Transcriptional Inhibition of Sp-IAG by Crustacean Female Sex Hormone in the Mud Crab, Scylla paramamosain
Abstract
:1. Introduction
2. Results
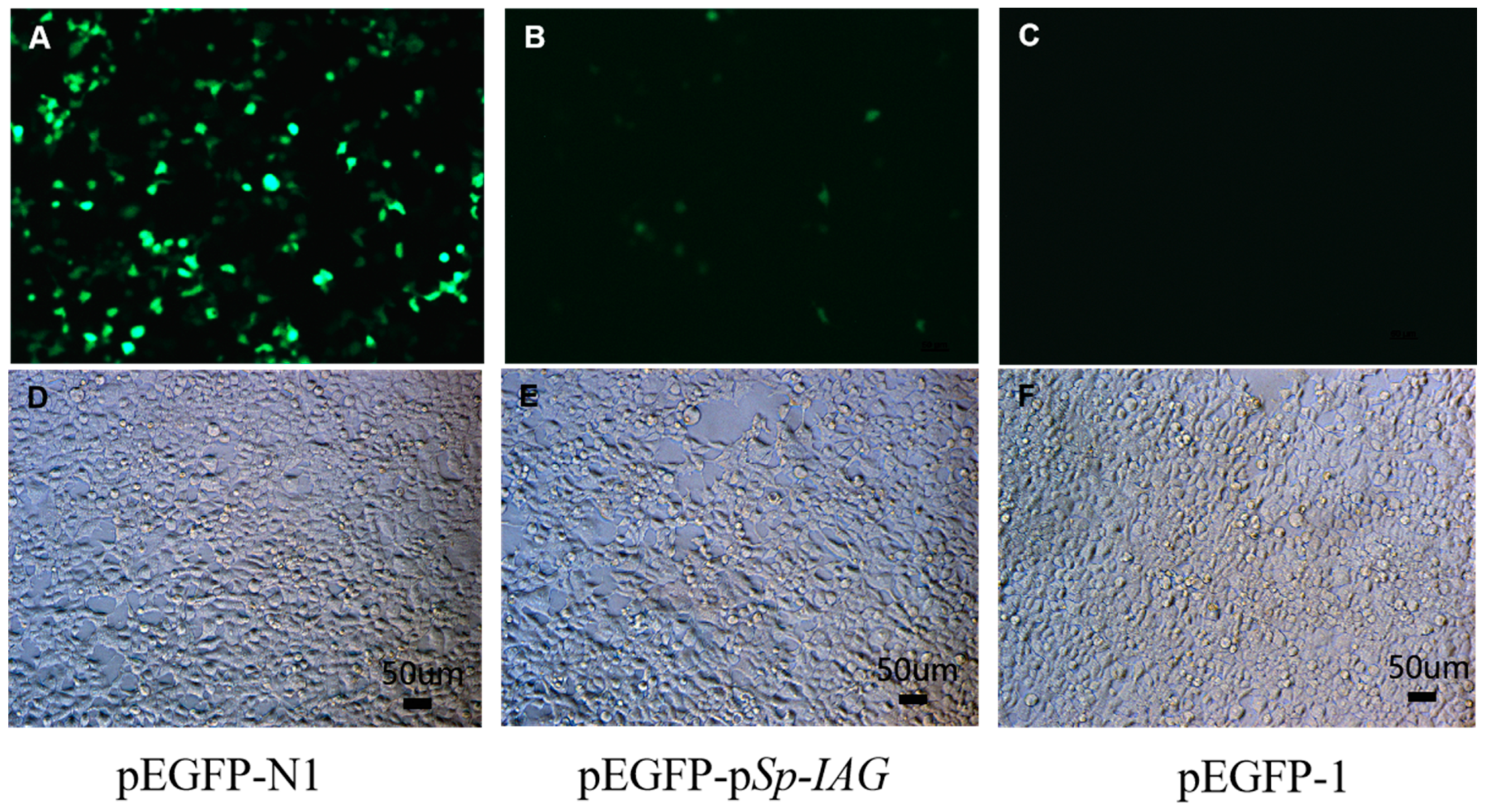
2.1. Sp-IAG Promoter Activity
2.2. Analysis of Promoter Activity by Serial Deletion of 5′-Flanking Region
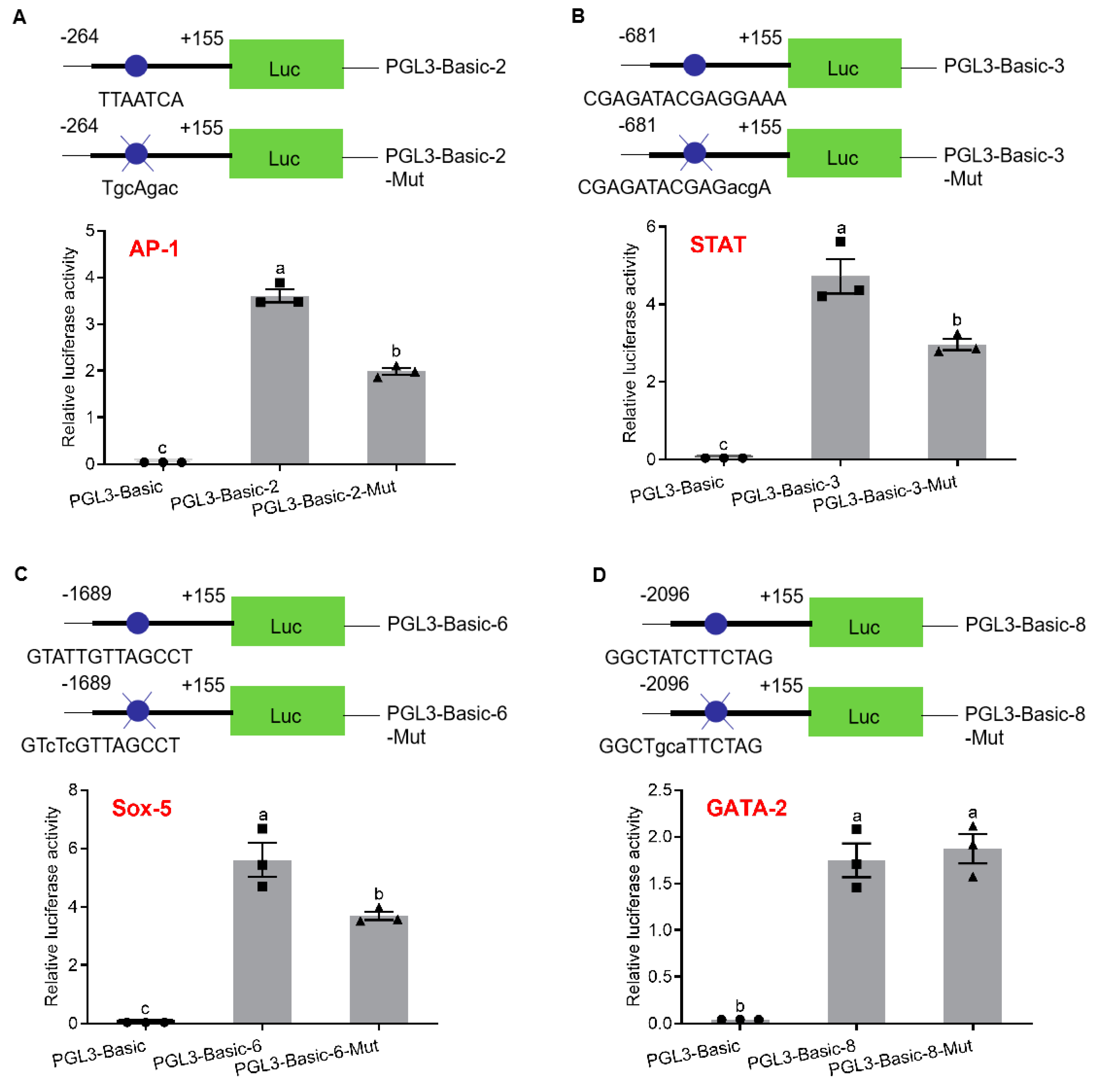
2.3. Analysis of Promoter Activity by Site-Directed Mutation (SDM)
2.4. Analysis of Gene Expression in AG Treated by rCFSH In Vitro
2.5. Knockdown of Sp-STAT Reducing Expression of Sp-IAG
3. Discussion
4. Materials and Methods
4.1. Animal Sources
4.2. Transcriptional Activity Analysis of Sp-IAG Promoter
4.3. Analysis of Promoter Activity by Serial Deletion of 5′ Region Constructs
4.4. Analysis of the Promoter Activity of the 5′ Region Flanking Sp-IAG with Site-Directed Mutagenesis (SDM)
4.5. Production of Recombinant CFSH
4.6. Analysis of Gene Expression in AG Treated by rCFSH in Vitro
4.7. Sp-STAT Knockdown in Vitro
4.8. Quantification of Gene Expression
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CFSH | crustacean female sex hormone |
| IAG | insulin-like androgenic hormone |
| SDM | site-directed mutation |
| STAT | signal transducers and activators of transcription |
| AP-1 | activator protein 1 |
| RNAi | RNA interfere |
| EGFP | enhanced green fluorescent protein |
| rCFSH | recombinant CFSH |
| AG | androgenic gland |
| AGH | androgenic gland hormone |
| IL-17 | interleukin-17 |
References
- Li, J.; Yu, H.; Wang, W.; Fu, C.; Zhang, W.; Han, F.; Wu, H. Genomic and transcriptomic insights into molecular basis of sexually dimorphic nuptial spines in Leptobrachium leishanense. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Q.; Lu, B.; Lin, D.; Huang, H.; Chen, X.; Ye, H. Role of crustacean female sex hormone (CFSH) in sex differentiation in early juvenile mud crabs, Scylla paramamosain. Gen. Comp. Endocrinol. 2020, 289, 113383. [Google Scholar] [CrossRef] [PubMed]
- Charniaux-Cotton, H. Arthropoda-Crustacea: Sexual differentiation. Reprod. Biol. Invert. 1992, 5, 281–323. [Google Scholar]
- Charniaux-Cotton, H. Discovery in, an amphipod crustacean (Orchestia gammarella) of an endocrine gland responsible for the differentiation of primary and secondary male sex characteristics. Cr. Acad. Bulg. Sci. 1954, 239, 780–782. [Google Scholar]
- Okuno, A.; Hasegawa, Y.; Ohira, T.; Katakura, Y.; Nagasawa, H. Characterization and cDNA Cloning of Androgenic Gland Hormone of the Terrestrial Isopod Armadillidium vulgare. Biochem. Biophys. Res. Commun. 1999, 264, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Khalaila, I.; Katz, T.; Abdu, U.; Yehezkel, G.; Sagi, A. Effects of Implantation of Hypertrophied Androgenic Glands on Sexual Characters and Physiology of the Reproductive System in the Female Red Claw Crayfish, Cherax quadricarinatus. Gen. Comp. Endocrinol. 2001, 121, 242–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagi, A.; Khalaila, I. The crustacean androgen: A hormone in an isopod and androgenic activity in decapods. Am. Zool. 2001, 41, 477–484. [Google Scholar] [CrossRef]
- Barki, A.; Karplus, I.; Khalaila, I.; Manor, R.; Sagi, A. Male-like behavioral patterns and physiological alterations induced by androgenic gland implantation in female crayfish. J. Exp. Boil. 2003, 206, 1791–1797. [Google Scholar] [CrossRef] [Green Version]
- Manor, R.; Aflalo, E.; Segall, C.; Weil, S.; Azulay, D.; Ventura, T.; Sagi, A. Androgenic gland implantation promotes growth and inhibits vitellogenesis in Cherax quadricarinatus females held in individual compartments. Invertebr. Reprod. Dev. 2004, 45, 151–159. [Google Scholar] [CrossRef]
- Rosen, O.; Manor, R.; Weil, S.; Gafni, O.; Linial, A.; Aflalo, E.; Ventura, T.; Sagi, A. A Sexual Shift Induced by Silencing of a Single Insulin-Like Gene in Crayfish: Ovarian Upregulation and Testicular Degeneration. PLoS ONE 2010, 5, e15281. [Google Scholar] [CrossRef] [Green Version]
- Ventura, T.; Rosen, O.; Sagi, A. From the discovery of the crustacean androgenic gland to the insulin-like hormone in six decades. Gen. Comp. Endocrinol. 2011, 173, 381–388. [Google Scholar] [CrossRef]
- Sagi, A.; Snir, E.; Khalaila, I. Sexual differentiation in decapod crustaceans: Role of the androgenic gland. Invertebr. Reprod. Dev. 1997, 31, 55–61. [Google Scholar] [CrossRef]
- Ventura, T.; Manor, R.; Aflalo, E.; Weil, S.; Raviv, S.; Glazer, L.; Sagi, A. Temporal Silencing of an Androgenic Gland-Specific Insulin-Like Gene Affecting Phenotypical Gender Differences and Spermatogenesis. Endocrinology 2009, 150, 1278–1286. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Ye, H.; Huang, H.; Yang, Y.; Gong, J. An insulin-like androgenic gland hormone gene in the mud crab, Scylla paramamosain, extensively expressed and involved in the processes of growth and female reproduction. Gen. Comp. Endocrinol. 2014, 204, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qiao, K.; Wang, S.; Peng, H.; Shan, Z.; Wang, K. Molecular identification of a new androgenic gland-specific insulin-like gene from the mud crab, Scylla paramamosain. Aquac. 2014, 433, 325–334. [Google Scholar] [CrossRef]
- Liu, A.; Liu, J.; Liu, F.; Huang, Y.; Wang, G.; Ye, H.-H. Crustacean Female Sex Hormone from the Mud Crab Scylla paramamosain is Highly Expressed in Prepubertal Males and Inhibits the Development of Androgenic Gland. Front. Physiol. 2018, 9, 924. [Google Scholar] [CrossRef] [Green Version]
- Denny, P.; Swift, S.; Brand, N.; Dabhade, N.; Barton, P.; Ashworth, A. A conserved family of genes related to the testis determining gene, SRY. Nucleic Acids Res. 1992, 20, 2887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uwanogho, D.; Rex, M.; Cartwright, E.J.; Pearl, G.; Healy, C.; Scotting, P.J.; Sharpe, P.T. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech. Dev. 1995, 49, 23–36. [Google Scholar] [CrossRef]
- Clarkson, M.; Harley, V.R. Sex with two SOX on: SRY and SOX9 in testis development. Trends Endocrinol. Metab. 2002, 13, 106–111. [Google Scholar] [CrossRef]
- Tevosian, S.G.; Albrecht, K.; Crispino, J.D.; Fujiwara, Y.; Eicher, E.M.; Orkin, S.H. Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development 2002, 129, 4627–4634. [Google Scholar]
- Robert, N.M.; Tremblay, J.J.; Viger, R.S. Friend of GATA (FOG)-1 and FOG-2 Differentially Repress the GATA-Dependent Activity of Multiple Gonadal Promoters. Endocrinology 2002, 143, 3963–3973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tremblay, J.J.; Viger, R.S. Novel roles for GATA transcription factors in the regulation of steroidogenesis. J. Steroid Biochem. Mol. Boil. 2003, 85, 291–298. [Google Scholar] [CrossRef]
- Viger, R.S.; Taniguchi, H.; Robert, N.M.; Tremblay, J.J.; And, N.M.R. The 25th Volume: Role of the GATA Family of Transcription Factors in Andrology. J. Androl. 2004, 25, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Chung, J.S. A Novel Hormone Is Required for the Development of Reproductive Phenotypes in Adult Female Crabs. Endocrinology 2014, 155, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Benderdour, M.; Tardif, G.; Pelletier, J.-P.; Di Battista, J.A.; Reboul, P.; Ranger, P.; Martel-Pelletier, J. Interleukin 17 (IL-17) induces collagenase-3 production in human osteoarthritic chondrocytes via AP-1 dependent activation: Differential activation of AP-1 members by IL-17 and IL-1beta. J. Rheumatol. 2002, 29, 1262–1272. [Google Scholar] [PubMed]
- Gao, C.; Liu, W.; Wang, X.R.; Liu, X.Z.; Zhao, S.G.; Fu, S.B. IL-17 stimulates migration of carotid artery vascular smooth muscle cells in an MMP-9 dependent manner via p38 MAPK and ERK1/2-dependent NF-κB and AP-1 activation. Cell. Mol. Neurobiol. 2009, 29, 1161–1168. [Google Scholar]
- Wu, J.; Guo, J.; Cao, Q.; Wang, Y.; Chen, J.; Wang, Z.; Yuan, Z. Autophagy impacts on oxaliplatin-induced hepatocarcinoma apoptosis via the IL-17/IL-17R-JAK2/STAT3 signaling pathway. Oncol. Lett. 2017, 13, 770–776. [Google Scholar] [CrossRef] [Green Version]
- Du, S.; Li, Z.; Xie, X.; Xu, C.; Shen, X.; Wang, N.; Shen, Y. IL-17 stimulates the expression of CCL2 in cardiac myocytes via Act1/TRAF6/p38MAPK dependent AP-1 activation. Scand. J. Immunol. 2020, 91, e12840. [Google Scholar] [CrossRef]
- Khalaila, I.; Manor, R.; Weil, S.; Granot, Y.; Keller, R.; Sagi, A. The eyestalk–androgenic gland–testis endocrine axis in the crayfish Cherax quadricarinatus. Gen. Comp. Endocrinol. 2002, 127, 147–156. [Google Scholar] [CrossRef]
- Sroyraya, M.; Chotwiwatthanakun, C.; Stewart, M.J.; Soonklang, N.; Kornthong, N.; Phoungpetchara, I.; Hanna, P.; Sobhon, P. Bilateral eyestalk ablation of the blue swimmer crab, Portunus pelagicus, produces hypertrophy of the androgenic gland and an increase of cells producing insulin-like androgenic gland hormone. Tissue Cell 2010, 42, 293–300. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, D.; Jia, X.; Zou, Z.; Wang, Y.; Zhang, Z. Functional analysis of the promoter of the molt-inhibiting hormone (mih) gene in mud crab Scylla paramamosain. Gen. Comp. Endocrinol. 2018, 259, 131–140. [Google Scholar] [CrossRef]
- Liu, C.Y.; Jia, X.W.; Zou, Z.H.; Wang, X.W.; Wang, Y.L.; Zhang, Z.P. VIH from the mud crab is specifically expressed in the eyestalk and potentially regulated by transactivator of Sox9/Oct4/Oct1. Gen. Comp. Endocr. 2018, 255, 1–11. [Google Scholar] [CrossRef]
- Wallis, M.C.; Waters, P.D.; Graves, J.M. Sex determination in mammals—Before and after the evolution of SRY. Cell. Mol. Life Sci. 2008, 65, 3182–3195. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Qiu, G.; Feng, J.; Li, J. Transcriptome Analysis of the Oriental River Prawn, Macrobrachium nipponense Using 454 Pyrosequencing for Discovery of Genes and Markers. PLoS ONE 2012, 7, e39727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, S.; Zhang, T.; Zhou, X.; Zhao, Y.; Li, Q.; Guo, Y.; Cheng, H.; Zhou, R. Molecular cloning, expression ofSox5 and its down-regulation ofDmrt1 transcription in Zebrafish. J. Exp. Zoöl. Part B Mol. Dev. Evol. 2005, 304, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Schartl, M.; Schories, S.; Wakamatsu, Y.; Nagao, Y.; Hashimoto, H.; Bertin, C.; Mourot, B.; Schmidt, C.; Wilhelm, D.; Centanin, L.; et al. Sox5 is involved in germ-cell regulation and sex determination in medaka following co-option of nested transposable elements. BMC Boil. 2018, 16, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Li, F.; Sun, Z.; Xiang, J. Two spliced variants of insulin-like androgenic gland hormone gene in the Chinese shrimp, Fenneropenaeus chinensis. Gen. Comp. Endocrinol. 2012, 177, 246–255. [Google Scholar] [CrossRef]
- Shabgah, A.G.; Fattahi, E.; Shahneh, F.Z. Interleukin-17 in human inflammatory diseases. Adv. Dermatol. Allergol. 2014, 31, 256–261. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Monin, L.; Castillo, P.; Elsegeiny, W.; Horne, W.; Eddens, T.; Vikram, A.; Good, M.; Schoenborn, A.A.; Bibby, K.; et al. Intestinal Interleukin-17 Receptor Signaling Mediates Reciprocal Control of the Gut Microbiota and Autoimmune Inflammation. Immunity 2016, 44, 659–671. [Google Scholar] [CrossRef] [Green Version]
- Subramaniam, S.; Cooper, R.S.; Adunyah, S.E. Evidence for the Involvement of JAK/STAT Pathway in the Signaling Mechanism of Interleukin-17. Biochem. Biophys. Res. Commun. 1999, 262, 14–19. [Google Scholar] [CrossRef]
- You, T.; Bi, Y.; Zhang, M.; Chen, X.; Zhang, K.; Li, J. IL-17 induces reactive astrocytes and up-regulation of vascular endothelial growth factor (VEGF) through JAK/STAT signaling. Sci. Rep. 2017, 7, 41779. [Google Scholar] [CrossRef] [PubMed]
- Jochum, W.; Passegué, E.; Wagner, E.F. AP-1 in mouse development and tumorigenesis. Oncogene 2001, 20, 2401–2412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grötsch, B.; Brachs, S.; Lang, C.; Luther, J.; Derer, A.; Schlötzer-Schrehardt, U.; Bozec, A.; Fillatreau, S.; Berberich, I.; Hobeika, E.; et al. The AP-1 transcription factor Fra1 inhibits follicular B cell differentiation into plasma cells. J. Exp. Med. 2014, 211, 2199–2212. [Google Scholar] [CrossRef]
- Li, J.K.; Nie, L.; Zhao, Y.P.; Zhang, Y.Q.; Wang, X.; Wang, S.S.; Liu, Y.; Zhao, H.; Cheng, L. IL-17 mediates inflammatory reactions via p38/c-Fos and JNK/c-Jun activation in an AP-1-dependent manner in human nucleus pulposus cells. J. Transl. Med. 2016, 14, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, S.; Minton, A.Z.; Ma, H.-Y.; Stankowska, D.L.; Sun, X.; Krishnamoorthy, R.R. Involvement of AP-1 and C/EBPβ in Upregulation of Endothelin B (ETB) Receptor Expression in a Rodent Model of Glaucoma. PLoS ONE 2013, 8, e79183. [Google Scholar] [CrossRef] [Green Version]
- Baksa, K.; Parke, T.; Dobens, L.L.; Dearolf, C.R. The Drosophila STAT Protein, Stat92E, Regulates Follicle Cell Differentiation during Oogenesis. Dev. Boil. 2002, 243, 166–175. [Google Scholar] [CrossRef] [Green Version]
- Arbouzova, N.I.; Zeidler, M.P. JAK/STAT signalling in Drosophila: Insights into conserved regulatory and cellular functions. Dev. 2006, 133, 2605–2616. [Google Scholar] [CrossRef] [Green Version]
- Li, W.X. Canonical and non-canonical JAK–STAT signaling. Trends Cell Boil. 2008, 18, 545–551. [Google Scholar] [CrossRef] [Green Version]
- Katsuyama, T.; Comoglio, F.; Seimiya, M.; Cabuy, E.; Paro, R. During Drosophila disc regeneration, JAK/STAT coordinates cell proliferation with Dilp8-mediated developmental delay. Proc. Natl. Acad. Sci. USA 2015, 112, E2327–E2336. [Google Scholar] [CrossRef] [Green Version]
- Kiger, A.A.; Tulina, N.; Matunis, E. Stem Cell Self-Renewal Specified by JAK-STAT Activation in Response to a Support Cell Cue. Science. 2001, 294, 2542–2545. [Google Scholar] [CrossRef]
- Tulina, N. Control of Stem Cell Self-Renewal in Drosophila Spermatogenesis by JAK-STAT Signaling. Science 2001, 294, 2546–2549. [Google Scholar] [CrossRef]
- Li, J.; Xia, F.; Li, W.X. Coactivation of STAT and Ras Is Required for Germ Cell Proliferation and Invasive Migration in Drosophila. Dev. Cell 2003, 5, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.; Zeidler, M.P.; Hombría, J.C.-G. JAK/STAT signalling in Drosophila controls cell motility during germ cell migration. Dev. Dyn. 2006, 235, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Zhang, W.; Li, J.; Li, J.; Hu, L.; Yan, W.; Liu, S.; He, J.; Weng, S. A signal transducers and activators of transcription (STAT) gene from Scylla paramamosain is involved in resistance against mud crab reovirus. Fish Shellfish. Immunol. 2019, 94, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Ye, H.; Xie, Y.; Yang, Y.; Huang, H.; Li, S.; Zeng, C. Ecdysone receptor in the mud crab Scylla paramamosain: A possible role in promoting ovarian development. J. Endocrinol. 2015, 224, 273–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Ye, H.; Feng, B.; Huang, H. Insights into insulin-like peptide system in invertebrates from studies on IGF binding domain-containing proteins in the female mud crab, Scylla paramamosain. Mol. Cell. Endocrinol. 2015, 416, 36–45. [Google Scholar] [CrossRef]
- Yang, Y.N.; Shu, L.; Jiang, Q.L.; Huang, H.Y.; Ye, H.H. Does the bone morphogenetic protein 7 inhibit oocyte maturation by autocrine/paracrine in mud crab? Gen. Comp. Endocr. 2018, 266, 119–125. [Google Scholar] [CrossRef]



| Name | Sequence (5′→ 3′) | Application |
|---|---|---|
| SpIAG-PF | cggGGTACCTCCAGCTCTTAGGATGCGTCCA | 5′-flanking region sequence |
| SpIAG-PR | cgcGGATCCAGGGAGGCCAAAGAAAACGTAG | |
| pspIAG-1F | cggGGTACC CTGAAACTTGTCAACATGCGGAG | serial deletion of 5′-flanking region |
| pspIAG-2F | cggGGTACC CTCTAAGTAAAAATTAGCAAAACAGACG | |
| pspIAG-3F | cggGGTACC GTTCCTGCCTCGTCATATCGC | |
| pspIAG-4F | cggGGTACCACAAATAAAGCGTGTATTCTGTGGTA | |
| pspIAG-5F | cggGGTACC CCGCAGCCAGAACCAATCT | |
| pspIAG-6F | cggGGTACC TGACTTGAACTGTTTCAAGGGAGAG | |
| pspIAG-7F | cggGGTACC TGATTCTAGCGGCGGACTGT | |
| pspIAG-8F | cggGGTACCCACTAGACCTTTTGGTGCTCGC | |
| pspIAG-R | ccgCTCGAGAGGGAGGCCAAAGAAAACGTAG | |
| Sp-AP-F-mut | GTAGAGGTGTGCAGACAGCCCTGACCGAGC | SDM |
| Sp-AP-R-mut | GCTCGGTCAGGGCTGTCTGCACACCTCTAC | |
| Sp-Sox-F-mut | CAGCCGAATACAGGTCTCGTTAGCCTAACC | |
| Sp-Sox-R-mut | GGTTAGGCTAACGAGACCTGTATTCGGCTG | |
| Sp-GATA-F-mut | GGGTTGTGTGTCTGGCTGCATTCTAGTAAG | |
| Sp-GATA-R-mut | CTTACTAGAATGCAGCCAGACACACAACCC | |
| Sp-STAT-F-mut | CACGCAGTCGAGATACGAGACGACCGCTGA | |
| Sp-STAT-R-mut | TCAGCGGTCGTCTCGTATCTCGACTGCGTG | |
| Sp-STATF | CACCAGATCAAGGAGTGTGAGCGACA | qPCR |
| Sp-STATR | GGTGACAAGTGAGGACAGCAAGCGA | |
| Sp-IAGF | ATCCTTTTCCTCCGTTTGCC | |
| Sp-IAGR | TCGGGTCTTCGTCTTGTTCC | |
| Sp-cfshF | CGTGTCCAGCATTTCTTGCAGTACC | |
| Sp-cfshR | TCATGTGTCCTATGATGGAGGAACG | |
| Sp-akF | TTCCTCCACCCTGTCCAACC | |
| Sp-akR | GAAGCGGTCACCCTCCTTGA | |
| Sp-actinF | CACACTTCACAGACCTTC | |
| Sp-actinR | CACAATGCCATCCTCTAC | |
| Sp-STAT-dsF | CTTGGTGCTCCACACACAACTAAT | Sp-STAT knockdown |
| Sp-STAT-dsR | CCATGTGGGGTTATTGGTATCTT | |
| Sp-GFP-dsF | TGGGCGTGGATAGCGGTTTG | Sp-GFP knockdown |
| Sp-GFP-dsR | GGTCGGGGTAGCGGCTGAAG | |
| CFSH-EF | CGCGGATCCTCCTCCATCATAGGACACATGAATTC | rCFSH expression |
| CFSH-ER | GGACTAGTTTTATTCTCGCTTAAGTCGATGTAG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Q.; Lu, B.; Wang, G.; Ye, H. Transcriptional Inhibition of Sp-IAG by Crustacean Female Sex Hormone in the Mud Crab, Scylla paramamosain. Int. J. Mol. Sci. 2020, 21, 5300. https://doi.org/10.3390/ijms21155300
Jiang Q, Lu B, Wang G, Ye H. Transcriptional Inhibition of Sp-IAG by Crustacean Female Sex Hormone in the Mud Crab, Scylla paramamosain. International Journal of Molecular Sciences. 2020; 21(15):5300. https://doi.org/10.3390/ijms21155300
Chicago/Turabian StyleJiang, Qingling, Bei Lu, Guizhong Wang, and Haihui Ye. 2020. "Transcriptional Inhibition of Sp-IAG by Crustacean Female Sex Hormone in the Mud Crab, Scylla paramamosain" International Journal of Molecular Sciences 21, no. 15: 5300. https://doi.org/10.3390/ijms21155300
APA StyleJiang, Q., Lu, B., Wang, G., & Ye, H. (2020). Transcriptional Inhibition of Sp-IAG by Crustacean Female Sex Hormone in the Mud Crab, Scylla paramamosain. International Journal of Molecular Sciences, 21(15), 5300. https://doi.org/10.3390/ijms21155300






