Bovine Organospecific Microvascular Endothelial Cell Lines as New and Relevant In Vitro Models to Study Viral Infections
Abstract
:1. Introduction
2. Results
2.1. Establishment of Bovine Endothelial Cell Lines
2.2. Characterization of Bovine Endothelial Cell Lines
2.2.1. Angiogenesis Assays
2.2.2. Identification of RNA Endothelial Cell Markers by RT-qPCR
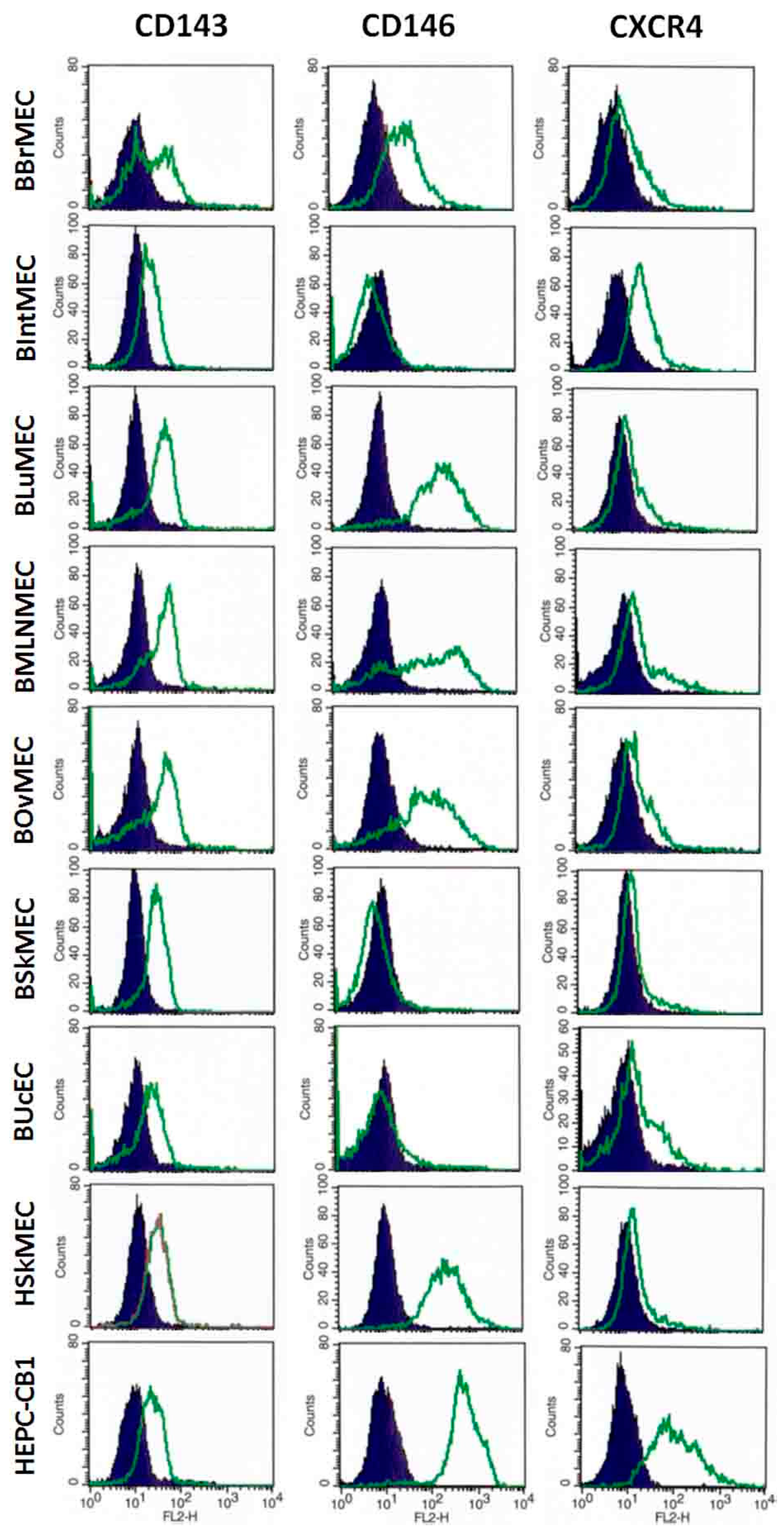
2.2.3. Identification of Endothelial Cell Markers by Flow Cytometry
2.2.4. Detection of VEGFA by ELISA
2.3. Permissive Assay to Viral Infection by Two Viruses
2.4. Ability of Endothelial Cell Lines to Express the NS3 Viral Protein upon DNA Transfection
3. Discussion
4. Materials and Methods
4.1. Endothelial Cells Used as Control Cells
4.2. Establishment of Bovine Endothelial Cell Lines
4.3. Characterization of Bovine Endothelial Cell Lines
4.3.1. Angiogenesis Assays
4.3.2. Identification of Endothelial Cell Markers by RT-qPCR
4.3.3. Identification of Endothelial Cell Markers by Flow Cytometry
4.3.4. Measurement of VEGFA by ELISA
4.4. Viral Infections of Bovine Endothelial Cells
4.4.1. BTV Infections
4.4.2. FMDV Infections
4.5. BTV8-NS3 Transfection Assay
5. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACE | Angiotensin-converting enzyme |
| APC | Allophycocyanin |
| BAEC | Bovine aortic primary endothelial cells |
| BBrMEC | Bovine brain microvascular endothelial cells |
| BIntMEC | Bovine intestine microvascular endothelial cells |
| BLuMEC | Bovine lung microvascular endothelial cells |
| BK | Buffalo kidney |
| BMLNMEC | Bovine mesenteric lymph node microvascular endothelial cells |
| BOvMEC | Bovine ovary microvascular endothelial cells |
| BSA | Bovine serum albumin |
| BSkMEC | Bovine skin microvascular endothelial cells |
| BT | Bluetongue |
| BTV | Bluetongue virus |
| BUcEC | Bovine umbilical cord endothelial cells |
| CPAE | Calf pulmonary artery endothelial cells |
| CD | Cluster of differentiation |
| CPE | Cytopathic effect |
| CXCR4 | Chemokine receptor type 4 |
| DMEM | Dulbecco’s modified Eagle’s medium |
| EC | Endothelial cells |
| EDTA | Ethylenediaminetetraacetic acid |
| ELISA | Enzyme-linked immunosorbent assay |
| eNOS | Endothelial nitric oxide synthase |
| FBS | Fetal bovine serum |
| FITC | Fluorescein isothiocyanate |
| FMD | Foot-and-mouth disease |
| FMDV | Foot-and-mouth disease virus |
| GFP | Green fluorescent protein |
| GUSB | Beta-glucuronidase |
| HEPC-CB1 | Human endothelial progenitor cell line–cord blood |
| hpi | Hours post-infection |
| HPRT1 | Hypoxanthine-guanine phosphoribosyltransferase |
| HSkMEC | Human skin microvascular endothelial cells |
| HUVEC | Human umbilical vein endothelial cell |
| IMDM | Iscove’s modified Dulbecco’s media |
| KDR | Kinase insert domain receptor |
| KIT | Proto-oncogene c-Kit |
| LYVE-1 | Endothelial hyaluronan receptor 1 |
| MCAM | Melanoma cell adhesion molecule |
| MDBK | Madin-Darby bovine kidney |
| MOI | Multiplicity of infection |
| OptiMEM | Improved Minimal Essential Medium |
| PBS | Phosphate-buffered saline |
| PCR | Polymerase chain reaction |
| PE | Phycoerythrin |
| PECAM1 | Platelet endothelial cell adhesion molecule |
| PFA | Paraformaldehyde |
| PFU | Plaque forming unit |
| PPIA | Peptidylprolyl isomerase A |
| PROM1 | Prominin-1 |
| RNA | Ribonucleic Acid |
| SD | Standard deviation |
| VEGFA | Vascular endothelial growth factor A |
| VEGFR | Vascular endothelial growth factor receptor |
| vWF | von Willebrand factor |
References
- Sahni, A.; Narra, H.P.; Patel, J.; Sahni, S.K. MicroRNA-Regulated Rickettsial Invasion into Host Endothelium via Fibroblast Growth Factor 2 and Its Receptor FGFR1. Cells 2018, 7, 240. [Google Scholar] [CrossRef] [Green Version]
- Bazzoni, G.; Dejana, E. Endothelial Cell-to-Cell Junctions: Molecular Organization and Role in Vascular Homeostasis. Physiol. Rev. 2004, 84, 869–901. [Google Scholar] [CrossRef] [Green Version]
- Mehta, D.; Ravindran, K.; Kuebler, W.M. Novel regulators of endothelial barrier function. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2014, 307, L924–L935. [Google Scholar] [CrossRef] [Green Version]
- Cai, Z.; Zhang, Y.; Zhang, Y.; Miao, X.; Li, S.; Yang, H.; Ling, Q.; Hoffmann, P.R.; Huang, Z. Use of a Mouse Model and Human Umbilical Vein Endothelial Cells to Investigate the Effect of Arsenic Exposure on Vascular Endothelial Function and the Associated Role of Calpains. Environ. Health Perspect. 2019, 127, 077003. [Google Scholar] [CrossRef] [PubMed]
- Talavera-Adame, D.; Ng, T.T.; Gupta, A.; Kurtovic, S.; Wu, G.D.; Dafoe, D.C. Characterization of microvascular endothelial cells isolated from the dermis of adult mouse tails. Microvasc. Res. 2011, 82, 97–104. [Google Scholar] [CrossRef]
- Munderloh, U.G.; Lynch, M.J.; Herron, M.J.; Palmer, A.T.; Kurtti, T.J.; Nelson, R.D.; Goodman, J.L. Infection of endothelial cells with Anaplasma marginale and A. phagocytophilum. Vet. Microbiol. 2004, 101, 53–64. [Google Scholar] [CrossRef]
- Khaiboullina, S.; Uppal, T.; Kletenkov, K.; St. Jeor, S.C.; Garanina, E.; Rizvanov, A.; Verma, S.C. Transcriptome Profiling Reveals Pro-Inflammatory Cytokines and Matrix Metalloproteinase Activation in Zika Virus Infected Human Umbilical Vein Endothelial Cells. Front. Pharmacol. 2019, 10, 642. [Google Scholar] [CrossRef]
- Stevens, L.M.; Moffat, K.; Cooke, L.; Nomikou, K.; Mertens, P.P.C.; Jackson, T.; Darpel, K.E. A low-passage insect-cell isolate of bluetongue virus uses a macropinocytosis-like entry pathway to infect natural target cells derived from the bovine host. J. Gen. Virol. 2019, 100, 568–582. [Google Scholar] [CrossRef] [PubMed]
- Drew, C.P.; Gardner, I.A.; Mayo, C.E.; Matsuo, E.; Roy, P.; MacLachlan, N.J. Bluetongue virus infection alters the impedance of monolayers of bovine endothelial cells as a result of cell death. Vet. Immunol. Immunopathol. 2010, 136, 108–115. [Google Scholar] [CrossRef] [Green Version]
- Darpel, K.E.; Monaghan, P.; Simpson, J.; Veronesi, E.; Brooks, H.W.; Brownlie, J.; Takamatsu, H.-H.; Mellor, P.S.; Mertens, P.P. Involvement of the skin during bluetongue virus infection and replication in the ruminant host. Vet. Res. 2012, 43, 40. [Google Scholar] [CrossRef] [Green Version]
- Beatty, P.R.; Puerta-Guardo, H.; Killingbeck, S.S.; Glasner, D.R.; Hopkins, K.; Harris, E. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci. Transl. Med. 2015, 7, 304ra141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allsopp, B.A. Natural history of Ehrlichia ruminantium. Vet. Parasitol. 2010, 167, 123–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hägglund, S.; Laloy, E.; Näslund, K.; Pfaff, F.; Eschbaumer, M.; Romey, A.; Relmy, A.; Rikberg, A.; Svensson, A.; Huet, H.; et al. Model of persistent foot-and-mouth disease virus infection in multilayered cells derived from bovine dorsal soft palate. Transbound. Emerg. Dis. 2020, 67, 133–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arzt, J.; Pacheco, J.M.; Rodriguez, L.L. The early pathogenesis of foot-and-mouth disease in cattle after aerosol inoculation. Identification of the nasopharynx as the primary site of infection. Vet. Pathol. 2010, 47, 1048–1063. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Tian, L.; Hu, G.; Teng, K.; Mu, X. Microvascular endothelial cells play potential immunoregulatory roles in the immune response to foot-and-mouth disease vaccines. Cell Biochem. Funct. 2011, 29, 394–399. [Google Scholar] [CrossRef]
- Crola Da Silva, C.; Lamerant-Fayel, N.; Paprocka, M.; Mitterrand, M.; Gosset, D.; Dus, D.; Kieda, C. Selective human endothelial cell activation by chemokines as a guide to cell homing. Immunology 2009, 126, 394–404. [Google Scholar] [CrossRef]
- Kieda, C.; Paprocka, M.; Krawczenko, A.; Załecki, P.; Dupuis, P.; Monsigny, M.; Radzikowski, C.; Duś, D. New human microvascular endothelial cell lines with specific adhesion molecules phenotypes. Endothel. J. Endothel. Cell Res. 2002, 9, 247–261. [Google Scholar] [CrossRef]
- Bizouarne, N.; Denis, V.; Legrand, A.; Monsigny, M.; Kieda, C. A SV-40 immortalized murine endothelial cell line from peripheral lymph node high endothelium expresses a new α-L-fucose binding protein. Biol. Cell 1993, 79, 209–218. [Google Scholar] [CrossRef]
- Berrich, M.; Kieda, C.; Grillon, C.; Monteil, M.; Lamerant, N.; Gavard, J.; Boulouis, H.J.; Haddad, N. Differential Effects of Bartonella henselae on Human and Feline Macro- and Micro-Vascular Endothelial Cells. PLoS ONE 2011, 6, e20204. [Google Scholar] [CrossRef]
- Zweygarth, E.; Vogel, S.W.; Josemans, A.I.; Horn, E. In vitro isolation and cultivation of Cowdria ruminantium under serum-free culture conditions. Res. Vet. Sci. 1997, 63, 161–164. [Google Scholar] [CrossRef]
- Marcelino, I. Characterization of Ehrlichia ruminantium replication and release kinetics in endothelial cell cultures. Vet. Microbiol. 2005, 110, 87–96. [Google Scholar] [CrossRef]
- Pruneau, L.; Emboulé, L.; Gely, P.; Marcelino, I.; Mari, B.; Pinarello, V.; Sheikboudou, C.; Martinez, D.; Daigle, F.; Lefrançois, T.; et al. Global gene expression profiling of Ehrlichia ruminantium at different stages of development. FEMS Immunol. Med. Microbiol. 2012, 64, 66–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craig, L.E.; Nealen, M.L.; Strandberg, J.D.; Zink, M.C. Differential Replication of Ovine Lentivirus in Endothelial Cells Cultured from Different Tissues. Virology 1997, 238, 316–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gospodarowicz, D.; Mescher, A.L.; Birdwell, C.R. Stimulation of corneal endothelial cell proliferation in vitro by fibroblast and epidermal growth factors. Exp. Eye Res. 1977, 25, 75–89. [Google Scholar] [CrossRef]
- Stins, M.F.; Prasadarao, N.V.; Zhou, J.; Arditi, M.; Kim, K.S. Bovine brain microvascular endothelial cells transfected with SV40-large T antigen: Development of an immortalized cell line to study pathophysiology of CNS disease. In Vitro Cell. Dev. Biol. Anim. 1997, 33, 243–247. [Google Scholar] [CrossRef]
- Sobue, K.; Yamamoto, N.; Yoneda, K.; Hodgson, M.E.; Yamashiro, K.; Tsuruoka, N.; Tsuda, T.; Katsuya, H.; Miura, Y.; Asai, K.; et al. Induction of blood–brain barrier properties in immortalized bovine brain endothelial cells by astrocytic factors. Neurosci. Res. 1999, 35, 155–164. [Google Scholar] [CrossRef]
- Buser, R.; Montesano, R.; Garcia, I.; Dupraz, P.; Pepper, M.S. Bovine microvascular endothelial cells immortalized with human telomerase. J. Cell. Biochem. 2006, 98, 267–286. [Google Scholar] [CrossRef]
- Korzekwa, A.J.; Bodek, G.; Bukowska, J.; Blitek, A.; Skarzynski, D.J. Characterization of bovine immortalized luteal endothelial cells: Action of cytokines on production and content of arachidonic acid metabolites. Reprod. Biol. Endocrinol. RBE 2011, 9, 27. [Google Scholar] [CrossRef] [Green Version]
- Wechsler, S.J.; McHolland, L.E. Susceptibilities of 14 cell lines to bluetongue virus infection. J. Clin. Microbiol. 1988, 26, 2324–2327. [Google Scholar] [CrossRef] [Green Version]
- Kopliku, L.; Relmy, A.; Romey, A.; Gorna, K.; Zientara, S.; Bakkali-Kassimi, L.; Blaise-Boisseau, S. Establishment of persistent foot-and-mouth disease virus (FMDV) infection in MDBK cells. Arch. Virol. 2015, 160, 2503–2516. [Google Scholar] [CrossRef]
- Schwartz, B.; Vicart, P.; Delouis, C.; Paulin, D. Mammalian cell lines can be efficiently established in vitro upon expression of the SV40 large T antigen driven by a promoter sequence derived from the human vimentin gene. Biol. Cell 1991, 73, 7–14. [Google Scholar] [CrossRef]
- Yeager, T.R.; Reddel, R.R. Constructing immortalized human cell lines. Curr. Opin. Biotechnol. 1999, 10, 465–469. [Google Scholar] [CrossRef]
- Paprocka, M.; Krawczenko, A.; Dus, D.; Kantor, A.; Carreau, A.; Grillon, C.; Kieda, C. CD133 positive progenitor endothelial cell lines from human cord blood. Cytometry A 2011, 79A, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Gordon, E.J.; Gale, N.W.; Harvey, N.L. Expression of the hyaluronan receptor LYVE-1 is not restricted to the lymphatic vasculature; LYVE-1 is also expressed on embryonic blood vessels. Dev. Dyn. 2008, 237, 1901–1909. [Google Scholar] [CrossRef] [PubMed]
- Klimkiewicz, K.; Weglarczyk, K.; Collet, G.; Paprocka, M.; Guichard, A.; Sarna, M.; Jozkowicz, A.; Dulak, J.; Sarna, T.; Grillon, C.; et al. A 3D model of tumour angiogenic microenvironment to monitor hypoxia effects on cell interactions and cancer stem cell selection. Cancer Lett. 2017, 396, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Collet, G.; Szade, K.; Nowak, W.; Klimkiewicz, K.; El Hafny-Rahbi, B.; Szczepanek, K.; Sugiyama, D.; Weglarczyk, K.; Foucault-Collet, A.; Guichard, A.; et al. Endothelial precursor cell-based therapy to target the pathologic angiogenesis and compensate tumor hypoxia. Cancer Lett. 2016, 370, 345–357. [Google Scholar] [CrossRef]
- Chopra, H.; Hung, M.K.; Kwong, D.L.; Zhang, C.F.; Pow, E.H.N. Insights into Endothelial Progenitor Cells: Origin, Classification, Potentials, and Prospects. Stem Cells Int. 2018, 2018, 9847015. [Google Scholar] [CrossRef]
- Russell, H.; O’Toole, D.T.; Bardsley, K.; Davis, W.C.; Ellis, J.A. Comparative Effects of Bluetongue Virus Infection of Ovine and Bovine Endothelial Cells. Vet. Pathol. 1996, 33, 319–331. [Google Scholar] [CrossRef]
- Drew, C.P.; Heller, M.C.; Mayo, C.; Watson, J.L.; MacLachlan, N.J. Bluetongue virus infection activates bovine monocyte-derived macrophages and pulmonary artery endothelial cells. Vet. Immunol. Immunopathol. 2010, 136, 292–296. [Google Scholar] [CrossRef] [Green Version]
- DeMaula, C.D.; Jutila, M.A.; Wilson, D.W.; MacLachlan, N.J. Infection kinetics, prostacyclin release and cytokine-mediated modulation of the mechanism of cell death during bluetongue virus infection of cultured ovine and bovine pulmonary artery and lung microvascular endothelial cells. J. Gen. Virol. 2001, 82, 787–794. [Google Scholar] [CrossRef] [Green Version]
- DeMaula, C.D.; Leutenegger, C.M.; Bonneau, K.R.; MacLachlan, N.J. The Role of Endothelial Cell-Derived Inflammatory and Vasoactive Mediators in the Pathogenesis of Bluetongue. Virology 2002, 296, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeMaula, C.D.; Leutenegger, C.M.; Jutila, M.A.; MacLachlan, N.J. Bluetongue virus-induced activation of primary bovine lung microvascular endothelial cells. Vet. Immunol. Immunopathol. 2002, 86, 147–157. [Google Scholar] [CrossRef]
- McLaughlin, B.E.; DeMaula, C.D.; Wilson, W.C.; Boyce, W.M.; MacLachlan, N.J. Replication of bluetongue virus and epizootic hemorrhagic disease virus in pulmonary artery endothelial cells obtained from cattle, sheep, and deer. Am. J. Vet. Res. 2003, 64, 860–865. [Google Scholar] [CrossRef] [PubMed]
- Howerth, E.W. Cytokine release and endothelial dysfunction: A perfect storm in orbivirus pathogenesis. Vet. Ital. 2015, 51, 275–281. [Google Scholar] [PubMed]
- Maree, F.; De Klerk-Lorist, L.-M.; Gubbins, S.; Zhang, F.; Seago, J.; Pérez-Martín, E.; Reid, L.; Scott, K.; Van Schalkwyk, L.; Bengis, R.; et al. Differential Persistence of Foot-and-Mouth Disease Virus in African Buffalo Is Related to Virus Virulence. J. Virol. 2016, 90, 5132–5140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brehm, K.E.; Ferris, N.P.; Lenk, M.; Riebe, R.; Haas, B. Highly sensitive fetal goat tongue cell line for detection and isolation of foot-and-mouth disease virus. J. Clin. Microbiol. 2009, 47, 3156–3160. [Google Scholar] [CrossRef] [Green Version]
- Arzt, J.; Baxt, B.; Grubman, M.J.; Jackson, T.; Juleff, N.; Rhyan, J.; Rieder, E.; Waters, R.; Rodriguez, L.L. The pathogenesis of foot-and-mouth disease II: Viral pathways in swine, small ruminants, and wildlife; myotropism, chronic syndromes, and molecular virus-host interactions. Transbound. Emerg. Dis. 2011, 58, 305–326. [Google Scholar] [CrossRef]
- Afonso, P.V.; Ozden, S.; Cumont, M.-C.; Seilhean, D.; Cartier, L.; Rezaie, P.; Mason, S.; Lambert, S.; Huerre, M.; Gessain, A.; et al. Alteration of Blood–Brain Barrier Integrity by Retroviral Infection. PLoS Pathog. 2008, 4, e1000205. [Google Scholar] [CrossRef]
- Carreau, A.; Kieda, C.; Grillon, C. Nitric oxide modulates the expression of endothelial cell adhesion molecules involved in angiogenesis and leukocyte recruitment. Exp. Cell Res. 2011, 317, 29–41. [Google Scholar] [CrossRef]
- Le Gal, M.C.; Dufour, B.; Geoffroy, E.; Zanella, G.; Moutou, F.; Millemann, Y.; Rieffel, J.N.; Pouilly, F. Bluetongue virus serotype 8 in the Ardennes in 2007. Vet. Rec. 2008, 163, 668. [Google Scholar] [CrossRef]





| Organs | Cell Line Names | Meaning | Doubling Time (h) |
|---|---|---|---|
| Brain | BBrMEC | Bovine brain microvascular endothelial cells | 48 |
| Intestine | BIntMEC | Bovine intestine microvascular endothelial cells | 30 |
| Lung | BLuMEC | Bovine lung microvascular endothelial cells | 36 |
| Mesenteric lymph node | BMLNMEC | Bovine mesenteric lymph node microvascular endothelial cells | 24 |
| Ovary | BOvMEC | Bovine ovary microvascular endothelial cells | 30 |
| Skin | BSkMEC | Bovine skin microvascular endothelial cells | 24 |
| Umbilical cord | BUcEC | Bovine umbilical cord endothelial cells | 36 |
| Markers | Normalized Relative Expression | BAEC | BBrMEC | BIntMEC | BLuMEC | BMLNMEC | BOvMEC | BSkMEC | BUcEC |
|---|---|---|---|---|---|---|---|---|---|
| Specific markers for mature endothelial cells | |||||||||
| CD31-PECAM1 | mean | 2641.39 | 0.14 | 0.26 | 0.04 | 0.04 | 0.03 | 2.41 | 0.40 |
| SD | 770.92 | 0.06 | 0.11 | 0.01 | 0.001 | 0.003 | 0.06 | 0.04 | |
| CD143-ACE | mean | 23.42 | 2.23 | 0.74 | 2.46 | 0.75 | 0.05 | 0.24 | 0.19 |
| SD | 8.35 | 1.55 | 0.32 | 0.03 | 0.29 | 0.01 | 0.06 | 0.07 | |
| CD146-MCAM | mean | 5.55 | 1.61 | 0.81 | 2.24 | 6.77 | 0.66 | 0.07 | 0.16 |
| SD | 0.44 | 0.57 | 0.51 | 0.20 | 1.63 | 0.04 | 0.01 | 0.01 | |
| VEGFA | mean | 0.08 | 0.79 | 3.70 | 1.58 | 1.05 | 1.18 | 0.13 | 0.68 |
| SD | 0.01 | 0.27 | 1.79 | 0.16 | 0.35 | 0.06 | 0.01 | 0.04 | |
| VEGFR-1-Flt1 | mean | 0.002 | 0.86 | 0.09 | 0.81 | 1.17 | 0.03 | 1.42 | 0.47 |
| SD | 0.0009 | 0.18 | 0.07 | 0.09 | 0.25 | 0.003 | 0.02 | 0.01 | |
| VEGFR-2-KDR-CD309 | mean | 163.16 | 14.32 | 0.04 | 0.11 | 0.92 | 0.01 | 0.01 | 0.65 |
| SD | 45.71 | 2.33 | 0.03 | 0.04 | 0.24 | 0.01 | 0.003 | 0.09 | |
| vWF | mean | 33.09 | 1.39 | 2.08 | 0.13 | 0.21 | 0.12 | 0.06 | 0.22 |
| SD | 26.91 | 1.67 | 1.11 | 0.03 | 0.06 | 0.11 | 0.01 | 0.07 | |
| Specific markers for lymphatic endothelial cells | |||||||||
| LYVE-1 | mean | 0.002 | 36.40 | 37.31 | 495.99 | 2.55 | 2 461.72 | 1.92 | 1.08 |
| SD | 0.001 | 32.36 | 23.17 | 36.94 | 0.51 | 547.28 | 0.17 | 0.12 | |
| Specific markers for progenitor endothelial cells | |||||||||
| CD117-KIT | mean | 0.04 | 19.08 | 0.0002 | 0.01 | 0.001 | 0.0003 | 0.0003 | 0.0001 |
| SD | 0.03 | 6.32 | 0.00004 | 0.01 | 0.0001 | 0.0001 | 0.0001 | ¤ | |
| CD133-PROM1 | mean | 0.01 | 1.72 | 2.13 | 0.12 | 4.18 | 0.41 | 0.71 | 0.82 |
| SD | 0.001 | 0.92 | 1.29 | 0.01 | 1.19 | 0.02 | 0.08 | 0.09 | |
| CXCR4-CD184 | mean | 172.99 | 1.37 | 1.97 | 0.40 | 1.49 | 0.02 | 0.18 | 2.23 |
| SD | 121.75 | 0.86 | 0.22 | 0.01 | 0.08 | 0.004 | 0.01 | 0.28 | |
| Specific marker for non-endothelial cells (hematopoietic cells) | |||||||||
| CD38 | mean | 0.008 | 0.22 | 4.60 | 0.05 | 0.03 | 0.01 | 0.66 | 0.02 |
| SD | 0.006 | 0.12 | 4.57 | 0.01 | 0.01 | 0.01 | 0.05 | 0.0004 | |
| Relative Fluorescence Intensity (Arbitrary Units) | BBrMEC | BIntMEC | BLuMEC | BMLNMEC | BOvMEC | BSkMEC | BUcEC | BAEC | HEPC-CB1 | HSkMEC |
|---|---|---|---|---|---|---|---|---|---|---|
| Specific markers for mature endothelial cells | ||||||||||
| CD31 (bovine) | <1 | <1 | <1 | <1 | <1 | <1 | <1 | 4 | <1 | <1 |
| CD143/ACE | 20 | 9 | 25 | 25 | 30 | 17 | 10 | 1 | 13 | 14 |
| CD146/MCAM | 44 | <1 | 291 | 278 | 172 | <1 | 2 | 1006 | 540 | 265 |
| CD105/endoglin | nt | nt | <1 | nt | nt | 3 | 1 | 5 | 190 | 358 |
| eNOS | nt | nt | 14 | nt | nt | 7 | 7 | 451 | nt | 108 |
| Specific markers for progenitor endothelial cells | ||||||||||
| CD133 | 3 | 4 | <1 | 7 | 2 | 2 | <1 | 1 | 190 | 1 |
| CD271 | 5 | 5 | <1 | < 1 | 3 | 20 | <1 | 5 | 525 | 4 |
| CXCR4/CD184 | 14 | 29 | 13 | 27 | 12 | 8 | 14 | 9 | 154 | 12 |
| Specific markers for non-endothelial cells (hematopoietic cells) | ||||||||||
| CD34 | nt | nt | 10 | nt | nt | 3 | 3 | 3 | 2 | 2 |
| CD45 | nt | nt | 3 | nt | nt | 2 | 2 | 3 | 2 | 2 |
| Inoculum | ZZ-R127 | BIntMEC | BLuMEC | BMLNMEC | BSkMEC | BUcEC |
|---|---|---|---|---|---|---|
| FMDV 10-3 MOI 20 hpi | CPE +++ | Neg | CPE + | CPE +++ | Neg | Neg |
| FMDV 10-5 MOI 20 hpi | Neg | Neg | Neg | CPE+ | Neg | Neg |
| FMDV 10-3 MOI 48 hpi | CPE +++ | Doubtful | CPE +++ | CPE +++ | Neg | CPE +++ |
| FMDV 10-5 MOI 48 hpi | CPE +++ | Neg | Neg | CPE+++ | Neg | CPE +++ |
| Primer Names | Associated Genes | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|---|
| gusb | Beta-glucuronidase | CTGGTTACTACTTCAAGACG | CTGCTTCATAGTTGGTGTTG |
| hprt1 | Hypoxanthine-guanine phosphoribosyltransferase | ATCCATTCCTATGACTGTGG | ACTTTTATGTCGCCTGTTG |
| ppia | Peptidylprolyl isomerase A | AAGACTGAGTGGTTGGATG | GTCAGCAATGGTGATCTTC |
| cd31 | Platelet endothelial cell adhesion molecule | GAAGACATTATCGGATGCC | TTAATGGCTTCATTGCATGG |
| cd143 | Angiotensin-converting enzyme | GAAATGAAACCCACTTTGAC | TCACGAAGTACCTGATATACG |
| cd146 | melanoma cell adhesion molecule | CTGGTTTTCTGTCCACAAG | CAGAGTAGTCCCTTTGTCC |
| vegfa | Vascular endothelial growth factor A | GCTGTAATGACGAAAGTCTG | GGAAGCTCATCTCTCCTATG |
| vegfr1 | Vascular endothelial growth factor receptor 1 | CAACCACAAAATACAGCAAG | GTGACTCTCTCGATAAACAG |
| vegfr2 | Vascular endothelial growth factor receptor 2 | TGATGAGGAATTTTGTAGGC | ATGGTCTGGTACATTTCTGG |
| vwf | von Willebrand Factor | CAGACACTTCAACAAGACC | TTCCTTGAGTCCTGAAGTC |
| lyve-1 | Lymphatic vessel endothelial hyaluronan receptor 1 | TACTGCCACAACTCATCTG | GTTGAATAAGGGATCATCGG |
| cd117 | proto-oncogene c-Kit | TAGTTCCGTGGACTCTATG | GATGCCAGCTATTCTTCTTC |
| cd133 | prominin-1 | CGACAGAAGAAAAGTGGTC | CGATGCTTATGAACACACAG |
| cxcr4 | chemokine receptor type 4 | GATCCGTATATTCACTTCCG | AAGATGATGGAGTAGACAGTG |
| cd38 | cyclic ADP ribose hydrolase | TTCATGAGTGCCTTCATTTC | TTTTCGCGTATTCATGAGC |
| Primary Antibody | Corresponding Isotype | Secondary Antibody | |||
|---|---|---|---|---|---|
| Target | Reference and Provider | Isotype | Reference and Provider | Antibody | Reference and Provider |
| CD31 (bovine cross reactivity) | MA3100, Invitrogen (Cergy Pontoise, France) | Mouse IgG (serum) | I5381, Sigma-Aldrich (Saint Quentin Fallavier, France) | Goat anti-mouse IgG-PE | Sc-3738, Santa Cruz (Heidelberg, Germany) |
| CD31(ovine) | MCA1097GA, Bio-Rad (Marnes-la-Coquette, France) | Mouse IgG (serum) | I5381, Sigma-Aldrich (Saint Quentin Fallavier, France) | Goat anti-mouse IgG-PE | Sc-3738, Santa Cruz (Heidelberg, Germany) |
| Mouse IgG2a | MCA1210 Bio-Rad (Marnes-la-Coquette, France) | ||||
| CD31-APC | 130-110-808, Miltenyi Biotec (Paris, France) | Mouse IgG1-APC | 555751, BD Biosciences (Le Pont de Claix, France) | N.A. | N.A. |
| CD143-PE | FAB929P, R&D Systems (Minneapolis, MN, USA) | Mouse IgG1-PE | IC002P, R&D Systems (Minneapolis, MN, USA) | N.A. | N.A. |
| CD146-PE | FAB932P, R&D Systems (Minneapolis, MN, USA) | Mouse IgG1-PE | IC002P, R&D Systems (Minneapolis, MN, USA) | N.A. | N.A. |
| CD133-PE | 130-090-853, Miltenyi Biotec (Paris, France) | Mouse IgG2b-PE | 130-092-215, Miltenyi Biotec (Paris, France) | N.A. | N.A. |
| CD271-FITC | 130-091-917, Miltenyi Biotec (Paris, France) | Mouse IgG1-FITC | 130-092-213, Miltenyi Biotec (Paris, France) | N.A. | N.A. |
| CXCR4-PE | FAB170P, R&D Systems (Minneapolis, MN, USA) | Mouse IgG2a-PE | IC003P, R&D Systems (Minneapolis, MN, USA) | N.A. | N.A. |
| CD34-FITC | 130-081-001, Miltenyi Biotec (Paris, France) | Mouse IgG2a-FITC | 130-091-837, Miltenyi Biotec (Paris, France) | N.A. | N.A. |
| CD45-FITC | 130-080-202, Miltenyi Biotec (Paris, France) | Mouse IgG2a-FITC | 130-091-837, Miltenyi Biotec (Paris, France) | N.A. | N.A. |
| CD105-PE | FAB10971P, R&D Systems (UK) | Mouse IgG1-PE | IC002P, R&D Systems (UK) | N.A. | N.A. |
| eNOS | 160880, Cayman chemical (Hamburg, Germany) | Rabbit IgG (serum) | I5006, Sigma-Aldrich (Saint Quentin Fallavier, France) | Goat anti-rabbit IgG-PE | Sc-3739, Santa Cruz (Heidelberg, Germany) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lagrée, A.-C.; Fasani, F.; Rouxel, C.; Pivet, M.; Pourcelot, M.; Fablet, A.; Romey, A.; Caignard, G.; Vitour, D.; Blaise-Boisseau, S.; et al. Bovine Organospecific Microvascular Endothelial Cell Lines as New and Relevant In Vitro Models to Study Viral Infections. Int. J. Mol. Sci. 2020, 21, 5249. https://doi.org/10.3390/ijms21155249
Lagrée A-C, Fasani F, Rouxel C, Pivet M, Pourcelot M, Fablet A, Romey A, Caignard G, Vitour D, Blaise-Boisseau S, et al. Bovine Organospecific Microvascular Endothelial Cell Lines as New and Relevant In Vitro Models to Study Viral Infections. International Journal of Molecular Sciences. 2020; 21(15):5249. https://doi.org/10.3390/ijms21155249
Chicago/Turabian StyleLagrée, Anne-Claire, Fabienne Fasani, Clotilde Rouxel, Marine Pivet, Marie Pourcelot, Aurore Fablet, Aurore Romey, Grégory Caignard, Damien Vitour, Sandra Blaise-Boisseau, and et al. 2020. "Bovine Organospecific Microvascular Endothelial Cell Lines as New and Relevant In Vitro Models to Study Viral Infections" International Journal of Molecular Sciences 21, no. 15: 5249. https://doi.org/10.3390/ijms21155249
APA StyleLagrée, A.-C., Fasani, F., Rouxel, C., Pivet, M., Pourcelot, M., Fablet, A., Romey, A., Caignard, G., Vitour, D., Blaise-Boisseau, S., Kieda, C., Boulouis, H.-J., Haddad, N., & Grillon, C. (2020). Bovine Organospecific Microvascular Endothelial Cell Lines as New and Relevant In Vitro Models to Study Viral Infections. International Journal of Molecular Sciences, 21(15), 5249. https://doi.org/10.3390/ijms21155249





