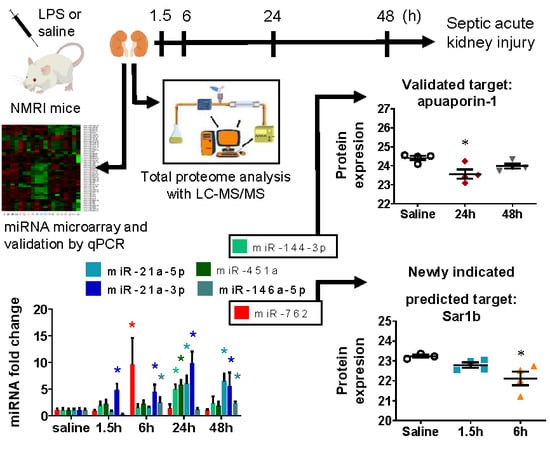
Time-Dependent miRNA Profile during Septic Acute Kidney Injury in Mice
Abstract
1. Introduction
2. Results
2.1. LPS-Induced Renal Pro-Inflammatory Cytokine Production
2.2. LPS-Induced Reversible Acute Kidney Injury
2.3. miRNA Array Profiling Revealed Three Differently Expressed Clusters
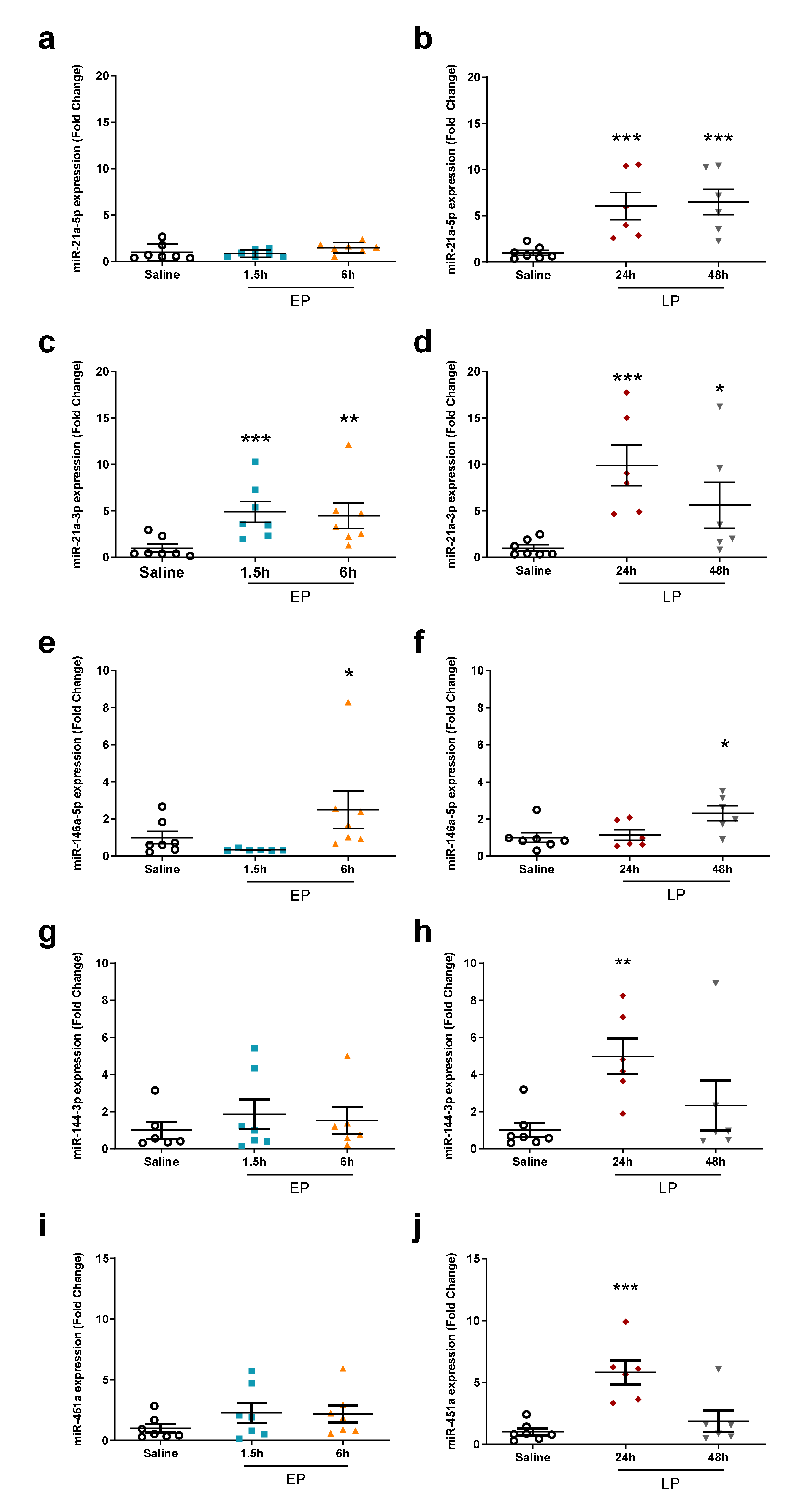
2.4. miRNA Microarray Validation
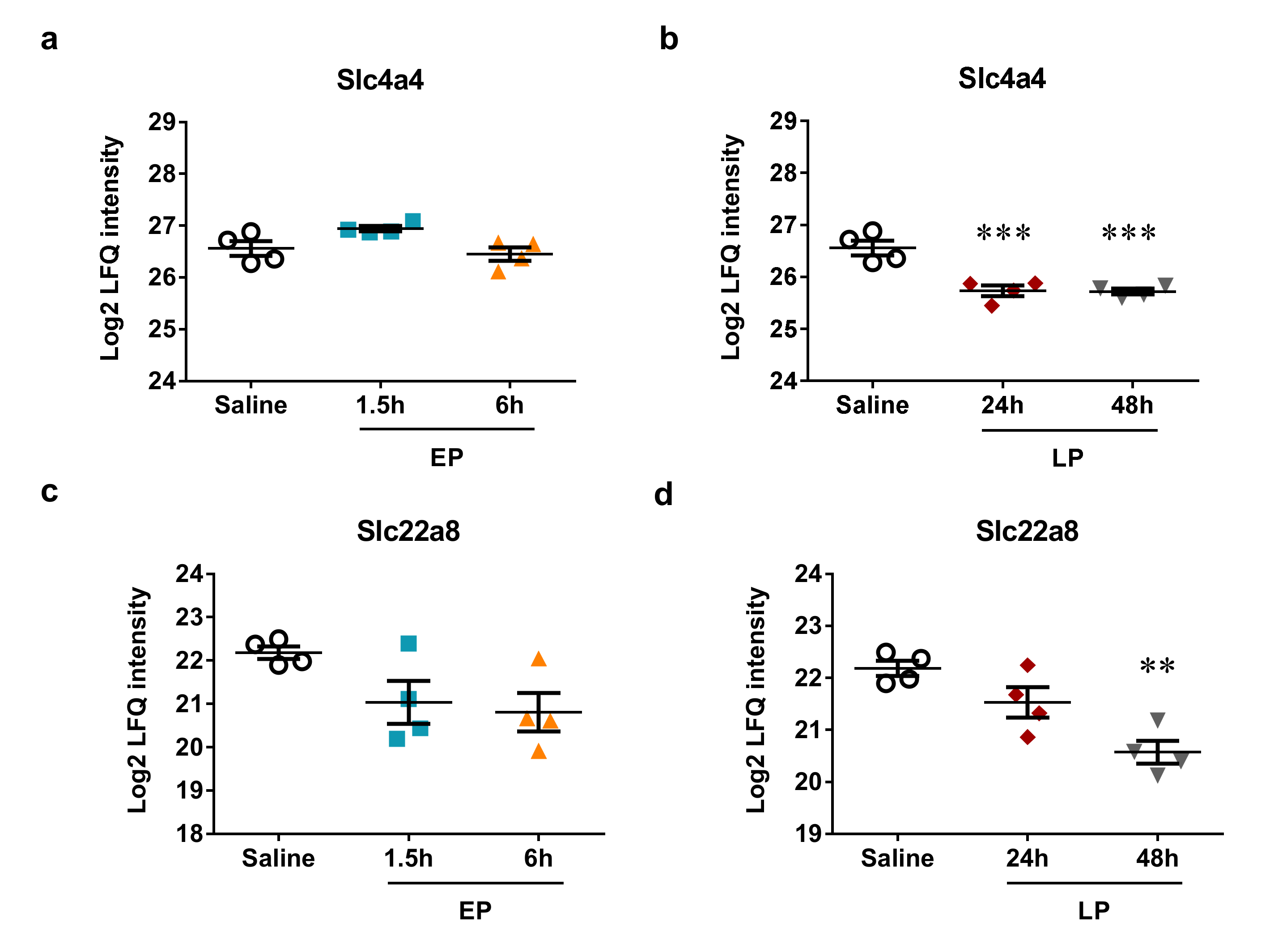
2.5. MicroRNA Targets Identified by Mass Spectrometry
3. Discussion
4. Methods
4.1. Mice
4.2. Endotoxin Preparations and Injection
4.3. Organ Harvest
4.4. Plasma Urea Determination
4.5. Total RNA Extraction and mRNA Real-Time PCR
4.6. MicroRNA Microarray Profiling
4.7. MicroRNA Real-Time PCR
4.8. Mass Spectrometry
4.9. Data Analysis
4.10. MicroRNA Target Prediction
4.11. Statistics
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sawhney, S.; Marks, A.; Fluck, N.; Levin, A.; Prescott, G.; Black, C. Intermediate and Long-term Outcomes of Survivors of Acute Kidney Injury Episodes: A Large Population-Based Cohort Study. Am. J. Kidney Dis. 2017, 69, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Makris, K.; Spanou, L. Acute Kidney Injury: Definition, Pathophysiology and Clinical Phenotypes. Clin. Biochem. Rev. 2016, 37, 85–98. [Google Scholar]
- Parmar, A.; Langenberg, C.; Wan, L.; May, C.N.; Bellomo, R.; Bagshaw, S.M. Epidemiology of septic acute kidney injury. Curr. Drug Targets 2009, 10, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Beutler, B.; Rietschel, E.T. Innate immune sensing and its roots: The story of endotoxin. Nat. Rev. Immunol. 2003, 3, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Kellum, J.A.; Prowle, J.R. Paradigms of acute kidney injury in the intensive care setting. Nat. Rev. Nephrol. 2018, 14, 217–230. [Google Scholar] [CrossRef]
- Ding, Y.; Guo, F.; Zhu, T.; Li, J.; Gu, D.; Jiang, W.; Lu, Y.; Zhou, D. Mechanism of long non-coding RNA MALAT1 in lipopolysaccharide-induced acute kidney injury is mediated by the miR-146a/NF-κB signaling pathway. Int. J. Mol. Med. 2018, 41, 446–454. [Google Scholar] [CrossRef]
- Meurer, M.; Höcherl, K. Deregulated renal magnesium transport during lipopolysaccharide-induced acute kidney injury in mice. Pflugers Arch. 2019, 471, 619–631. [Google Scholar] [CrossRef]
- Feng, X.; Guan, W.; Zhao, Y.; Wang, C.; Song, M.; Yao, Y.; Yang, T.; Fan, H. Dexmedetomidine ameliorates lipopolysaccharide-induced acute kidney injury in rats by inhibiting inflammation and oxidative stress via the GSK-3β/Nrf2 signaling pathway. J. Cell. Physiol. 2019, 234, 18994–19009. [Google Scholar] [CrossRef]
- Heemann, U.; Szabo, A.; Hamar, P.; Müller, V.; Witzke, O.; Lutz, J.; Philipp, T. Lipopolysaccharide pretreatment protects from renal ischemia/reperfusion injury: Possible connection to an interleukin-6-dependent pathway. Am. J. Pathol. 2000, 156, 287–293. [Google Scholar] [CrossRef]
- Colbert, J.F.; Ford, J.A.; Haeger, S.M.; Yang, Y.; Dailey, K.L.; Allison, K.C.; Neudecker, V.; Evans, C.M.; Richardson, V.L.; Brodsky, K.S.; et al. A model-specific role of microRNA-223 as a mediator of kidney injury during experimental sepsis. Am. J. Physiol. Physiol. 2017, 313, F553–F559. [Google Scholar] [CrossRef]
- Wang, X.; Huang, W.; Yang, Y.; Wang, Y.; Peng, T.; Chang, J.; Caldwell, C.C.; Zingarelli, B.; Fan, G.C. Loss of duplexmiR-223 (5p and 3p) aggravates myocardial depression and mortality in polymicrobial sepsis. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Qiao, Y.; Wang, P.; Li, S.; Zhao, W.; Gao, C. MicroRNA-210 negatively regulates LPS-induced production of proinflammatory cytokines by targeting NF-κB1 in murine macrophages. FEBS Lett. 2012, 586, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wu, C.; Zhang, Q. microRNA-34a participates in lipopolysaccharide mediated sepsis related renal function impairment via Kruppel-like factor 4. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 2018, 30, 351–354. [Google Scholar] [CrossRef]
- Fu, D.; Dong, J.; Li, P.; Tang, C.; Cheng, W.; Xu, Z.; Zhou, W.; Ge, J.; Xia, C.; Zhang, Z. MiRNA-21 has effects to protect kidney injury induced by sepsis. Biomed. Pharmacother. 2017, 94, 1138–1144. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Benovoy, D.; Kwan, T.; Majewski, J. Effect of polymorphisms within probe-target sequences on olignonucleotide microarray experiments. Nucleic Acids Res. 2008, 36, 4417–4423. [Google Scholar] [CrossRef]
- Tsai, C.A.; Hsueh, H.M.; Chen, J.J. Estimation of False Discovery Rates in Multiple Testing: Application to Gene Microarray Data. Biometrics 2003, 59, 1071–1081. [Google Scholar] [CrossRef]
- Pawitan, Y.; Michiels, S.; Koscielny, S.; Gusnanto, A.; Ploner, A. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics 2005, 21, 3017–3024. [Google Scholar] [CrossRef]
- Wright, G.W.; Simon, R.M. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 2003, 19, 2448–2455. [Google Scholar] [CrossRef]
- Reiner, A.; Yekutieli, D.; Benjamini, Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 2003, 19, 368–375. [Google Scholar] [CrossRef]
- Ueno, K.; Samura, M.; Nakamura, T.; Tanaka, Y.; Takeuchi, Y.; Kawamura, D.; Takahashi, M.; Hosoyama, T.; Morikage, N.; Hamano, K. Increased plasma VEGF levels following ischemic preconditioning are associated with downregulation of miRNA-762 and miR-3072-5p. Sci. Rep. 2016, 6, 36758. [Google Scholar] [CrossRef] [PubMed]
- Takai, Y.; Sasaki, T.; Matozaki, T. Small GTP-binding proteins. Physiol. Rev. 2001, 81, 153–208. [Google Scholar] [CrossRef] [PubMed]
- Sané, A.; Ahmarani, L.; Delvin, E.; Auclair, N.; Spahis, S.; Levy, E. SAR1B GTPase is necessary to protect intestinal cells from disorders of lipid homeostasis, oxidative stress, and inflammation. J. Lipid Res. 2019, 60, 1755–1764. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, X.; Zhu, H.; Zhu, C.; Wang, Y.; Pu, W.T.; Jegga, A.G.; Fan, G.C. Synergistic effects of the GATA-4-mediated miR-144/451 cluster in protection against simulated ischemia/reperfusion-induced cardiomyocyte death. J. Mol. Cell. Cardiol. 2010, 49, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Palmer, L.E.; Lechauve, C.; Zhao, G.; Yao, Y.; Luan, J.; Vourekas, A.; Tan, H.; Peng, J.; Schuetz, J.D.; et al. Regulation of gene expression by miR-144/451 during mouse erythropoiesis. Blood 2019, 133, 2518–2528. [Google Scholar] [CrossRef]
- Li, J.; Rohailla, S.; Gelber, N.; Rutka, J.; Sabah, N.; Gladstone, R.A.; Wei, C.; Hu, P.; Kharbanda, R.K.; Redington, A.N. MicroRNA-144 is a circulating effector of remote ischemic preconditioning. Basic Res. Cardiol. 2014, 109. [Google Scholar] [CrossRef]
- Fan, G.; Jiang, X.; Wu, X.; Fordjour, P.A.; Miao, L.; Zhang, H.; Zhu, Y.; Gao, X. Anti-Inflammatory Activity of Tanshinone IIA in LPS-Stimulated RAW264.7 Macrophages via miRNAs and TLR4–NF-κB Pathway. Inflammation 2016, 39, 375–384. [Google Scholar] [CrossRef]
- Li, H.; Shi, H.; Gao, M.; Ma, N.; Sun, R. Long non-coding RNA CASC2 improved acute lung injury by regulating miR-144-3p/AQP1 axis to reduce lung epithelial cell apoptosis. Cell Biosci. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Yu, G.; Liu, Q.; Dong, X.; Tang, K.; Li, B.; Liu, C.; Zhang, W.; Wang, Y.; Jin, Y. Inhibition of inflammation using diacerein markedly improved renal function in endotoxemic acute kidney injured mice. Cell. Mol. Biol. Lett. 2018, 23, 1–12. [Google Scholar] [CrossRef]
- Li, B.; Liu, C.; Tang, K.; Dong, X.; Xue, L.; Su, G.; Zhang, W.; Jin, Y. Aquaporin-1 attenuates macrophage-mediated inflammatory responses by inhibiting p38 mitogen-activated protein kinase activation in lipopolysaccharide-induced acute kidney injury. Inflamm. Res. 2019, 68, 1035–1047. [Google Scholar] [CrossRef]
- Kieran, N.E.; Doran, P.P.; Connolly, S.B.; Greenan, M.C.; Higgins, D.F.; Leonard, M.; Godson, C.; Taylor, C.T.; Henger, A.; Kretzler, M.; et al. Modification of the transcriptomic response to renal ischemia/reperfusion injury by lipoxin analog. Kidney Int. 2003, 64, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Asvapromtada, S.; Sonoda, H.; Kinouchi, M.; Oshikawa, S.; Takahashi, S.; Hoshino, Y.; Sinlapadeelerdkul, T.; Yokota-Ikeda, N.; Matsuzaki, T.; Ikeda, M. Characterization of urinary exosomal release of aquaporin-1 and -2 after renal ischemia-reperfusion in rats. Am. J. Physiol. Physiol. 2018, 314, F584–F601. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, C.; Summer, S.N.; Falk, S.; Wang, W.; Ljubanovic, D.; Schrier, R.W. Role of AQP1 in endotoxemia-induced acute kidney injury. Am. J. Physiol. Ren. Physiol. 2008, 294, 1473–1480. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, W.; Yu, G.; Liu, Q.; Jin, Y. Cytoprotective effect of aquaporin 1 against lipopolysaccharide-induced apoptosis and inflammation of renal epithelial hk-2 cells. Exp. Ther. Med. 2018, 15, 4243–4252. [Google Scholar] [CrossRef] [PubMed]
- Höcherl, K.; Schmidt, C.; Bucher, M. COX-2 inhibition attenuates endotoxin-induced downregulation of organic anion transporters in the rat renal cortex. Kidney Int. 2009, 75, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Huh, Y.; Keep, R.F.; Smith, D.E. Impact of Lipopolysaccharide-Induced Inflammation on the Disposition of the Aminocephalosporin Cefadroxil. Antimicrob. Agents Chemother. 2013, 57, 6171–6178. [Google Scholar] [CrossRef]
- Xu, X.; Kriegel, A.J.; Liu, Y.; Usa, K.; Mladinov, D.; Liu, H.; Fang, Y.; Ding, X.; Liang, M. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int. 2012, 82, 1167–1175. [Google Scholar] [CrossRef]
- Kaucsár, T.; Révész, C.; Godó, M.; Krenács, T.; Albert, M.; Szalay, C.I.; Rosivall, L.; Benyó, Z.; Bátkai, S.; Thum, T.; et al. Activation of the miR-17 Family and miR-21 During Murine Kidney Ischemia-Reperfusion Injury. Nucleic Acid Ther. 2013, 23, 344–354. [Google Scholar] [CrossRef]
- Pan, T.; Jia, P.; Chen, N.; Fang, Y.; Liang, Y.; Guo, M.; Ding, X. Delayed remote ischemic preconditioning confers renoprotection against septic acute kidney injury via exosomal miR-21. Theranostics 2019, 9, 405–423. [Google Scholar] [CrossRef]
- Jia, P.; Wu, X.; Dai, Y.; Teng, J.; Fang, Y.; Hu, J.; Zou, J.; Liang, M.; Ding, X. MicroRNA-21 Is Required for Local and Remote Ischemic Preconditioning in Multiple Organ Protection Against Sepsis. Crit. Care Med. 2017, 45, e703–e710. [Google Scholar] [CrossRef]
- Boldin, M.P.; Taganov, K.D.; Rao, D.S.; Yang, L.; Zhao, J.L.; Kalwani, M.; Garcia-Flores, Y.; Luong, M.; Devrekanli, A.; Xu, J.; et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011, 208, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Jia, P.; Fang, Y.; Liu, H.; Jiao, X.; He, J.C.; Ding, X. miR-146a is essential for lipopolysaccharide (LPS)-induced cross-tolerance against kidney ischemia/reperfusion injury in mice. Sci. Rep. 2016, 6, 27091. [Google Scholar] [CrossRef] [PubMed]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.D.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef]
- Wang, J.; Yu, M.; Yu, G.; Bian, J.; Deng, X.; Wan, X.; Zhu, K. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem. Biophys. Res. Commun. 2010, 394, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-H.; Xu, C.-F.; Li, Y.-M. Association of MicroRNA-223 Expression with Hepatic Ischemia/Reperfusion Injury in Mice. Dig. Dis. Sci. 2009, 54, 2362–2366. [Google Scholar] [CrossRef]
- Wang, J.; Bai, X.; Song, Q.; Fan, F.; Hu, Z.; Cheng, G.; Zhang, Y. miR-223 inhibits lipid deposition and inflammation by suppressing toll-like receptor 4 signaling in macrophages. Int. J. Mol. Sci. 2015, 16, 24965–24982. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, X.; Wang, T.; Xing, J. MiR-223-3p promotes cell proliferation and metastasis by downregulating SLC4A4 in clear cell renal cell carcinoma. Aging 2019, 11, 615–633. [Google Scholar] [CrossRef]
- Róka, B.; Tod, P.; Kaucsár, T.; Vizovišek, M.; Vidmar, R.; Turk, B.; Fonović, M.; Szénási, G.; Hamar, P. The Acute Phase Response Is a Prominent Renal Proteome Change in Sepsis in Mice. Int. J. Mol. Sci. 2019, 21, 200. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Csordas, A.; Sun, Z.; Jarnuczak, A.; Perez-Riverol, Y.; Ternent, T.; Campbell, D.S.; Bernal-Llinares, M.; Okuda, S.; Kawano, S.; et al. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017, 45, D1100–D1106. [Google Scholar] [CrossRef]
- Chou, C.-H.; Shrestha, S.; Yang, C.-D.; Chang, N.-W.; Lin, Y.-L.; Liao, K.-W.; Huang, W.-C.; Sun, T.-H.; Tu, S.-J.; Lee, W.-H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2007, 36, D149–D153. [Google Scholar] [CrossRef]
- Motulsky, H.J.; Brown, R.E. Detecting outliers when fitting data with nonlinear regression—A new method based on robust nonlinear regression and the false discovery rate. BMC Bioinform. 2006, 7, 123. [Google Scholar] [CrossRef]







| Cluster I (EP: 6 h) | Cluster II (LP) | Cluster III (LP) |
|---|---|---|
| miR-21a-3p | miR-20a-5p | miR-34c-3p |
| miR-204-3p | miR-21a-5p | miR-150-5p |
| miR-291a-3p | miR-144-3p | miR-1839-3p |
| miR-361-3p | miR-146a-5p | |
| miR-466n-3p | miR-155 | |
| miR-665-3p | miR-451a | |
| miR-711 | ||
| miR-762 | ||
| miR-763 | ||
| miR-1934-5p | ||
| miR-1971 | ||
| miR-2137 | ||
| miR-2861 | ||
| miR-3090-5p | ||
| miR-3100-3p | ||
| miR-3102-5p | ||
| miR-5116 | ||
| miR-5126 | ||
| miR-5129-5p | ||
| miR-5626-3p | ||
| miR-5627-3p |
| miRNA | 1.5 h | 6 h | 24 h | 48 h |
|---|---|---|---|---|
| miR-204-3p | 2.82 ± 0.20 *** | 1.88 ± 0.28 *** | 1.08 ± 0.10 (ns) | 1.21 ± 0.25 (ns) |
| miR-3102-5p | 1.11 ± 0.23 (ns) | 2.77 ± 0.52 *** | 1.05 ± 0.32 (ns) | 1.15 ± 0.25 (ns) |
| miR-762 | 1.19 ± 0.27 (ns) | 2.69 ± 0.34 *** | 1.16 ± 0.32 (ns) | 1.24 ± 0.22 (ns) |
| miR-2137 | 0.96 ± 0.01 (ns) | 2.61 ± 0.26 *** | 1.41 ± 0.13 (ns) | 1.42 ± 0.63 (ns) |
| miR-3090-5p | 1.21 ± 0.16 (ns) | 2.50 ± 0.61 * | 1.11 ± 0.35 (ns) | 1.36 ± 0.42 (ns) |
| miR-2861 | 1.06 ± 0.29 (ns) | 2.46 ± 0.40 ** | 2.22 ± 0.72 ** | 1.49 ± 0.48 (ns) |
| miR-665-3p | 1.25 ± 0.22 (ns) | 2.32 ± 0.22 *** | 0.93 ± 0.13 (ns) | 0.98 ± 0.11 (ns) |
| miR-5129-5p | 1.17 ± 0.34 (ns) | 2.25 ± 0.69 * | 0.94 ± 0.37 (ns) | 0.95 ± 0.15 (ns) |
| miR-21a-3p | 1.36 ± 0.12 (ns) | 2.00 ± 0.20 *** | 1.88 ± 0.18 *** | 1.34 ± 0.17 (ns) |
| miR-5116 | 1.05 ± 0.07 (ns) | 1.76 ± 0.17 *** | 1.20 ± 0.18 (ns) | 1.00 ± 0.15 (ns) |
| miR-3100-3p | 1.03 ± 0.08 (ns) | 1.67 ± 0.18 *** | 1.28 ± 0.22 (ns) | 1.10 ± 0.11 (ns) |
| miR-711 | 1.00 ± 0.08 (ns) | 1.66 ± 0.24 ** | 1.23 ± 0.08 (ns) | 1.31 ± 0.47 (ns) |
| miR-466n-3p | 1.01 ± 0.06 (ns) | 1.61 ± 0.25 * | 1.02 ± 0.20 (ns) | 0.90 ± 0.08 (ns) |
| miR-223-3p | 1.40 ± 0.20 (ns) | 1.60 ± 0.10 ** | 1.33 ± 0.12 * | 1.23 ± 0.16 (ns) |
| miR-3474 | 1.01 ± 0.01 (ns) | 1.60 ± 0.24 ** | 1.12 ± 0.09 (ns) | 1.02 ± 0.13 (ns) |
| miR-21a-5p | 1.07 ± 0.2 (ns) | 1.39 ± 0.18 * | 4.44 ± 0.66 *** | 4.59 ± 1.34 ** |
| miR-451a | 1.63 ± 0.98 (ns) | 1.08 ± 0.33 (ns) | 3.76 ± 1.56 ** | 2.22 ± 2.41 (ns) |
| miR-144-3p | 1.37 ± 0.87 (ns) | 1.10 ± 0.24 (ns) | 2.56 ± 1.04 ** | 1.65 ± 1.43 (ns) |
| miR-2861 | 1.06 ± 0.29 (ns) | 2.46 ± 0.40 ** | 2.22 ± 0.72 ** | 1.49 ± 0.48 (ns) |
| miR-21a-3p | 1.36 ± 0.12 (ns) | 2.00 ± 0.20 *** | 1.88 ± 0.18 *** | 1.34 ± 0.17 (ns) |
| miR-146a-5p | 1.10 ± 0.08 (ns) | 0.98 ± 0.08 (ns) | 1.51 ± 0.14 ** | 1.58 ± 0.21 ** |
| miR-1839-3p | 1.02 ± 0.21 (ns) | 0.92 ± 0.24 (ns) | 0.57 ± 0.09 ** | 0.74 ± 0.16 ** |
| miR-34c-3p | 0.96 ± 0.19 (ns) | 0.87 ± 0.24 (ns) | 0.57 ± 0.17 ** | 0.68 ± 0.16 ** |
| miR-150-5p | 1.01 ± 0.20 (ns) | 0.79 ± 0.13 (ns) | 0.59 ± 0.06 *** | 0.63 ± 0.13 *** |
| miR-129-1-3p | 0.90 ± 0.08 (ns) | 0.81 ± 0.06 * | 0.70 ± 0.05 *** | 0.65 ± 0.10 *** |
| miR-34b-3p | 1.01 ± 0.11 (ns) | 0.85 ± 0.16 (ns) | 0.74 ± 0.04 ** | 0.81 ± 0.12 (ns) |
| miR-3070a-5p/ miR-3070b-5p | 1.05 ± 0.11 (ns) | 0.85 ± 0.09 * | 0.78 ± 0.08 (ns) | 0.71 ± 0.07 ** |
| Target Gene | Forward Primer | Reverse Primer |
|---|---|---|
| TNF-α | AAATGGCCTCCCTCTCATCA | AGATAGCAAATCGGCTGACG |
| IL-6 | CAAAGCCAGAGTCCTTCAGAGA | GGTCTTGGTCCTTAGCCACTC |
| Lcn-2 | ACGGACTACAACCAGTTCGC | AATGCATTGGTCGGTGGGG |
| GAPDH | TTCACCACCATGGAGAGGGC | GGCATGGACTGTGGTCATGA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tod, P.; Róka, B.; Kaucsár, T.; Szatmári, K.; Vizovišek, M.; Vidmar, R.; Fonovič, M.; Szénási, G.; Hamar, P. Time-Dependent miRNA Profile during Septic Acute Kidney Injury in Mice. Int. J. Mol. Sci. 2020, 21, 5316. https://doi.org/10.3390/ijms21155316
Tod P, Róka B, Kaucsár T, Szatmári K, Vizovišek M, Vidmar R, Fonovič M, Szénási G, Hamar P. Time-Dependent miRNA Profile during Septic Acute Kidney Injury in Mice. International Journal of Molecular Sciences. 2020; 21(15):5316. https://doi.org/10.3390/ijms21155316
Chicago/Turabian StyleTod, Pál, Beáta Róka, Tamás Kaucsár, Krisztina Szatmári, Matej Vizovišek, Robert Vidmar, Marko Fonovič, Gábor Szénási, and Péter Hamar. 2020. "Time-Dependent miRNA Profile during Septic Acute Kidney Injury in Mice" International Journal of Molecular Sciences 21, no. 15: 5316. https://doi.org/10.3390/ijms21155316
APA StyleTod, P., Róka, B., Kaucsár, T., Szatmári, K., Vizovišek, M., Vidmar, R., Fonovič, M., Szénási, G., & Hamar, P. (2020). Time-Dependent miRNA Profile during Septic Acute Kidney Injury in Mice. International Journal of Molecular Sciences, 21(15), 5316. https://doi.org/10.3390/ijms21155316