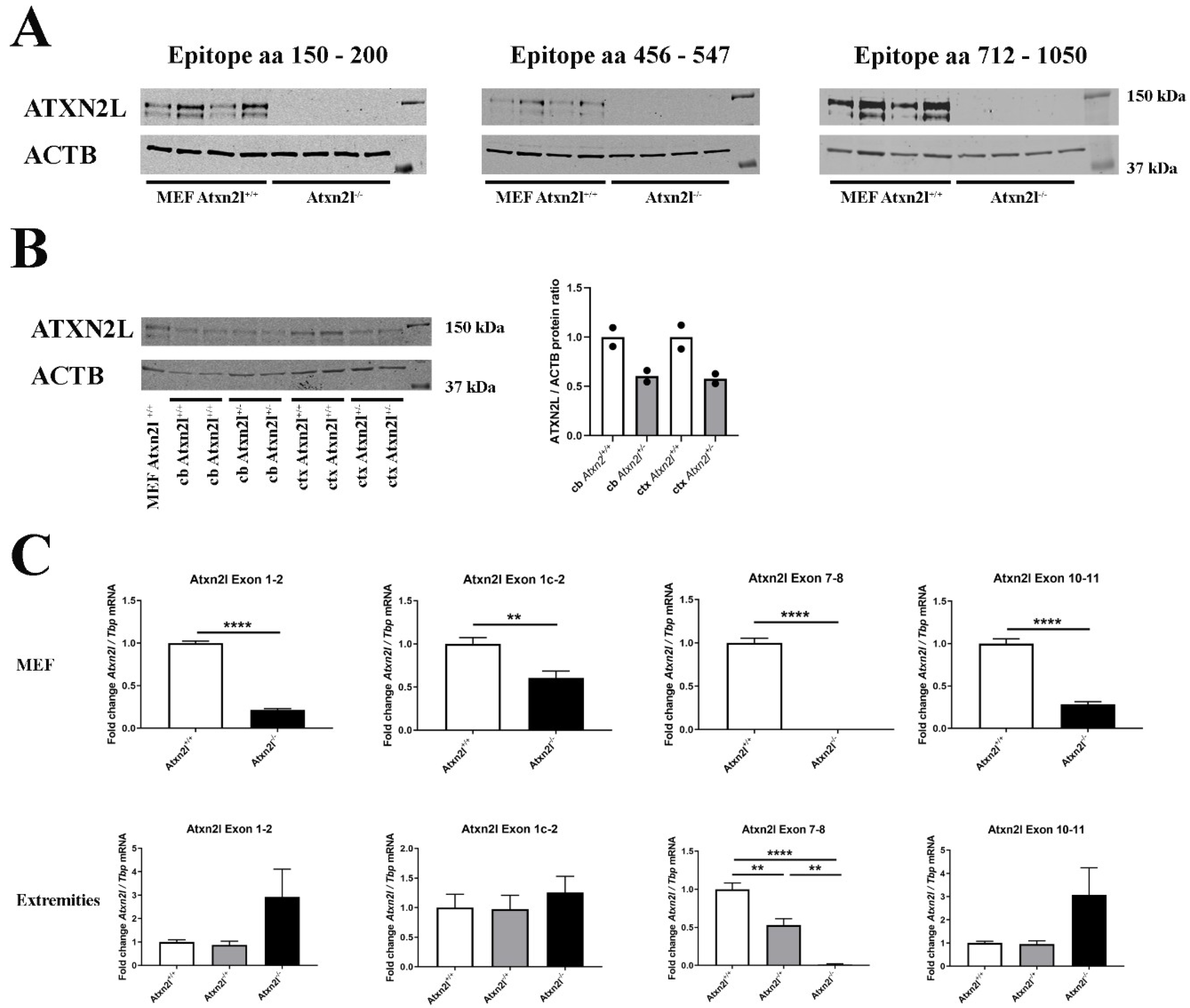
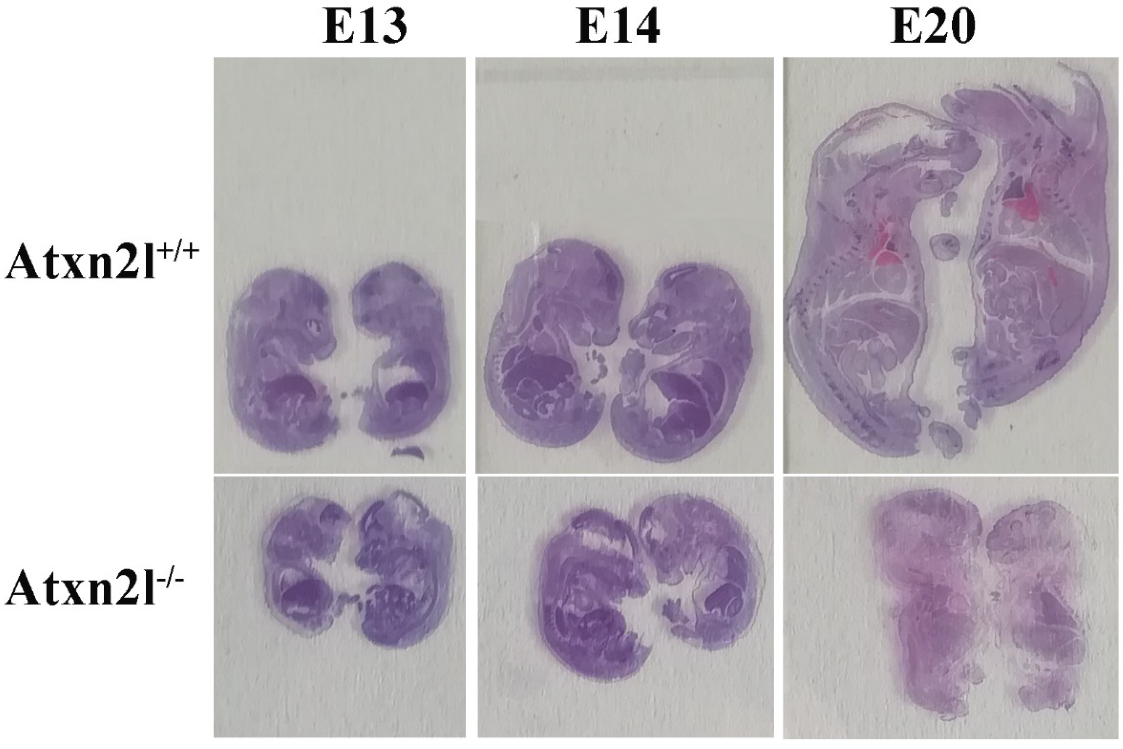
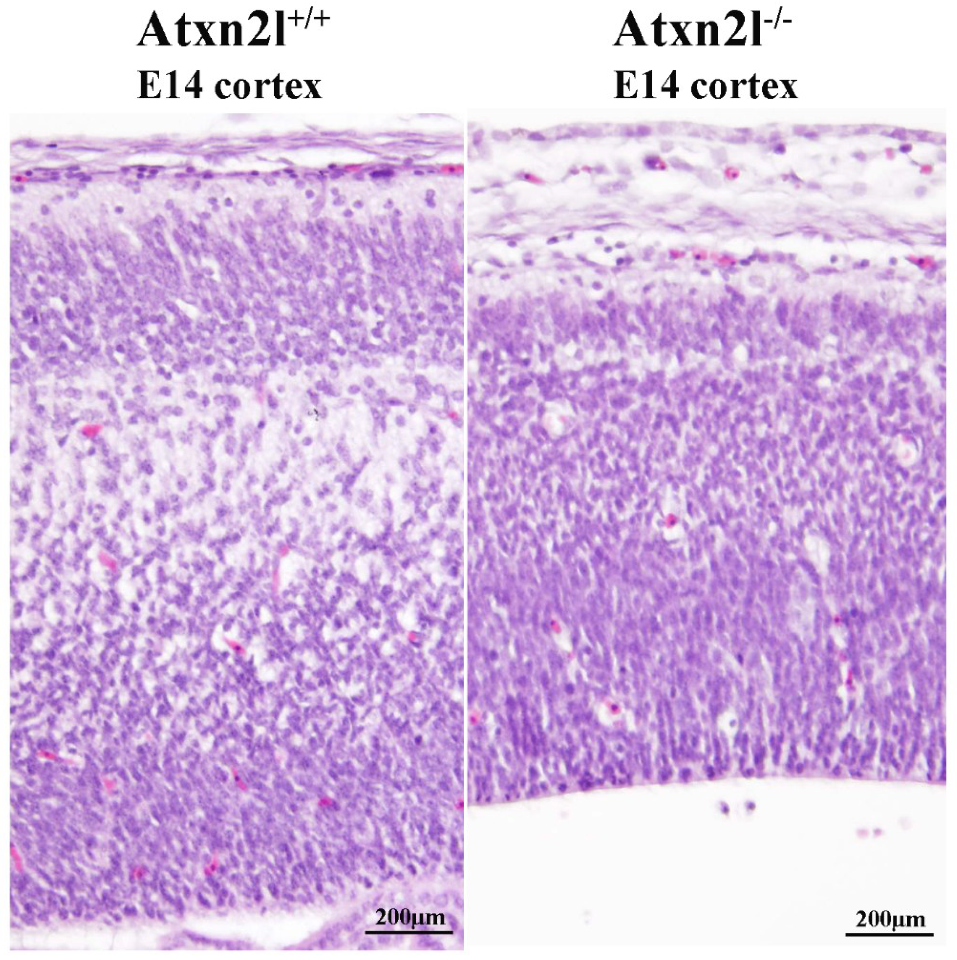
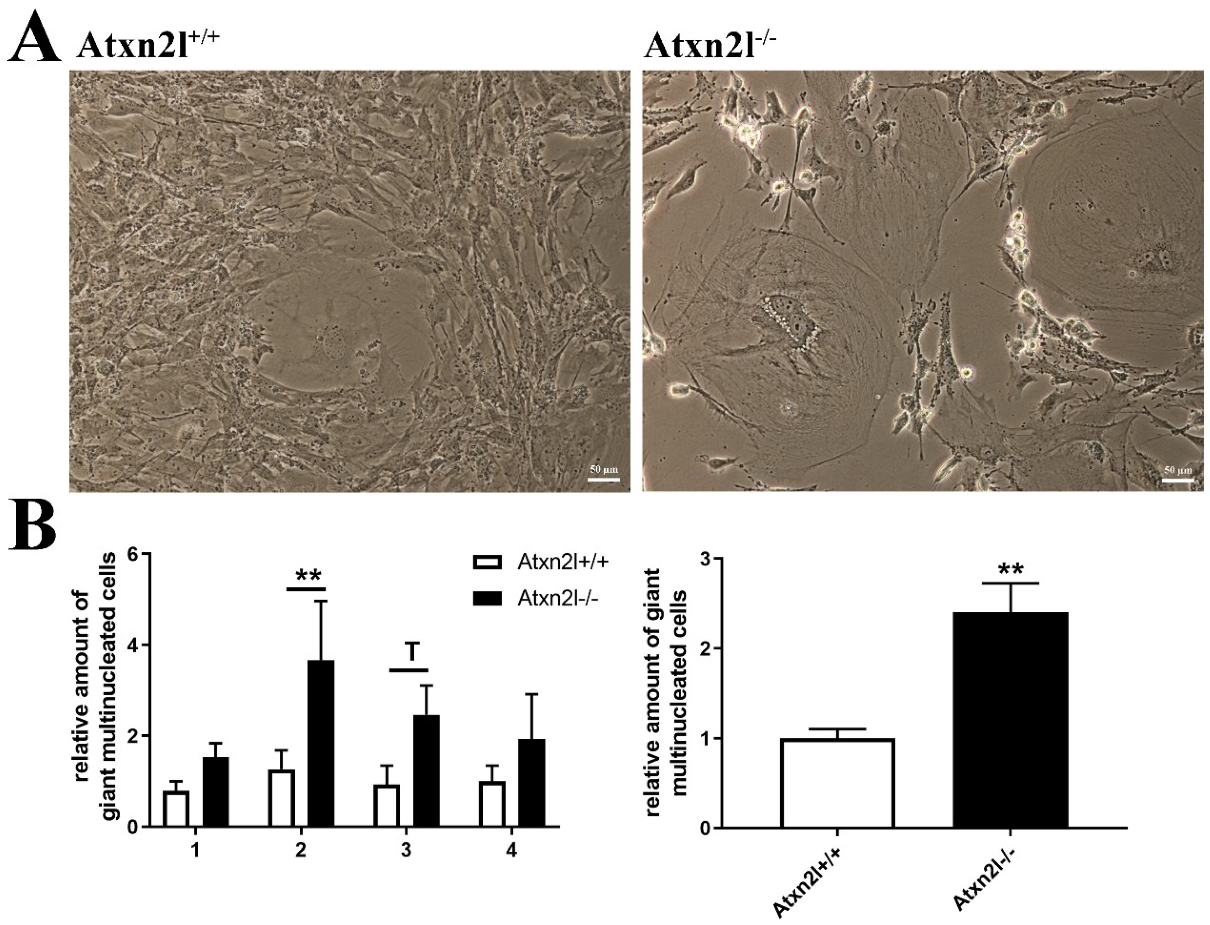
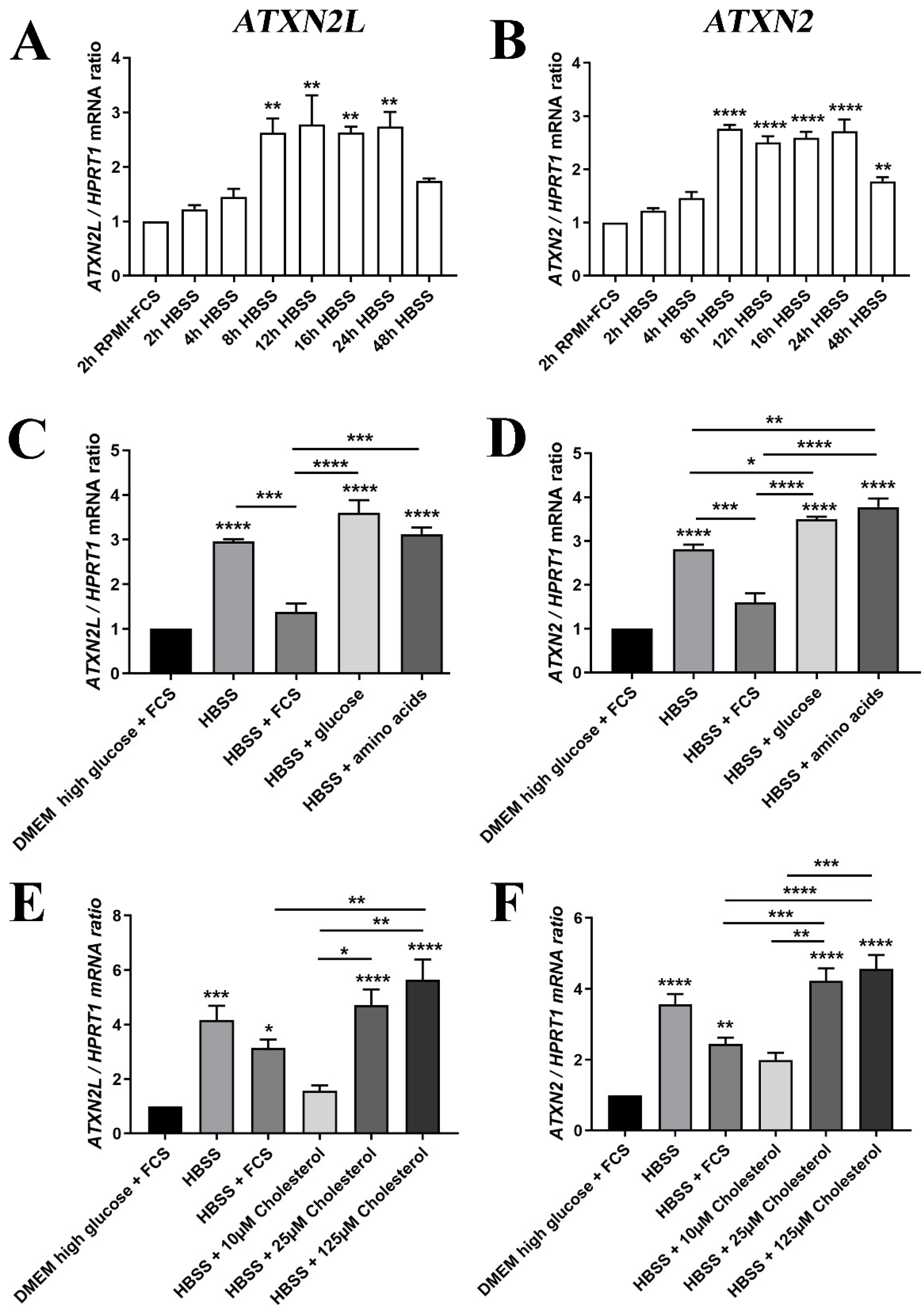
2. Results
3. Discussion
4. Materials and Methods
4.1. Generation, Breeding, Maintenance, and Dissection of Atxn2l−/− Mice
4.2. Genotyping
4.3. Behavioral Tests
4.4. Generation and Cell Culture of MEF Cells
4.5. Poly(I:C) Treatment
4.6. RNA Extraction and Expression Analysis
4.7. Immunoblotting
4.8. Sections and Staining
4.9. Microscopy and Cell Counting of MEF
4.10. Cell Culture Experiments
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| °C | Degrees Celsius (temperature) |
| aa | amino acid |
| ACTB | beta-actin |
| Adnp | Activity-Dependent Neuroprotector Homeobox Protein |
| ALS | Amyotrophic Lateral Sclerosis |
| ANOVA | Analysis of variance |
| ARHGEF11 | Rho Guanine Nucleotide Exchange Factor (GEF) 11 |
| ATF1 | Activating Transcription Factor 1 |
| ATG | Start codon Adenine-Thymidine/Uracil-Guanine |
| ATX-2 | the single nematode ortholog of mammalian Ataxin-2 and Ataxin-2-like |
| ATXN2 | Ataxin-2 |
| ATXN2L | Ataxin-2-like, aka: Ataxin-2-Related Protein, A2LP, A2RP or A2D |
| BDNF | Brain-Derived Neurotrophic Factor |
| bp | Base pair |
| BSA | Bovine serum albumin |
| Carm1 | Coactivator Associated Arginine Methyltransferase 1 |
| Cart1 | Cartilage paired class homeoprotein 1 (aka Alx1) |
| Cb | Cerebellum |
| Cbp | cAMP-response element binding protein (Creb) binding protein (aka Crebbp) |
| cDNA | complementary Deoxyribo-Nucleic Acid |
| CID3 | CTC-interacting domain 3 protein in Arabidopsis thaliana |
| CID4 | CTC-interacting domain 4 protein in Arabidopsis thaliana |
| Cited2 | Cbp/P300 Interacting Transactivator With Glu/Asp Rich Carboxy-Terminal Domain 2 |
| Crispr/Cas9 | Clustered Regularly Interspaced Short Palindromic Repeats/Crispr-associated protein 9 |
| C-term | C-terminal end of the protein |
| Ctx | Cortex |
| DAPK1 | Death-Associated Protein Kinase 1 |
| DDX6 | DEAD/H (Asp-Glu-Ala-Asp/His) Box Polypeptide 6 (RNA Helicase, 54kD) |
| DENND1A | DENN/MADD Domain Containing 1A (RAB35-activating guanine nucleotide exchange factor) |
| DMEM | Dulbecco’s modified essential medium |
| DNA | Desoxyribo-Nucleic-Acid chain |
| E | Embryonic development day |
| E2F1 | E2F Transcription Factor 1 |
| e.g., | Exemplo gratia (for example, in Latin language) |
| EGF | Epidermal growth factor |
| ELF1 | E74 Like ETS Transcription Factor 1 |
| Ep300 | E1A Binding Protein, Histone Acetyltransferase P300 |
| EST | Expressed sequence tag |
| Ex1 | Exon 1 |
| FCS | Fetal calf serum |
| FTLD | Fronto-temporal lobar dementia |
| Foxj2 | Forkhead Box Protein J2, a transcriptional activator |
| g | Gram |
| G3BP2 | Ras-GTPase Activating Protein SH3 Domain-Binding Protein 2 |
| GLAST GWAS | Sodium-Dependent Glutamate/Aspartate Transporter 1 (in astrocytes) Genome wide association studies |
| h | hour |
| H&E | Hematoxylin and Eosin staining |
| HBSS | Hank’s buffered salt solution |
| Het | Heterozygote |
| HNRNP | Heterogeneous Nuclear Ribonucleoprotein |
| Kb | kilo-base |
| IRF1 | Interferon Regulatory Factor 1 |
| ITS | Insulin-Transferrin-Selenium |
| KD | Knock-Down |
| KO | Knock-Out |
| L | liter |
| lncRNA | long non-coding RNA |
| Lsm | Like “Smith antigen” protein domain |
| LSM12 | Homolog of yeast cytosolic LSM12 protein |
| LsmAD | Lsm-associated domain |
| mg | milliGram |
| mm | milliMeter |
| mM | milliMolar |
| µg | microGram |
| µL | microLiter |
| µM | microMolar |
| MEF | Mouse embryonal fibroblasts |
| MEM | Minimal Essential Medium |
| MGI mGluR5 | Mouse Genome Informatics metabotropic Glutamate Receptor 5 |
| MPL | Interactor domain with Myelo-Proliferative Leukemia gene, encodes Thrombopoietin receptor |
| mRNA | Messenger ribonucleic acid |
| mTORC1 | Mechanistic “target of rapamycin” complex 1 |
| Neat1 | Nuclear Paraspeckle Assembly Transcript 1 |
| NFATC2IP | Nuclear Factor of Activated T-Cells, Cytoplasmic, Calcineurin-Dependent 2 Interacting Protein |
| ng | nanoGram |
| NHGRI-EBI N-term | American National Human Genetics Research Institute & European Bioinformatics Institute N-terminal end of the protein |
| NUFIP2 | Nuclear Fragile X Mental Retardation Protein 1 Interacting Protein 2 |
| ORF | open reading frame |
| p | probability value for the occurrence of a given observation by chance |
| PABPC1 | Poly(A)-binding-protein cytosolic 1 |
| PAM2 | Poly(A)-binding mediator domain 2 |
| PBS | Phosphate buffered saline |
| PFA | Paraformaldehyde |
| poly(A) tail | adenosine-repeat at the 3′ end of mRNA |
| Poly(I:C) | Poly-Inosinic: Poly-Cytidylic acid chain |
| Pro | Proline |
| PRD | Proline-rich domain |
| PSD-95 PSP | Postsynaptic Density Protein 95 (aka DLK4, Disks Large Homolog 4) progressive supranuclear palsy (aka Parkinson plus) |
| Q, 22Q | glutamine, repeat of 22 consecutive glutamines |
| RA | Retinoic acid |
| RT-qPCR | Reverse-Transcriptase quantitative Polymerase Chain Reaction |
| S6R | Small ribosomal subunit protein 6 |
| SCA1 | Spinocerebellar Ataxia type 1 |
| SCA2 | Spinocerebellar Ataxia type 2 |
| SDS | Sodium-Dodecyl-Sulfate detergent |
| s.e.m. | Standard error of the mean |
| Setd5 | Histone-Lysine N-Methyltransferase, SET domain containing 5 |
| SG | Stress granule |
| sgRNA | Single guide Ribo-Nucleic-Acid chain |
| Sh2b1 | Pro-Rich, PH And SH2 Domain-Containing Signaling Mediator gene |
| SH3 | Src-homology 3 domain |
| SH-SY5Y | Cell line subcloned from SK-N-SH line from 4-year-old female neuroblastoma patient |
| SITE+3 | Selenite, Insulin, Transferrin, Ethanolamine +3 |
| SLC9A3R2 SNP | Solute Carrier Family 9 (Sodium/Hydrogen Exchange) Member 3 Regulator 2 (aka NHERF2) Single nucleotide polymorphism |
| SPITE | Selenite, Pyruvate, Insulin, Transferrin, Ethanolamine +3 |
| SR-B1 Src | Scavenger Receptor Class B Member 1 (aka CD36, Thrombospondin Receptor-Like 1) V-Src Avian Sarcoma (Schmidt-Ruppin A-2) Viral Oncogene |
| Tbp | TATA-binding factor of transcription |
| TDP-43 | TAR (transcription active response element) DNA-Binding Protein 43 (encoded by TARDBP) |
| Tfap2a | Transcription Factor Activator-Protein-2 alpha |
| TIGM, TX | Texas Institute of Genomic Medicine, Texas, United States of America |
| TPM | Transcripts per million (expression unit as normalization method in RNA-sequencing) |
| Tufm | Tu Translation Elongation Factor, Mitochondrial gene |
| ULK1 | Unc-51 Like Autophagy Activating Kinase 1 |
| VPS54 | Vacuolar Protein Sorting-Associated Protein 54 (endosomal factor for retrograde transport) |
| WT | Wild-Type |
References
- Auburger, G.; Sen, N.E.; Meierhofer, D.; Basak, A.N.; Gitler, A.D. Efficient Prevention of Neurodegenerative Diseases by Depletion of Starvation Response Factor Ataxin-2. Trends Neurosci. 2017, 40, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Satterfield, T.F.; Pallanck, L.J. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum. Mol. Genet. 2006, 15, 2523–2532. [Google Scholar] [CrossRef] [PubMed]
- Yokoshi, M.; Li, Q.; Yamamoto, M.; Okada, H.; Suzuki, Y.; Kawahara, Y. Direct binding of Ataxin-2 to distinct elements in 3’ UTRs promotes mRNA stability and protein expression. Mol. Cell 2014, 55, 186–198. [Google Scholar] [CrossRef] [PubMed]
- van de Loo, S.; Eich, F.; Nonis, D.; Auburger, G.; Nowock, J. Ataxin-2 associates with rough endoplasmic reticulum. Exp. Neurol. 2009, 215, 110–118. [Google Scholar] [CrossRef]
- Fittschen, M.; Lastres-Becker, I.; Halbach, M.V.; Damrath, E.; Gispert, S.; Azizov, M.; Walter, M.; Muller, S.; Auburger, G. Genetic ablation of ataxin-2 increases several global translation factors in their transcript abundance but decreases translation rate. Neurogenetics 2015, 16, 181–192. [Google Scholar] [CrossRef]
- Lastres-Becker, I.; Nonis, D.; Eich, F.; Klinkenberg, M.; Gorospe, M.; Kotter, P.; Klein, F.A.; Kedersha, N.; Auburger, G. Mammalian ataxin-2 modulates translation control at the pre-initiation complex via PI3K/mTOR and is induced by starvation. Biochim. Biophys. Acta 2016, 1862, 1558–1569. [Google Scholar] [CrossRef]
- Nonhoff, U.; Ralser, M.; Welzel, F.; Piccini, I.; Balzereit, D.; Yaspo, M.L.; Lehrach, H.; Krobitsch, S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell 2007, 18, 1385–1396. [Google Scholar] [CrossRef]
- Jagdeo, J.M.; Dufour, A.; Klein, T.; Solis, N.; Kleifeld, O.; Kizhakkedathu, J.; Luo, H.; Overall, C.M.; Jan, E. N-Terminomics TAILS Identifies Host Cell Substrates of Poliovirus and Coxsackievirus B3 3C Proteinases That Modulate Virus Infection. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Nonis, D.; Schmidt, M.H.H.; van de Loo, S.; Eich, F.; Dikic, I.; Nowock, J.; Auburger, G. Ataxin-2 associates with the endocytosis complex and affects EGF receptor trafficking. Cell. Signal. 2008, 20, 1725–1739. [Google Scholar] [CrossRef]
- Drost, J.; Nonis, D.; Eich, F.; Leske, O.; Damrath, E.; Brunt, E.R.; Lastres-Becker, I.; Heumann, R.; Nowock, J.; Auburger, G. Ataxin-2 modulates the levels of Grb2 and SRC but not ras signaling. J. Mol. Neurosci. 2013, 51, 68–81. [Google Scholar] [CrossRef]
- Lastres-Becker, I.; Nonis, D.; Nowock, J.; Auburger, G. New alternative splicing variants of the ATXN2 transcript. Neurol. Res. Pract. 2019, 1, 22. [Google Scholar] [CrossRef]
- Prudencio, M.; Belzil, V.V.; Batra, R.; Ross, C.A.; Gendron, T.F.; Pregent, L.J.; Murray, M.E.; Overstreet, K.K.; Piazza-Johnston, A.E.; Desaro, P.; et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 2015, 18, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- DeMille, D.; Badal, B.D.; Evans, J.B.; Mathis, A.D.; Anderson, J.F.; Grose, J.H. PAS kinase is activated by direct SNF1-dependent phosphorylation and mediates inhibition of TORC1 through the phosphorylation and activation of Pbp1. Mol. Biol. Cell 2015, 26, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Bar, D.Z.; Charar, C.; Dorfman, J.; Yadid, T.; Tafforeau, L.; Lafontaine, D.L.; Gruenbaum, Y. Cell size and fat content of dietary-restricted Caenorhabditis elegans are regulated by ATX-2, an mTOR repressor. Proc. Natl. Acad. Sci. USA 2016, 113, E4620–E4629. [Google Scholar] [CrossRef]
- Takahara, T.; Maeda, T. Transient sequestration of TORC1 into stress granules during heat stress. Mol. Cell 2012, 47, 242–252. [Google Scholar] [CrossRef]
- Meierhofer, D.; Halbach, M.; Sen, N.E.; Gispert, S.; Auburger, G. Ataxin-2 (Atxn2)-Knock-Out Mice Show Branched Chain Amino Acids and Fatty Acids Pathway Alterations. Mol. Cell. Proteom 2016, 15, 1728–1739. [Google Scholar] [CrossRef] [PubMed]
- Sen, N.E.; Drost, J.; Gispert, S.; Torres-Odio, S.; Damrath, E.; Klinkenberg, M.; Hamzeiy, H.; Akdal, G.; Gulluoglu, H.; Basak, A.N.; et al. Search for SCA2 blood RNA biomarkers highlights Ataxin-2 as strong modifier of the mitochondrial factor PINK1 levels. Neurobiol. Dis. 2016, 96, 115–126. [Google Scholar] [CrossRef]
- Seidel, G.; Meierhofer, D.; Sen, N.E.; Guenther, A.; Krobitsch, S.; Auburger, G. Quantitative Global Proteomics of Yeast PBP1 Deletion Mutants and Their Stress Responses Identifies Glucose Metabolism, Mitochondrial, and Stress Granule Changes. J. Proteome Res. 2017, 16, 504–515. [Google Scholar] [CrossRef]
- Wang, X.; Chen, X.J. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 2015, 524, 481–484. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Elden, A.C.; Kim, H.J.; Hart, M.P.; Chen-Plotkin, A.S.; Johnson, B.S.; Fang, X.; Armakola, M.; Geser, F.; Greene, R.; Lu, M.M.; et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 2010, 466, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Li, Y.R.; Ingre, C.; Weber, M.; Grehl, T.; Gredal, O.; de Carvalho, M.; Meyer, T.; Tysnes, O.B.; Auburger, G.; et al. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum. Mol. Genet. 2011, 20, 1697–1700. [Google Scholar] [CrossRef] [PubMed]
- Gispert, S.; Kurz, A.; Waibel, S.; Bauer, P.; Liepelt, I.; Geisen, C.; Gitler, A.D.; Becker, T.; Weber, M.; Berg, D.; et al. The modulation of Amyotrophic Lateral Sclerosis risk by ataxin-2 intermediate polyglutamine expansions is a specific effect. Neurobiol. Dis. 2012, 45, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Becker, L.A.; Huang, B.; Bieri, G.; Ma, R.; Knowles, D.A.; Jafar-Nejad, P.; Messing, J.; Kim, H.J.; Soriano, A.; Auburger, G.; et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 2017, 544, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Lahut, S.; Omur, O.; Uyan, O.; Agim, Z.S.; Ozoguz, A.; Parman, Y.; Deymeer, F.; Oflazer, P.; Koc, F.; Ozcelik, H.; et al. ATXN2 and its neighbouring gene SH2B3 are associated with increased ALS risk in the Turkish population. PLoS ONE 2012, 7, e42956. [Google Scholar] [CrossRef] [PubMed]
- Ross, O.A.; Rutherford, N.J.; Baker, M.; Soto-Ortolaza, A.I.; Carrasquillo, M.M.; DeJesus-Hernandez, M.; Adamson, J.; Li, M.; Volkening, K.; Finger, E.; et al. Ataxin-2 repeat-length variation and neurodegeneration. Hum. Mol. Genet. 2011, 20, 3207–3212. [Google Scholar] [CrossRef]
- Shulman, J.M.; Feany, M.B. Genetic modifiers of tauopathy in Drosophila. Genetics 2003, 165, 1233–1242. [Google Scholar]
- Scoles, D.R.; Meera, P.; Schneider, M.D.; Paul, S.; Dansithong, W.; Figueroa, K.P.; Hung, G.; Rigo, F.; Bennett, C.F.; Otis, T.S.; et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 2017, 544, 362–366. [Google Scholar] [CrossRef]
- Al-Ramahi, I.; Perez, A.M.; Lim, J.; Zhang, M.; Sorensen, R.; de Haro, M.; Branco, J.; Pulst, S.M.; Zoghbi, H.Y.; Botas, J. dAtaxin-2 mediates expanded Ataxin-1-induced neurodegeneration in a Drosophila model of SCA1. PLoS Genet. 2007, 3, e234. [Google Scholar] [CrossRef]
- Mangus, D.A.; Amrani, N.; Jacobson, A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 1998, 18, 7383–7396. [Google Scholar] [CrossRef]
- Lastres-Becker, I.; Brodesser, S.; Lutjohann, D.; Azizov, M.; Buchmann, J.; Hintermann, E.; Sandhoff, K.; Schurmann, A.; Nowock, J.; Auburger, G. Insulin receptor and lipid metabolism pathology in ataxin-2 knock-out mice. Hum. Mol. Genet. 2008, 17, 1465–1481. [Google Scholar] [CrossRef] [PubMed]
- Kiehl, T.R.; Nechiporuk, A.; Figueroa, K.P.; Keating, M.T.; Huynh, D.P.; Pulst, S.M. Generation and characterization of Sca2 (ataxin-2) knockout mice. Biochem. Biophys. Res. Commun. 2006, 339, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Lopez, D.; Guzman, P. Insights into the evolution and domain structure of Ataxin-2 proteins across eukaryotes. BMC Res. Notes 2014, 7, 453. [Google Scholar] [CrossRef]
- Kaehler, C.; Isensee, J.; Nonhoff, U.; Terrey, M.; Hucho, T.; Lehrach, H.; Krobitsch, S. Ataxin-2-like is a regulator of stress granules and processing bodies. PLoS ONE 2012, 7, e50134. [Google Scholar] [CrossRef] [PubMed]
- Kaehler, C.; Guenther, A.; Uhlich, A.; Krobitsch, S. PRMT1-mediated arginine methylation controls ATXN2L localization. Exp. Cell Res. 2015, 334, 114–125. [Google Scholar] [CrossRef]
- Meunier, C.; Bordereaux, D.; Porteu, F.; Gisselbrecht, S.; Chretien, S.; Courtois, G. Cloning and characterization of a family of proteins associated with Mpl. J. Biol. Chem. 2002, 277, 9139–9147. [Google Scholar] [CrossRef]
- Figueroa, K.P.; Pulst, S.M. Identification and expression of the gene for human ataxin-2-related protein on chromosome 16. Exp. Neurol. 2003, 184, 669–678. [Google Scholar] [CrossRef]
- Lin, L.; Li, X.; Pan, C.; Lin, W.; Shao, R.; Liu, Y.; Zhang, J.; Luo, Y.; Qian, K.; Shi, M.; et al. ATXN2L upregulated by epidermal growth factor promotes gastric cancer cell invasiveness and oxaliplatin resistance. Cell Death Dis. 2019, 10, 173. [Google Scholar] [CrossRef]
- STRING Web Platform. Available online: https://string-db.org/ (accessed on 17 July 2020).
- Lee, J.; Yoo, E.; Lee, H.; Park, K.; Hur, J.H.; Lim, C. LSM12 and ME31B/DDX6 Define Distinct Modes of Posttranscriptional Regulation by ATAXIN-2 Protein Complex in Drosophila Circadian Pacemaker Neurons. Mol. Cell 2017, 66, 129–140.e127. [Google Scholar] [CrossRef]
- Dougherty, J.D.; Tsai, W.C.; Lloyd, R.E. Multiple Poliovirus Proteins Repress Cytoplasmic RNA Granules. Viruses 2015, 7, 6127–6140. [Google Scholar] [CrossRef]
- Tajirika, T.; Tokumaru, Y.; Taniguchi, K.; Sugito, N.; Matsuhashi, N.; Futamura, M.; Yanagihara, K.; Akao, Y.; Yoshida, K. DEAD-Box Protein RNA-Helicase DDX6 Regulates the Expression of HER2 and FGFR2 at the Post-Transcriptional Step in Gastric Cancer Cells. Int. J. Mol. Sci. 2018, 19, 2005. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Zeddies, S.; Meinders, M.; di Summa, F.; van Alphen, F.P.J.; Hoogendijk, A.J.; Moore, K.S.; Halbach, M.; Gutierrez, L.; van den Biggelaar, M.; et al. The RNA-Binding Protein ATXN2 is Expressed during Megakaryopoiesis and May Control Timing of Gene Expression. Int. J. Mol. Sci. 2020, 21, 967. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; He, L.; Zhou, Q.; Wang, M.; Shen, W.-J.; Azhar, S.; Pan, F.; Guo, Z.; Hu, Z. Nherf1 and nherf2 regulation of sr-b1 stability via ubiquitination and proteasome degradation. Biochem. Biophys. Res. Commun. 2017, 490, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Hryciw, D.H.; Jenkin, K.A.; Simcocks, A.C.; Grinfeld, E.; McAinch, A.J.; Poronnik, P. The interaction between megalin and clc-5 is scaffolded by the na+–h+ exchanger regulatory factor 2 (nherf2) in proximal tubule cells. Int. J. Biochem. Cell Biol. 2012, 44, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Rayfield, A.; Hryciw, D.H.; Ma, T.A.; Wang, D.; Pow, D.; Broer, S.; Yun, C.; Poronnik, P. Na+–h+ exchanger regulatory factor 1 is a pdz scaffold for the astroglial glutamate transporter glast. Glia 2007, 55, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Filippov, A.K.; Simon, J.; Barnard, E.A.; Brown, D.A. The scaffold protein nherf2 determines the coupling of p2y1 nucleotide and mglur5 glutamate receptor to different ion channels in neurons. J. Neurosci. 2010, 30, 11068–11072. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Jeon, E.S.; Lee, H.J.; Oh, Y.-S.; Suh, P.-G.; Jung, J.S.; Donowitz, M.; Kim, J.H. Nherf2 increases platelet-derived growth factor-induced proliferation through pi-3-kinase/akt-, erk-, and src family kinase-dependent pathway. Cell. Signal. 2004, 16, 791–800. [Google Scholar] [CrossRef]
- Allen Brain Atlas. Available online: https://mouse.brain-map.org/ (accessed on 17 July 2020).
- Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 17 July 2020).
- Coexpedia Database. Available online: www.coexpedia.org (accessed on 17 July 2020).
- Sen, N.E.; Canet-Pons, J.; Halbach, M.V.; Arsovic, A.; Pilatus, U.; Chae, W.H.; Kaya, Z.E.; Seidel, K.; Rollmann, E.; Mittelbronn, M.; et al. Generation of an Atxn2-CAG100 knock-in mouse reveals N-acetylaspartate production deficit due to early Nat8l dysregulation. Neurobiol. Dis. 2019, 132, 104559. [Google Scholar] [CrossRef]
- Kiehl, T.R.; Shibata, H.; Pulst, S.M. The ortholog of human ataxin-2 is essential for early embryonic patterning in C. elegans. J. Mol. Neurosci. 2000, 15, 231–241. [Google Scholar] [CrossRef]
- Ciosk, R.; DePalma, M.; Priess, J.R. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development 2004, 131, 4831–4841. [Google Scholar] [CrossRef]
- Gross, N.; Strillacci, M.G.; Penagaricano, F.; Khatib, H. Characterization and functional roles of paternal RNAs in 2-4 cell bovine embryos. Sci. Rep. 2019, 9, 20347. [Google Scholar] [CrossRef] [PubMed]
- Flores, L.E.; Hildebrandt, T.B.; Kuhl, A.A.; Drews, B. Early detection and staging of spontaneous embryo resorption by ultrasound biomicroscopy in murine pregnancy. Reprod. Biol. Endocrinol. 2014, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, V.E.; Behringer, R.R. Early embryonic lethality in genetically engineered mice: Diagnosis and phenotypic analysis. Vet. Pathol. 2012, 49, 64–70. [Google Scholar] [CrossRef]
- Armit, C.; Richardson, L.; Hill, B.; Yang, Y.; Baldock, R.A. eMouseAtlas informatics: Embryo atlas and gene expression database. Mamm. Genome 2015, 26, 431–440. [Google Scholar] [CrossRef]
- Dhenain, M.; Ruffins, S.W.; Jacobs, R.E. Three-dimensional digital mouse atlas using high-resolution MRI. Dev. Biol. 2001, 232, 458–470. [Google Scholar] [CrossRef] [PubMed]
- Bard, J.L.; Kaufman, M.H.; Dubreuil, C.; Brune, R.M.; Burger, A.; Baldock, R.A.; Davidson, D.R. An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech. Dev. 1998, 74, 111–120. [Google Scholar] [CrossRef]
- Adams, D.; Baldock, R.; Bhattacharya, S.; Copp, A.J.; Dickinson, M.; Greene, N.D.; Henkelman, M.; Justice, M.; Mohun, T.; Murray, S.A.; et al. Bloomsbury report on mouse embryo phenotyping: Recommendations from the IMPC workshop on embryonic lethal screening. Dis. Models Mech. 2013, 6, 571–579. [Google Scholar] [CrossRef]
- Perez-Garcia, V.; Fineberg, E.; Wilson, R.; Murray, A.; Mazzeo, C.I.; Tudor, C.; Sienerth, A.; White, J.K.; Tuck, E.; Ryder, E.J.; et al. Placentation defects are highly prevalent in embryonic lethal mouse mutants. Nature 2018, 555, 463–468. [Google Scholar] [CrossRef]
- Lee, T.C.; Threadgill, D.W. Generation and validation of mice carrying a conditional allele of the epidermal growth factor receptor. Genesis 2009, 47, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Dackor, J.; Caron, K.M.; Threadgill, D.W. Placental and embryonic growth restriction in mice with reduced function epidermal growth factor receptor alleles. Genetics 2009, 183, 207–218. [Google Scholar] [CrossRef]
- Teves, M.E.; Modi, B.P.; Kulkarni, R.; Han, A.X.; Marks, J.S.; Subler, M.A.; Windle, J.; Newall, J.M.; McAllister, J.M.; Strauss, J.F., 3rd. Human DENND1A.V2 Drives Cyp17a1 Expression and Androgen Production in Mouse Ovaries and Adrenals. Int. J. Mol. Sci. 2020, 21, 2545. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, P.; Droce, A.; Moser, J.M.; Cuhlmann, S.; Padilla, C.O.; Heimann, P.; Bartsch, J.W.; Fuchtbauer, A.; Fuchtbauer, E.M.; Schmitt-John, T. Loss of vps54 function leads to vesicle traffic impairment, protein mis-sorting and embryonic lethality. Int. J. Mol. Sci. 2013, 14, 10908–10925. [Google Scholar] [CrossRef]
- Li, M.; Zhao, X.; Wang, W.; Shi, H.; Pan, Q.; Lu, Z.; Perez, S.P.; Suganthan, R.; He, C.; Bjoras, M.; et al. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018, 19, 69. [Google Scholar] [CrossRef] [PubMed]
- Bae, B.; Miura, P. Emerging Roles for 3’ UTRs in Neurons. Int. J. Mol. Sci. 2020, 21, 3413. [Google Scholar] [CrossRef] [PubMed]
- Pham, J.; Keon, M.; Brennan, S.; Saksena, N. Connecting RNA-Modifying Similarities of TDP-43, FUS, and SOD1 with MicroRNA Dysregulation Amidst A Renewed Network Perspective of Amyotrophic Lateral Sclerosis Proteinopathy. Int. J. Mol. Sci. 2020, 21, 3464. [Google Scholar] [CrossRef] [PubMed]
- Holt, D.J.; Grainger, D.W. Multinucleated giant cells from fibroblast cultures. Biomaterials 2011, 32, 3977–3987. [Google Scholar] [CrossRef]
- Atilla-Gokcumen, G.E.; Muro, E.; Relat-Goberna, J.; Sasse, S.; Bedigian, A.; Coughlin, M.L.; Garcia-Manyes, S.; Eggert, U.S. Dividing cells regulate their lipid composition and localization. Cell 2014, 156, 428–439. [Google Scholar] [CrossRef]
- Fernandez, C.; Lobo Md Mdel, V.; Gomez-Coronado, D.; Lasuncion, M.A. Cholesterol is essential for mitosis progression and its deficiency induces polyploid cell formation. Exp. Cell Res. 2004, 300, 109–120. [Google Scholar] [CrossRef]
- Sen, N.E.; Arsovic, A.; Meierhofer, D.; Brodesser, S.; Oberschmidt, C.; Canet-Pons, J.; Kaya, Z.E.; Halbach, M.V.; Gispert, S.; Sandhoff, K.; et al. In Human and Mouse Spino-Cerebellar Tissue, Ataxin-2 Expansion Affects Ceramide-Sphingomyelin Metabolism. Int. J. Mol. Sci. 2019, 20, 5854. [Google Scholar] [CrossRef]
- Ng, M.M.; Chang, F.; Burgess, D.R. Movement of membrane domains and requirement of membrane signaling molecules for cytokinesis. Dev. Cell 2005, 9, 781–790. [Google Scholar] [CrossRef]
- Kettle, E.; Page, S.L.; Morgan, G.P.; Malladi, C.S.; Wong, C.L.; Boadle, R.A.; Marsh, B.J.; Robinson, P.J.; Chircop, M. A Cholesterol-Dependent Endocytic Mechanism Generates Midbody Tubules During Cytokinesis. Traffic 2015, 16, 1174–1192. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Schwarz, H.; Jesuthasan, S. Furrow-specific endocytosis during cytokinesis of zebrafish blastomeres. Exp. Cell Res. 2002, 279, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Neuwald, A.F.; Koonin, E.V. Ataxin-2, global regulators of bacterial gene expression, and spliceosomal snRNP proteins share a conserved domain. J. Mol. Med. 1998, 76, 3–5. [Google Scholar] [CrossRef] [PubMed]
- NHGRI-EBI Catalog of Human Genome-Wide Association Studies. Available online: https://www.ebi.ac.uk/gwas/ (accessed on 22 November 2019).
- Hoffmann, T.J.; Choquet, H.; Yin, J.; Banda, Y.; Kvale, M.N.; Glymour, M.; Schaefer, C.; Risch, N.; Jorgenson, E. A Large Multiethnic Genome-Wide Association Study of Adult Body Mass Index Identifies Novel Loci. Genetics 2018, 210, 499–515. [Google Scholar] [CrossRef]
- Graff, M.; Scott, R.A.; Justice, A.E.; Young, K.L.; Feitosa, M.F.; Barata, L.; Winkler, T.W.; Chu, A.Y.; Mahajan, A.; Hadley, D.; et al. Genome-wide physical activity interactions in adiposity—A meta-analysis of 200,452 adults. PLoS Genet. 2017, 13, e1006528. [Google Scholar] [CrossRef]
- Tachmazidou, I.; Suveges, D.; Min, J.L.; Ritchie, G.R.S.; Steinberg, J.; Walter, K.; Iotchkova, V.; Schwartzentruber, J.; Huang, J.; Memari, Y.; et al. Whole-Genome Sequencing Coupled to Imputation Discovers Genetic Signals for Anthropometric Traits. Am. J. Hum. Genet. 2017, 100, 865–884. [Google Scholar] [CrossRef]
- Lee, J.J.; Wedow, R.; Okbay, A.; Kong, E.; Maghzian, O.; Zacher, M.; Nguyen-Viet, T.A.; Bowers, P.; Sidorenko, J.; Karlsson Linner, R.; et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018, 50, 1112–1121. [Google Scholar] [CrossRef]
- Tikkanen, E.; Gustafsson, S.; Amar, D.; Shcherbina, A.; Waggott, D.; Ashley, E.A.; Ingelsson, E. Biological Insights Into Muscular Strength: Genetic Findings in the UK Biobank. Sci. Rep. 2018, 8, 6451. [Google Scholar] [CrossRef]
- Hill, W.D.; Marioni, R.E.; Maghzian, O.; Ritchie, S.J.; Hagenaars, S.P.; McIntosh, A.M.; Gale, C.R.; Davies, G.; Deary, I.J. A combined analysis of genetically correlated traits identifies 187 loci and a role for neurogenesis and myelination in intelligence. Mol. Psychiatry 2019, 24, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.; Trampush, J.W.; Yu, J.; Knowles, E.; Davies, G.; Liewald, D.C.; Starr, J.M.; Djurovic, S.; Melle, I.; Sundet, K.; et al. Large-Scale Cognitive GWAS Meta-Analysis Reveals Tissue-Specific Neural Expression and Potential Nootropic Drug Targets. Cell Rep. 2017, 21, 2597–2613. [Google Scholar] [CrossRef]
- Sniekers, S.; Stringer, S.; Watanabe, K.; Jansen, P.R.; Coleman, J.R.I.; Krapohl, E.; Taskesen, E.; Hammerschlag, A.R.; Okbay, A.; Zabaneh, D.; et al. Genome-wide association meta-analysis of 78,308 individuals identifies new loci and genes influencing human intelligence. Nat. Genet. 2017, 49, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.R.I.; Bryois, J.; Gaspar, H.A.; Jansen, P.R.; Savage, J.E.; Skene, N.; Plomin, R.; Munoz-Manchado, A.B.; Linnarsson, S.; Crawford, G.; et al. Biological annotation of genetic loci associated with intelligence in a meta-analysis of 87,740 individuals. Mol. Psychiatry 2019, 24, 182–197. [Google Scholar] [CrossRef]
- Okbay, A.; Beauchamp, J.P.; Fontana, M.A.; Lee, J.J.; Pers, T.H.; Rietveld, C.A.; Turley, P.; Chen, G.B.; Emilsson, V.; Meddens, S.F.; et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 2016, 533, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rietveld, C.A.; Medland, S.E.; Derringer, J.; Yang, J.; Esko, T.; Martin, N.W.; Westra, H.J.; Shakhbazov, K.; Abdellaoui, A.; Agrawal, A.; et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 2013, 340, 1467–1471. [Google Scholar] [CrossRef] [PubMed]
- Kanai, M.; Akiyama, M.; Takahashi, A.; Matoba, N.; Momozawa, Y.; Ikeda, M.; Iwata, N.; Ikegawa, S.; Hirata, M.; Matsuda, K.; et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018, 50, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Imielinski, M.; Baldassano, R.N.; Griffiths, A.; Russell, R.K.; Annese, V.; Dubinsky, M.; Kugathasan, S.; Bradfield, J.P.; Walters, T.D.; Sleiman, P.; et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat. Genet. 2009, 41, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.R.; Li, J.; Zhao, S.D.; Bradfield, J.P.; Mentch, F.D.; Maggadottir, S.M.; Hou, C.; Abrams, D.J.; Chang, D.; Gao, F.; et al. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat. Med. 2015, 21, 1018–1027. [Google Scholar] [CrossRef]
- Wills, A.M.; Hubbard, J.; Macklin, E.A.; Glass, J.; Tandan, R.; Simpson, E.P.; Brooks, B.; Gelinas, D.; Mitsumoto, H.; Mozaffar, T.; et al. Hypercaloric enteral nutrition in patients with amyotrophic lateral sclerosis: A randomised, double-blind, placebo-controlled phase 2 trial. Lancet 2014, 383, 2065–2072. [Google Scholar] [CrossRef]
- Dorst, J.; Cypionka, J.; Ludolph, A.C. High-caloric food supplements in the treatment of amyotrophic lateral sclerosis: A prospective interventional study. Amyotroph. Lateral Scler. Front. Degener. 2013, 14, 533–536. [Google Scholar] [CrossRef]
- Genotype-Tissue-Expression Project Database. Available online: https://gtexportal.org/home/ (accessed on 17 July 2020).
- Almaguer-Mederos, L.E.; Aguilera-Rodriguez, R.; Almaguer-Gotay, D.; Hechavarria-Barzaga, K.; Alvarez-Sosa, A.; Chapman-Rodriguez, Y.; Silva-Ricardo, Y.; Gonzalez-Zaldivar, Y.; Vazquez-Mojena, Y.; Cuello-Almarales, D.; et al. Testosterone Levels Are Decreased and Associated with Disease Duration in Male Spinocerebellar Ataxia Type 2 Patients. Cerebellum 2020. [Google Scholar] [CrossRef]
- Klockgether, T.; Ludtke, R.; Kramer, B.; Abele, M.; Burk, K.; Schols, L.; Riess, O.; Laccone, F.; Boesch, S.; Lopes-Cendes, I.; et al. The natural history of degenerative ataxia: A retrospective study in 466 patients. Brain 1998, 121 Pt 4, 589–600. [Google Scholar] [CrossRef]
- War, A.R.; Dang, K.; Jiang, S.; Xiao, Z.; Miao, Z.; Yang, T.; Li, Y.; Qian, A. Role of cancer stem cells in the development of giant cell tumor of bone. Cancer Cell Int. 2020, 20, 135. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, Y.; Lim, C.X.; Weichhart, T.; Valeyre, D.; Bentaher, A.; Calender, A. Sarcoidosis and the mTOR, Rac1, and Autophagy Triad. Trends Immunol. 2020, 41, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Feliciano, D.M.; Su, T.; Lopez, J.; Platel, J.C.; Bordey, A. Single-cell Tsc1 knockout during corticogenesis generates tuber-like lesions and reduces seizure threshold in mice. J. Clin. Investig. 2011, 121, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Key, J.; Mueller, A.K.; Gispert, S.; Matschke, L.; Wittig, I.; Corti, O.; Munch, C.; Decher, N.; Auburger, G. Ubiquitylome profiling of Parkin-null brain reveals dysregulation of calcium homeostasis factors ATP1A2, Hippocalcin and GNA11, reflected by altered firing of noradrenergic neurons. Neurobiol. Dis. 2019, 127, 114–130. [Google Scholar] [CrossRef] [PubMed]
- Tunster, S.J. Genetic sex determination of mice by simplex PCR. Biol. Sex Differ. 2017, 8, 31. [Google Scholar] [CrossRef]
- Damrath, E.; Heck, M.V.; Gispert, S.; Azizov, M.; Nowock, J.; Seifried, C.; Rub, U.; Walter, M.; Auburger, G. ATXN2-CAG42 sequesters PABPC1 into insolubility and induces FBXW8 in cerebellum of old ataxic knock-in mice. PLoS Genet. 2012, 8, e1002920. [Google Scholar] [CrossRef]
- Key, J.; Kohli, A.; Barcena, C.; Lopez-Otin, C.; Heidler, J.; Wittig, I.; Auburger, G. Global Proteome of LonP1(+/-) Mouse Embryonal Fibroblasts Reveals Impact on Respiratory Chain, but No Interdependence between Eral1 and Mitoribosomes. Int. J. Mol. Sci. 2019, 20, 4523. [Google Scholar] [CrossRef]
- Torres-Odio, S.; Key, J.; Hoepken, H.H.; Canet-Pons, J.; Valek, L.; Roller, B.; Walter, M.; Morales-Gordo, B.; Meierhofer, D.; Harter, P.N.; et al. Progression of pathology in PINK1-deficient mouse brain from splicing via ubiquitination, ER stress, and mitophagy changes to neuroinflammation. J. Neuroinflamm. 2017, 14, 154. [Google Scholar] [CrossRef]
- Gispert, S.; Parganlija, D.; Klinkenberg, M.; Drose, S.; Wittig, I.; Mittelbronn, M.; Grzmil, P.; Koob, S.; Hamann, A.; Walter, M.; et al. Loss of mitochondrial peptidase Clpp leads to infertility, hearing loss plus growth retardation via accumulation of CLPX, mtDNA and inflammatory factors. Hum. Mol. Genet. 2013, 22, 4871–4887. [Google Scholar] [CrossRef]


| Observed/Expected Number of Live Born Mice with Indicated Genotype | ||||
| +/+ | +/− | −/− | Number of offspring | |
| Live born female | 50/55 | 77/110 | 0/55 | 127/220 |
| Live born male | 60/55 | 83/110 | 0/55 | 143/220 |
| Live born total | 110/110 | 160/220 | 0/110 | 270/440 |
| Observed/Expected Number of Mice with Indicated Genotype | ||||
| +/+ | +/− | −/− | Number of offspring | |
| Dissected embryos female | 48/39 | 57/78 | 5/39 | 110/156 |
| Dissected embryos male | 30/39 | 71/78 | 16/39 | 117/156 |
| Dissected embryos total | 78/78 | 128/156 | 21/78 | 227/312 |
| Weight of Embryos [mg] | |||
|---|---|---|---|
| +/+ | +/− | −/− | |
| Day E14–15 | 322 | 306 | 226 |
| Day E15–16 | 447 | 444 | 313 |
| Day E20 | 1472 | 1481 | 372 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Key, J.; Harter, P.N.; Sen, N.-E.; Gradhand, E.; Auburger, G.; Gispert, S. Mid-Gestation lethality of Atxn2l-Ablated Mice. Int. J. Mol. Sci. 2020, 21, 5124. https://doi.org/10.3390/ijms21145124
Key J, Harter PN, Sen N-E, Gradhand E, Auburger G, Gispert S. Mid-Gestation lethality of Atxn2l-Ablated Mice. International Journal of Molecular Sciences. 2020; 21(14):5124. https://doi.org/10.3390/ijms21145124
Chicago/Turabian StyleKey, Jana, Patrick N. Harter, Nesli-Ece Sen, Elise Gradhand, Georg Auburger, and Suzana Gispert. 2020. "Mid-Gestation lethality of Atxn2l-Ablated Mice" International Journal of Molecular Sciences 21, no. 14: 5124. https://doi.org/10.3390/ijms21145124
APA StyleKey, J., Harter, P. N., Sen, N.-E., Gradhand, E., Auburger, G., & Gispert, S. (2020). Mid-Gestation lethality of Atxn2l-Ablated Mice. International Journal of Molecular Sciences, 21(14), 5124. https://doi.org/10.3390/ijms21145124




