Abstract
β-Galactosidase from Arthrobacter sp. 32cB (ArthβDG) is a cold-adapted enzyme able to catalyze hydrolysis of β-d-galactosides and transglycosylation reaction, where galactosyl moiety is being transferred onto an acceptor larger than a water molecule. Mutants of ArthβDG: D207A and E517Q were designed to determine the significance of specific residues and to enable formation of complexes with lactulose and sucrose and to shed light onto the structural basis of the transglycosylation reaction. The catalytic assays proved loss of function mutation E517 into glutamine and a significant drop of activity for mutation of D207 into alanine. Solving crystal structures of two new mutants, and new complex structures of previously presented mutant E441Q enables description of introduced changes within active site of enzyme and determining the importance of mutated residues for active site size and character. Furthermore, usage of mutants with diminished and abolished enzymatic activity enabled solving six complex structures with galactose, lactulose or sucrose bounds. As a result, not only the galactose binding sites were mapped on the enzyme’s surface but also the mode of lactulose, product of transglycosylation reaction, and binding within the enzyme’s active site were determined and the glucopyranose binding site in the distal of active site was discovered. The latter two especially show structural details of transglycosylation, providing valuable information that may be used for engineering of ArthβDG or other analogous galactosidases belonging to GH2 family.
1. Introduction
β-d-Galactosidases (βDGs), enzymes catalyzing hydrolysis of β-d-galactosides, belong to five different glycosyl hydrolase (GH) families: GH1, GH2, GH35, GH42, and GH59, as the classification was made based on protein fold not its primary activity. Their common feature is the presence of a TIM barrel fold catalytic domain; however, they differ in the number of surrounding domains possessing β architecture [1].
β-d-Galactosidases belonging to the GH2 family are large proteins, usually ~100 kDa, of which the active forms are: tetramers [2,3,4], hexamers [5], and as recently discovered dimers [6,7]. Their catalytic site can be divided into a galactose binding subsite and a platform for weak binding of different moieties [8]. Their primary mode of action is to catalyze the hydrolysis of lactose to d-galactose and d-glucose. Some of GH2 β-d-galactosidases are able to catalyze transglycosylation reactions when the galactosyl moiety is transferred onto an acceptor larger than a water molecule. The best studied example is lacZ β-d-galactosidase from Escherichia coli, which produces allolactose, disaccharide composed of d-galactose, and d-glucose moieties linked through a β-(1→6)-glycosidic bond, if excess of galactose occurs [9,10].
Prebiotics are non-digestible food ingredients that support development and functioning of human organisms through selective assistance of growth or activity of one or a number of bacteria species in the lower intestine [11,12,13]. Ones that are used to enrich a daily diet are chosen to stimulate the growth of bacteria from Bifidobacteria or Lactobacilli families [14,15,16,17]. The importance of prebiotics, especially galactooligosaccharides, as an additive to infant formula has been widely tested. They support the colonization of intestines by beneficial bacteria. In consequence, they strengthen immunity by prevention of bacterial adhesion at an early stage of infection [18,19,20,21,22,23,24,25]. Morerover, some oligosaccharides are a rich source of sialic acid, indispensable for proper development of the brain [26,27]. Prebiotics are also important in adults’ nutrition, as they may support absorption of minerals [28,29,30], support recovery after influenza, reduce stress-related digestive problems [31], support lipid metabolism, and also counteract the development of tumors. They augment prevention in liver encephalopathy, glycemia/insulinemia, and also have a positive effect on immunomodulation [12,32].
Even though prebiotics such as galactooligosaccharides (GOS) and heterooligosaccharides (HOS) have been successfully synthesized, their production in the course of enzymatic catalysis proved to be more beneficial due to the higher specificity of product and milder reaction conditions. For this purpose, glycosyltransferases (EC 2.4) or glycosidic hydrolases (EC 3.2.1) are used—enzymes that have the ability to catalyze the transfer of a galactosyl moiety to a sugar acceptor.
Nowadays, both for lactose removal and GOS/HOS synthesis, the β-d-galactosidase from Kluyveromyces lactis is predominantly used [33,34,35,36]. Introduction of new enzyme into food production must be preceded by a number of studies that will not only ensure its high efficiency but will also ensure the consumers’ safety. Enzyme immobilization may be the way to ensure that it will not induce allergic reactions in humans.
Nonetheless, an implementation of cold-adapted enzymes would reduce the overall process temperature bringing numerous benefits [37,38]. Lowering the temperature of the process eliminates need for heating. Therefore, not only are costs being cut, but the process becomes more environmentally friendly. Furthermore, final product quality may be improved, as conducting enzymatic reactions at low temperature in food processing prevents the loss of valuable substances and formation of undesired products of heat conversions. What is more, cold-adapted enzymes are perfect candidates to perform catalysis in organic solvent environment which is especially interesting for the pharma industry.
This broad range of benefits that can be achieved by implementation of cold-adapted enzymes provides rationale for the efforts to engineer enhanced activity cold-adapted enzymes tailored for an exact industrial applications [39,40]. The natural way of research is to provide as a first step structural information that can be further used for knowledge base enzyme engineering. However, due to the difficulty of cold-adapted enzymes’ crystallization the amount of crystal structures available in Protein Data Bank is surprisingly low. Only structures of 11 cold-adapted glycosyl hydrolases are deposited there: α-amylase from Alteromonas haloplanctis [41], endo-1,4-beta-glucanase from Eisenia fetida [42], psychrophilic endo-beta-1,4-xylanase [43], β-1,4-d-mannanase from Cryptopygus antarcticus [44], chitinase from Moritella marina [45], endo-beta-1,4-xylanase from Aegilops speltoides subsp. speltoides [43], β-glucosidase from Exiguobacterium antarcticum B7 [46], xylanase from Pseudoalteromonas haloplanktis [47], β-galactosidase from Rahnella sp. R3 [48], β-d-galactosidase from Paracoccus sp. 32d [6], and further discussed here β-d-galactosidase from Arthrobacter sp. 32 cB [7]. Hence, detailed information on the structure of β-d-galactoside processing cold-adapted enzyme that could be used for prebiotics production at the industrial scale is still in demand. The modification of transglycosylase activity specificity and efficiency may be achieved by controlling reaction equilibrium or by enzyme engineering. Studies concentrated on introducing mutations into subsites of GHs showed that the modulation of hydrolysis and transglycosylation activities can be achieved by means of knowledge-based enzyme engineering. However, the role of individual amino acids in the active site must be discovered as basis for successful design of an enzyme with specific, desired activities [49].
Psychrophilic β-d-galactosidase from Arthrobacter sp. 32cB is an interesting candidate for industrial use, as it can hydrolyze lactose at rate comparable to mesophilic βDG from Kluyveromyces lactis. Furthermore, it exhibits additional transglycosylation activity, it catalyzes synthesis of GOS and HOS, for example: lactulose and alkyl glycosides. [50] Architecture of its five domains [7], structural details of hydrolysis of lactose catalyzed by this enzyme [51], and its structural features responsible for the enzyme’s cold adaptation [52] were investigated and discussed previously. Upon the solution of native structure, E441 and E517 were assigned as catalytic residues based on superposition of ArthβDG structure with lacZ E. coli β-d-galactosidase [7]. The role of residue E441 was previously proven: a loss-of-function mutant E441Q was designed, a biochemical activity assay was performed, and the protein was crystallized. The introduction of point mutation E441Q did not disrupt the structure of protein, yet no catalytic activity was exhibited anymore. The complex structures ArthβDG_LACs and ArthβDG_LACd, with natural substrate-lactose bound in shallow and deep mode revealed residues directly involved in the binding of galactosyl group in two modes [51]. Especially interesting was the D207 residue, as it contributes in the deep binding of substrate by stabilizing O4 hydroxyl group but also creates a bottom of the active site limiting its size. The question of the effect of point mutation of the other catalytic residue, E517, has not been answered before. Similarly, structural elements implicated in the enzyme’s secondary activity, transglycosylation, remain elusive.
This is why, two new mutants, ArthβDG_D207A and ArthβDG_E517Q, were designed to elucidate the impact of these two residues on protein’s structure and activity. Furthermore, here we attempted to shed a light on the structural side of transglycosylation catalyzed by cold-adapted GH2 ArthβDG through usage of the loss and depletion of the function mutants for crystallization and formation of complexes with selected reaction substrates or products, especially lactulose.
2. Results and Discussion
2.1. Activity of ArthβDG Mutants
The hydrolytic activity of ArthβDG_D207A and ArthβDG_E517Q was measured using ONPG as a substrate. The ArthβDG_E517Q exhibited no catalytic activity, whereas activity of mutant ArthβDG_D207A was severely depleted (Table 1).

Table 1.
Activity of ArthβDG, ArthβDG_D207A, and ArthβDG_E517Q in hydrolysis reaction. The activity was measured with ONPG as a substrate at 28 °C and pH 8.0.
The results of the activity assays show that the hydrolytic activity of the enzyme was affected not only when catalytic residues were mutated but also when the mutation of the amino acids that play a key role in building the active site was introduced. As expected of the mutation of the catalytic residue, E517 into glutamine caused loss of function of the enzyme. The mutation of the residue involved in the stabilization of galactosyl moiety within active site, D207, diminished the enzyme’s activity.
Such a drastic drop of activity related to the mutation of one of the residues contributing to binding and positioning of the substrate in the active site of enzyme suggests that the net of interactions within the ArthβDG’s active center was in a very delicate balance. This cold-adapted enzyme was already characterized by a high turnover rate, and losing contacts within its active site cripples its activity instead of further enhancement.
2.2. Thermofluor Shift Assay
Results of the TSA (thermofluor shift assay) of ArthβDG_D207A did not exhibit significant differences compared with ones obtained for wild-type protein ArthβDG [52]. The highest melting temperature (44 °C) was obtained in conditions containing 50 mM sodium phosphate pH 6.0. Similar stability was obtained for protein samples in buffers, such as PIPES, HEPES, MES BIS-TRIS propane, ADA, and MOPS, within a pH range 6.0–6.7, independently, on addition of 250 mM NaCl. Using the same set of buffers in the pH ranges 5.0–6.0 and 7.0–8.0 the melting temperature was on average 3 °C lower. Also, the stabilization effect of previously established for ArthβDG crystallization solution (37% TascimateTM pH 8.0) augments stability of ArthβDG_D207A by 7 °C compared to 50 mM sodium phosphate pH 6.0 (Figure S1).
Mutation of catalytic residue E517 into glutamine resulted in the elevation of the melting temperature of the mutant in respect to ArthβDG by 3.5–5 °C, depending on the investigated buffer solution. In the case of 50 mM sodium phosphate pH 6.0, it attained 49 °C, which means that it was higher by 4.5 °C compared to wild-type protein. The trend of increasing the melting temperature of the mutant ArthβDG_D207A by approximately 4 °C was maintained for all tested buffers in the pH range 6.0–7.0. In the pH ranges 5.0–6.0 and 7.0–8.0, the melting temperature was elevated on average by 3 °C compared to ArthβDG. Nonetheless, the impact of crystallization solution on protein stability compare to 50 mM sodium phosphate pH 6.0 was higher than in the case of ArthβDG_N440D, by 5 °C (Figure S2).
The results of TSA showed that the introduced mutation did not lead to a dramatic drop of mutant’s melting temperature that would be expected if mutations would have a destabilizing effect on the protein’s structure. It also showed that they exhibited similar features to wild-type protein; thus, crystallization was attempted in conditions previously established for wild-type protein without initial screening. Such an approach allowed us to obtain mutants’ crystal almost readily without need for much of a protein sample.
2.3. ArthβDG Mutants
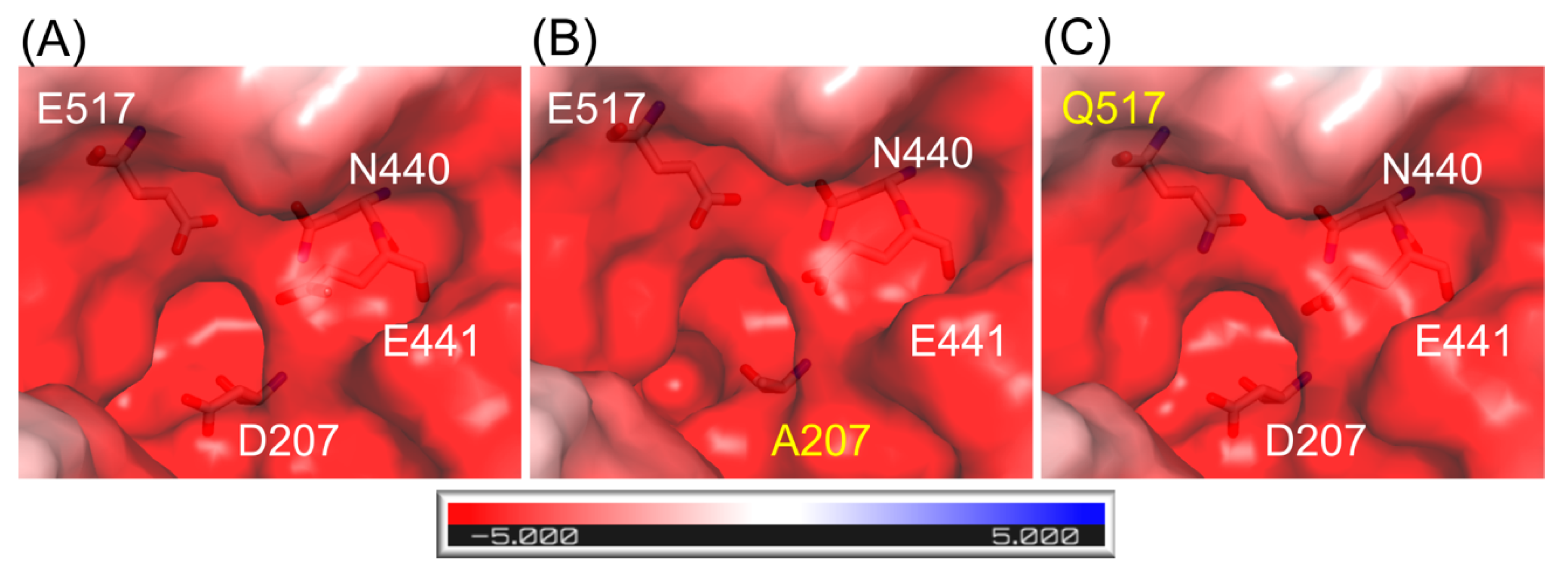
The structures of ArthβDG mutants: ArthβDG_D207A and ArthβDG_E517Q, were solved proving that the point mutations within the active site of ArthβDG have no destabilizing effect on protein’s structure. In the case of ArthβDG_E517Q, only a slight change in character of the active site was observed (Figure 1C)—similar to previously studied loss of function mutant ArthβDG_E441Q [51].
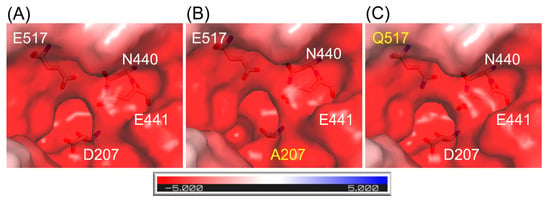
Figure 1.
Representation of surface and charge distribution within the active site of ArthβDG (A) and its mutants: ArthβDG_D207A (B) and ArthβDG_E517Q (C). The residues under scrutiny are shown as stick and mutations are indicated with yellow. The charge distribution was calculated using the APBS [53] plugin to PyMOL.
In the case of mutant ArthβDG_D207A, where a larger side chain was substituted with alanine residue, the change in active site size and shape was introduced (Figure 1B).
2.4. Influence of Mutations within Active Site on Binding of Galactosyl Residue
The functional dimer of ArthβDG has two independent active sites. Each of them is located on the top of TIM-barrel Domain 3 and is constituted of residues from both subunits of the dimer. It can be described as a deep acidic funnel with an antechamber.
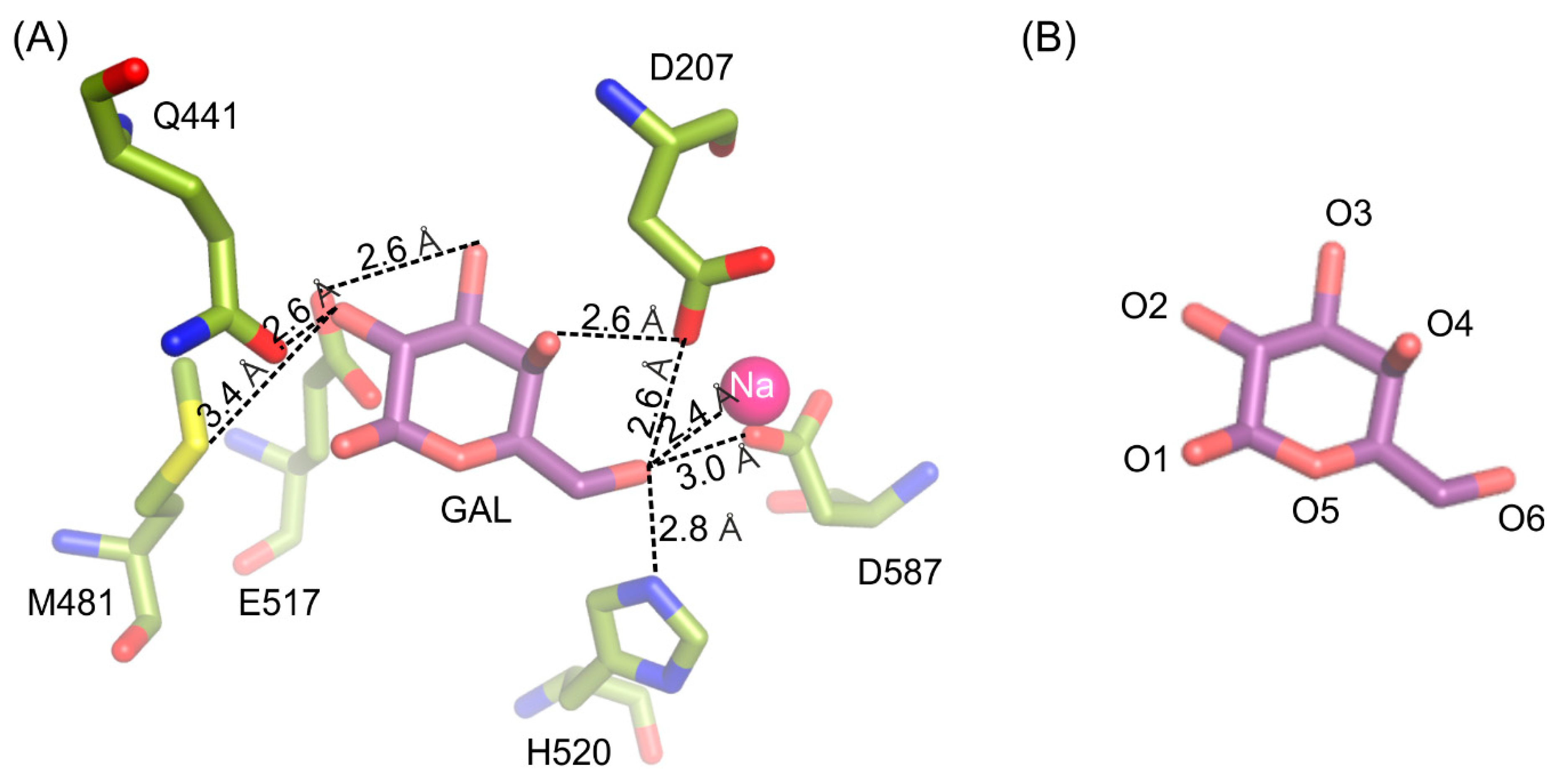
The galactosyl moiety is stabilized in the deep binding by a net of hydrogen interactions between O6 and H520 (2.8 Å), D587 (3.0 Å), D207 (2.6 Å) and sodium ion (2.4 Å); O4 and D207 (2.6 Å); O3 and E517 (2.6 Å); O2 and M481 (3.3 Å) and Q441 (2.6 Å) (Figure 2).
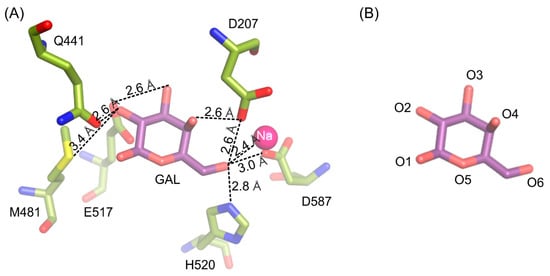
Figure 2.
Interactions stabilizing galactose molecule in the active site of ArthβDG_E441Q/gal complex structure (A). Naming convention of galactose’s oxygen atoms presented for easier interpretation (B).
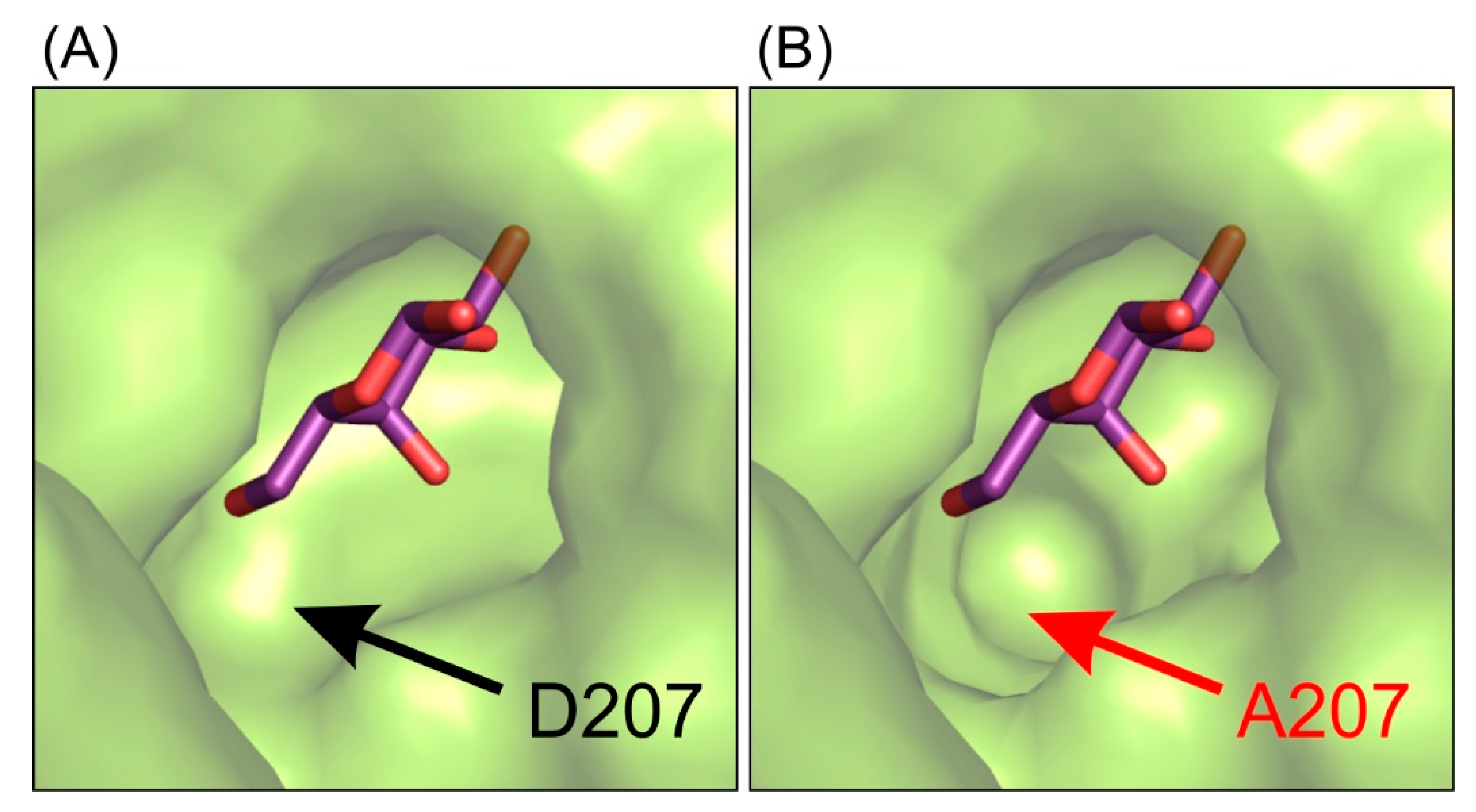
This interaction pattern between the galactosyl moiety bound in the deep mode in the enzyme’s active site remains uninterrupted. Single-point mutations of residues E441 and E517 introduced in the enzyme’s active site were not sufficient to change spatial arrangement of galactosyl moiety bound in the deep binding mode but were enough to abolish the enzyme’s activity. Even though the mutant ArthβDG_D207A retained partial activity of the wild-type enzyme, no crystal structure of its complex with galactose bound within the binding site was obtained. The complex structure ArthβDG_D207A/gal showed a galactose bound only in G1 and G2 sites (galactose binding sites described in detail in part 2.5). Lack of even a residual electron density that could be attributed to galactose molecule presence in the active sites was consistent for more than 15 datasets obtained under different soaking conditions in triplicates. This may indicate that even though D207 is not a catalytic residue, its presence is essential for the enzyme function. D207 assures a correct position of the substrate by retaining the net of interactions and ensuring proper shape of the active site (Figure 3).
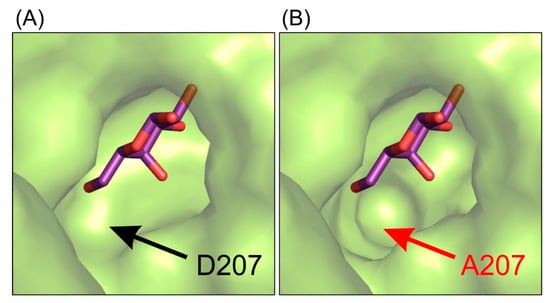
Figure 3.
The representation of the shape of active site of ArthβDG_E441Q/gal complex structure (A) and superposition of galactose molecule on the structure of ArthβDG_D207A (B) showing that this mutation introduce changes in the shape of active site, making it less selective for galactosyl moiety.
Combining the structural data with results of activity assays suggests that the mutation of D207 effects the enzyme’s activity by disturbing the hydroxyl group O4 stabilization of galactosyl moiety. It is caused by the cumulative effect of change in the shape of active site (Figure 1) and removing possible stabilizing contacts between carboxyl group of D207 and hydroxyl O4 of galactosyl moiety.
2.5. Galactose Binds on the Enzyme’s Surface
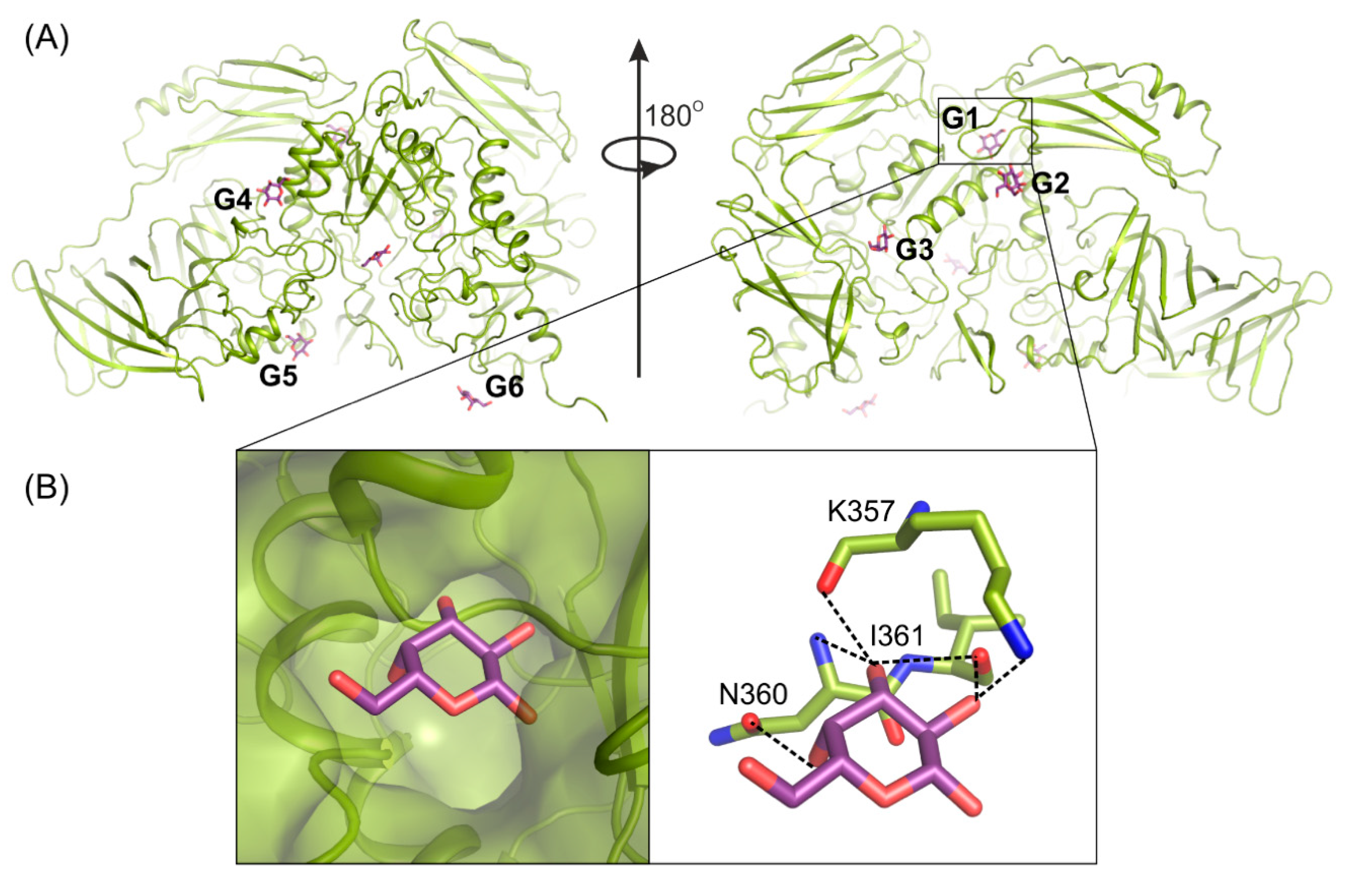
Determining the crystal structure of ArthβDG_E441Q/gal complex at atomic resolution, 1.5 Å, resulted in discovering six galactose binding sites at the surface of the protein (Figure 4A). Five of these (G2–G6) have a potential to bind larger galactosides, e.g., lactose. The question, why such a large enzyme was developed and retained by extremophilic bacteria for catalysis of relatively simple reactions, remains open. However such a strategy, to elevate the accessibility of a substrate by its weak binding on the enzyme’s surface, may be one of explanation together with the maximization of the energy gain from the surface residue–solvent interactions [52].
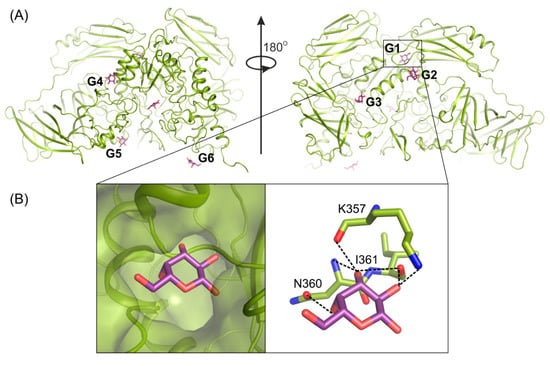
Figure 4.
Galactose (violet) binding sites G1–G6 on the surface of ArthβDG (A); structure of ArthβDG_E441Q in complex with galactose (B). The galactose molecule (violet) interacting with main chain atoms at G1 site; galactose hydroxyl group O2 interacts with oxygen K357 (2.8 Å), oxygen I361 (2.9 Å); O3 with oxygen I361 (3.4 Å), nitrogen N360 (3.5 Å) and oxygen K357 (2.7 Å); O4 with N360 (2.8 Å).
Previously obtained biochemical data showed that d-galactose, one of the products of enzyme’s hydrolytic activity, has an inhibitory effect on protein’s activity. Its presence at 50 mM concentration results in the reduction of the enzyme’s activity by half [50]. The analysis of atomic resolution ArthβDG_E441Q/gal complex revealed that galactose binding site G1 is a possible allosteric site of enzyme. What is worth noting, is the addition of galactose was used for soaking of multiple ArthβDG complex structures to inhibit the activity of enzyme and, thus, enabled complex formation. Structural analysis of these complex structures showed that the galactose molecule was present at the G1 site for all of them, namely, previously published ArthβDG_E441Q/LACd [51] and, discussed here, ArthβDG_E517Q/lact and ArthβDG_ E441Q/lact. It should be stated that such a complex could not be successfully obtained without galactose addition.
The G1 site is located in the cleft formed at the junction of tree domains: Domain 2, 3, and 4 (Figure 4A). It is highly selective toward galactose molecules as residues N360, I361, and K357, constituting its bottom, shape it in such a way that binding of galactosyl moiety is highly preferred. The correct conformation of hydroxyl groups O2, O3, O4, and O6 is necessary for monosaccharide molecule to enter and bind within G1 site. Its limited size prevents basically any molecule larger than galactose from entering (Figure 4B).
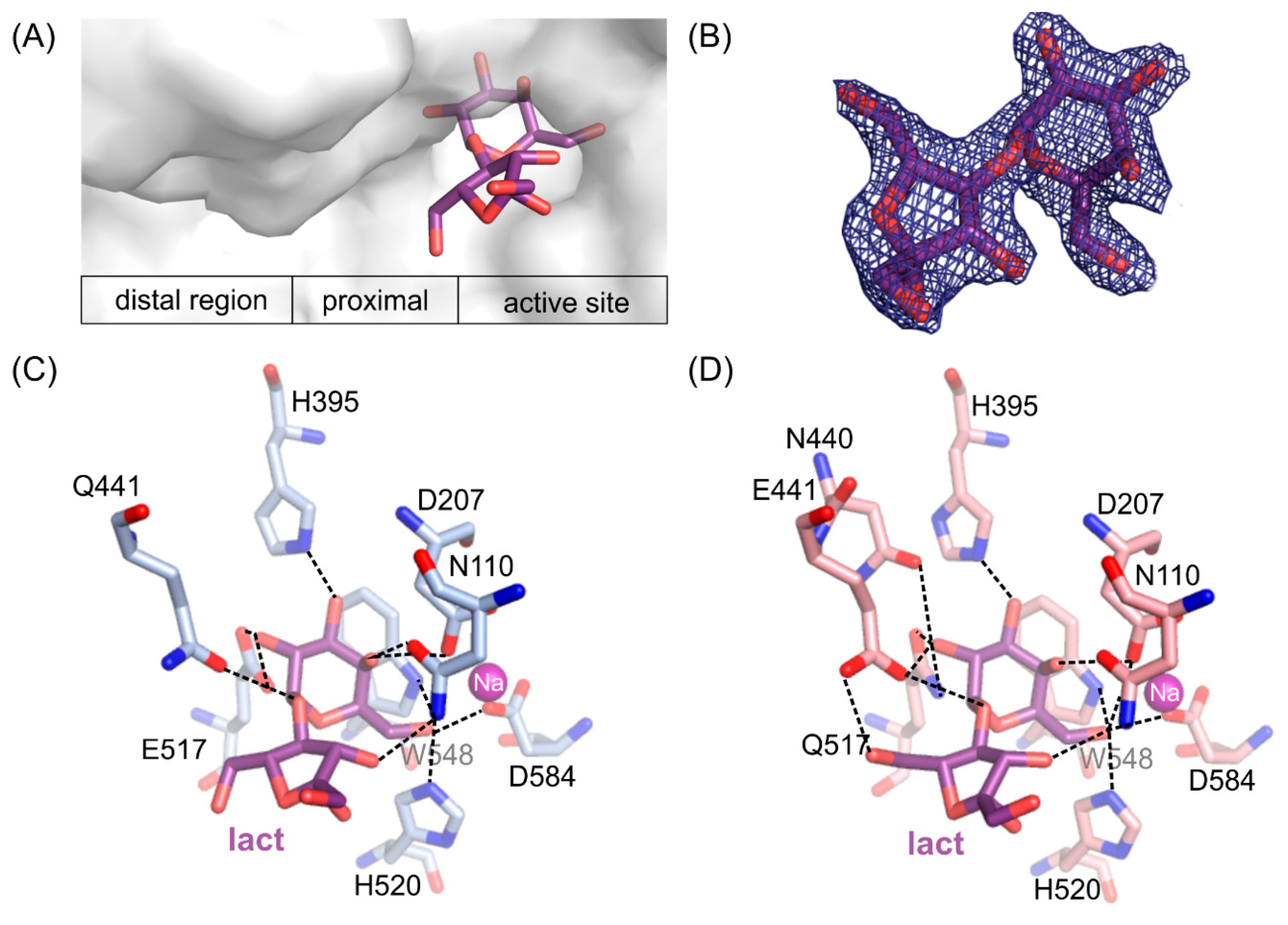
2.6. ArthβDG Mutants in Complexes with Lactulose
Crystal structures of loss of function ArthβDG_E441Q and ArthβDG_E517Q mutants in complexes with lactulose were determined both at 2.0 Å resolution. Lactulose molecule, a heterooligosaccharide composed of d-galactose and d-fructose linked through a β-(1→4)-glycosidic bond and is one of the products of transglycosylation reaction catalyzed by ArthβDG. It was bound in a deep mode in both structures (Figure 5A). The galactosyl moiety is stabilized by a net of interaction characteristic for binding of this group by ArthβDG, and described in detail in Section 2.4.
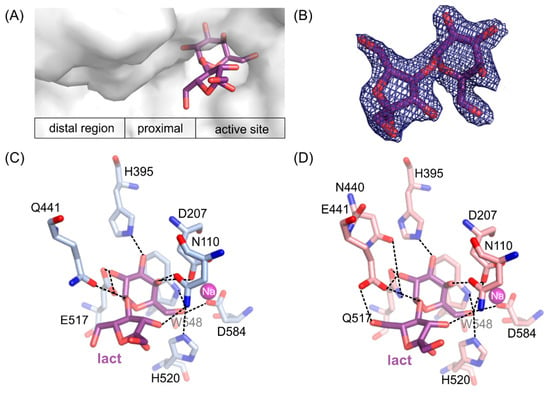
Figure 5.
The position of lactulose binding by the loss of function mutants (A). 2Fo-Fc electron-density map at the 2 σ level for the lactulose molecule in ArthβDG_E517Q/lact complex (B). Lactulose bound in the active site of ArthβDG_E441Q/lact (C) and ArthβDG_E517Q/lact (D) crystal structures with hydrogen bonds marked with dashed lines.
Fructose moiety of lactulose was stabilized relatively weakly in the position. In the case of a complex ArthβDG_E441Q/lact (Figure 5C), there was only one hydrogen bond between hydroxyl group O3′ of lactulose and N110 (3.2 Å). Whereas, in the case of ArthβDG_E517Q/lact (Figure 5D) there were two hydrogen bonds stabilizing fructose moiety: between O3′ of lactulose and N110 (3.3 Å) and O6’ and E441 (3.0 Å).
What is interesting, regardless of the relatively weak stabilization of the fructose moiety of lactulose, it creates more interactions within the active site of ArthβDG than glucosyl moiety of lactose, natural substrate [51]. The hydrogen bonds, forming between sugar moiety and the amino acids at the entrance to the active site, are crucial for sugar ring stabilization indispensable for it to become a galactosyl group acceptor. Such a delicate net of interactions suggests that residues E441, N110, and possibly N440 play important roles regarding transglycosylation activity of ArthβDG.
Insight into these two complex structures enables description of positioning and binding of fructose moiety, which may act as galactosyl group acceptor, in the catalytic center of ArthβDG mutants: ArthβDG_D207A and ArthβDG_E517Q. As both these mutations are distal to space occupied by fructose moiety, one may assume that similar interactions are in place in the case of wild-type enzyme. However, the substantially higher activity of wild-type enzyme makes it impossible to capture lactulose molecule bound in its active site.
2.7. Mapping the Binding Potential of the Distal Region of Active Site
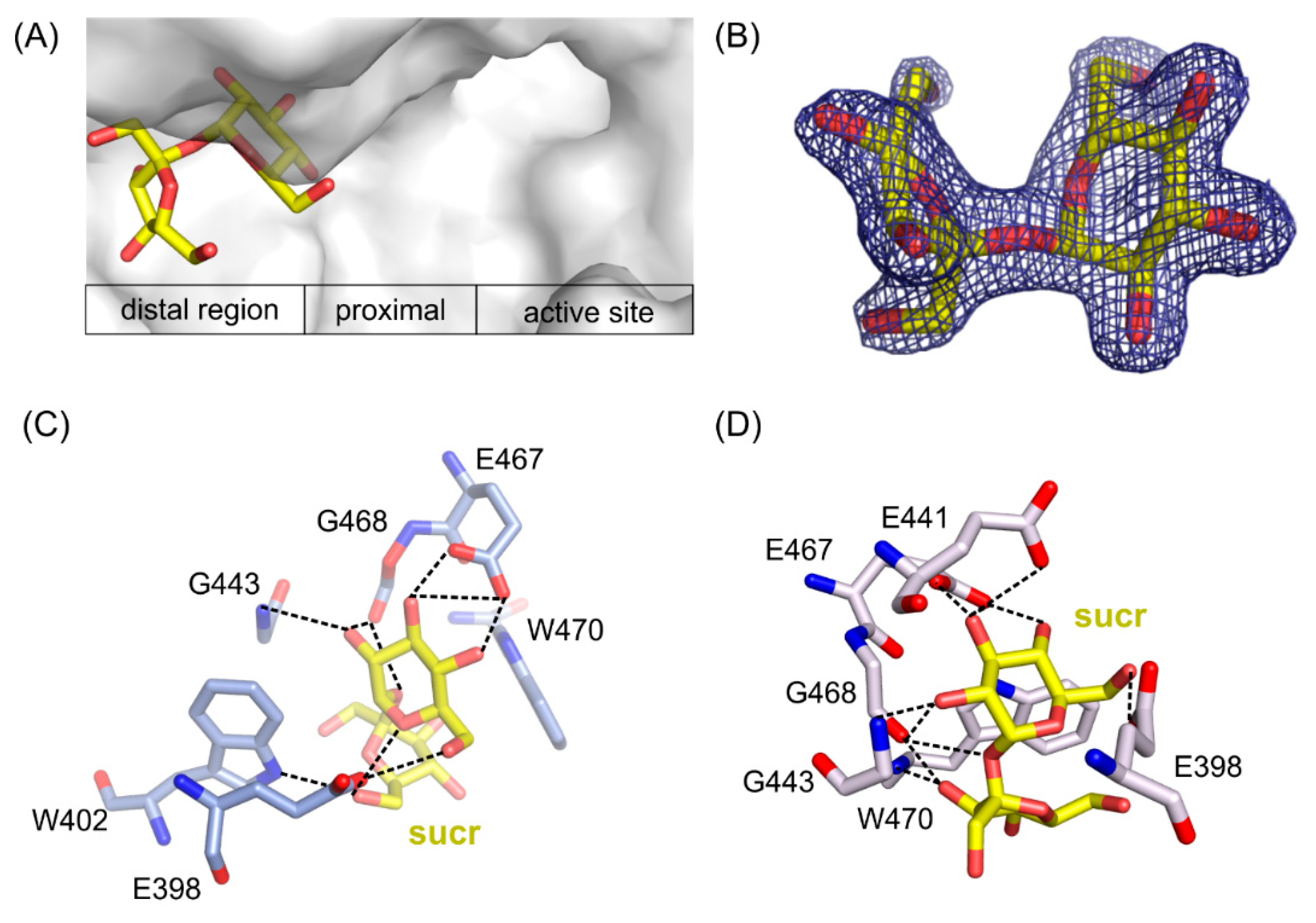
Two structures of the mutants’ complexes with sucrose, a common sugar composed of d-galoctose and d-fructose linked through a β-(1→2)-glycosidic bond, were determined with resolution 1.7 Å for ArthβDG_E441Q/suc and 2.2 Å for ArthβDG_D207A/sucr. In both cases, sucrose was bound in the same place outside of the active site of enzyme, revealing a glucopyranose binding site in its distal region (Figure 6A).
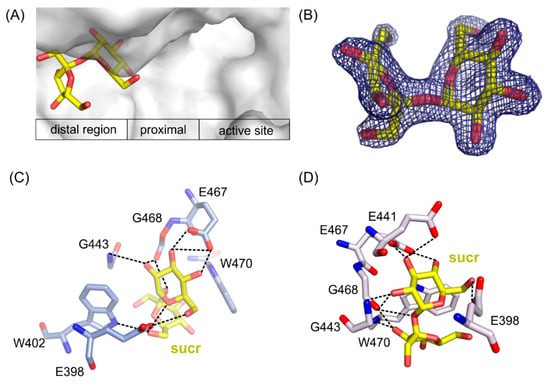
Figure 6.
Location of sucrose molecule in the distal region of the active site in ArthβDG_E441Q/sucr complex (A). 2Fo-Fc electron-density map at the 2 σ level for the saccharose molecule in ArthβDG_E441Q/sucr complex (B). Sucrose bound in the active site of ArthβDG_ E441Q/sucr (C) and ArthβDG_ D207A/sucr (D) crystal structures with hydrogen bonds marked with dashed lines.
In ArthβDG_D207A/sucr complex (Figure 6D), the sucrose molecule was stabilized predominantly by bonds between the glucosyl moiety interacting with residues E398, E467, E468, and G443. Fructosyl moiety was stabilized by only two hydrogen bonds created between hydroxyl O6’ and side chain oxygen of E398 (2.5 Å) and the same hydroxyl group O6’ with nitrogen of W402 indole ring (3.1 Å).
Sucrose molecule in complex structure of ArthβDG_E441Q/sucr moved a little toward the active site of enzyme (Figure 6C), and its hydroxyl 03 interacted via hydrogen bond with the mutated catalytic residue Q441 (3.1 Å). Nonetheless, it is stabilized by a similar net of interactions involving residues E398, E467, G443, and additionally E398. Hydroxyl group O3′ of fructosyl moiety was stabilized by a single hydrogen bond with indole ring nitrogen of W470.
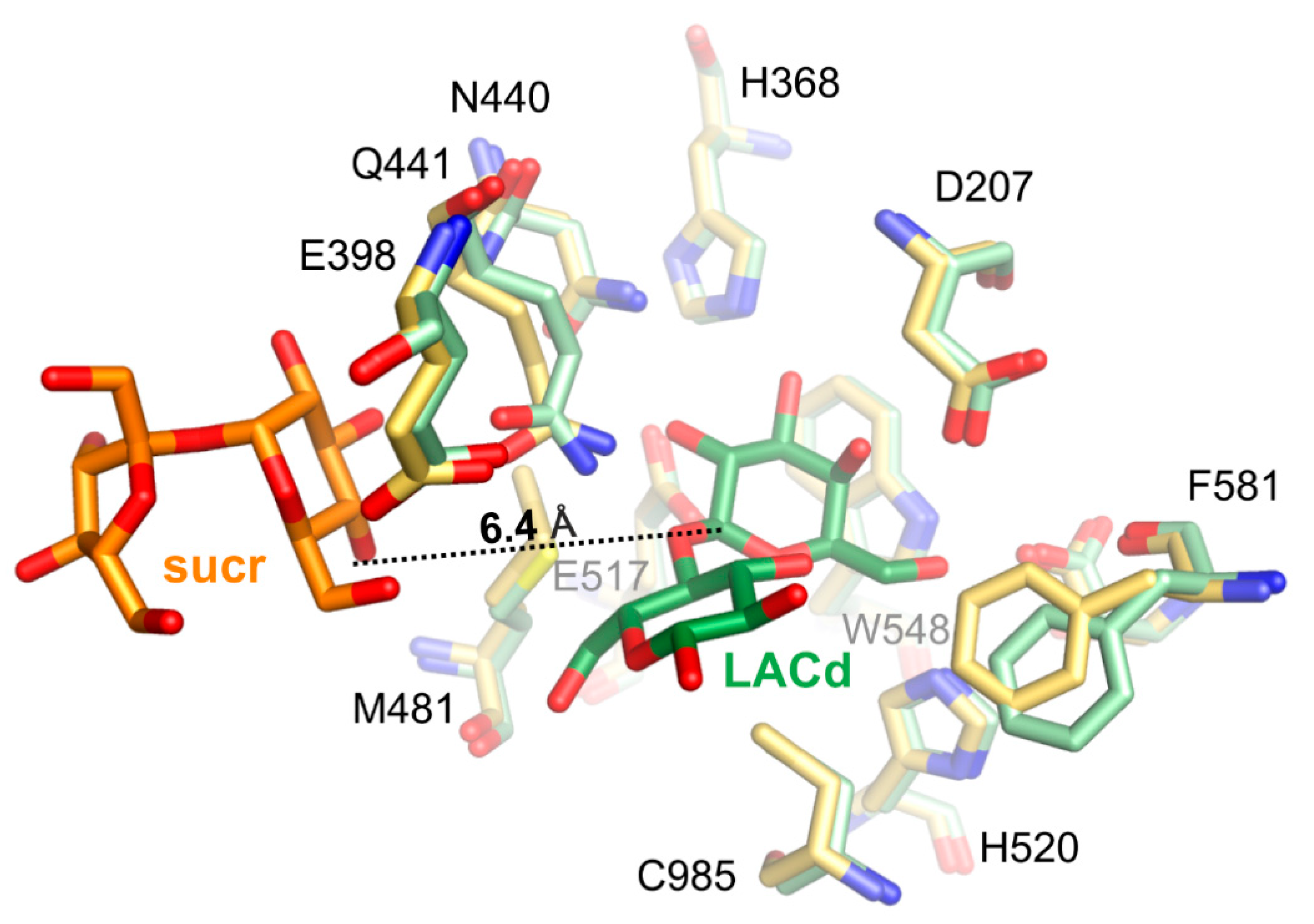
Superposition of ArthβDG_E441Q/sucr with complex structure of the same mutant with lactose bound in deep mode, ArthβDG_E441Q/LACd (PDB ID: 6SEA [51]) shows that the distance between anomeric carbon atom of galactosyl moiety bound in deep mode and hydroxyl group O4 of sucrose was 6.4 Å (Figure 7). This insight into sucrose binding in the distal region of active site shows how the glycosyl moiety is positioned in the vicinity of the active site, so it can become galactosyl group acceptor.
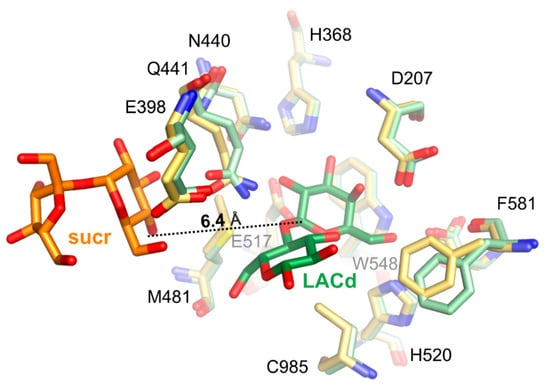
Figure 7.
Superposition of ArthβDG_E441Q/sucr (yellow/orange) with complex structure of the same mutant with lactose bound in deep mode (pale green/green), ArthβDG_E441Q/LACd (PDB ID: 6SEA), showing the positioning of sucrose molecule toward galactosyl moiety.
3. Summary
Results of activity assays and analysis of a number of complex structures of new mutants of ArthβDG, ArthβDG_D207A and ArthβDG_E517Q, resulted in a better understanding of the importance of residues D207, E441, and E517 in the native enzyme. The inactive mutant E517Q was designed specially to enable binding of transglycosylation product—lactulose, in the active site of enzyme and thus elucidating the mode of its binding. Especially surprising was the gravity of mutation of D207 to alanine, which is part of Domain 1. It is situated on the top of a part of Domain 1, which inserts itself in a space between the top of Domain 3 TIM-barrel and highly flexible C-terminal region of Domain 3, and constitutes bottom of the active site, which means that its mutation to alanine has a significant impact on active site volume. As such, not only a stabilizing interaction between D207 and O4 hydroxyl of galactosyl moiety is lost but also the transfer of substrate from shallow to deep binding site of the enzyme is disrupted.
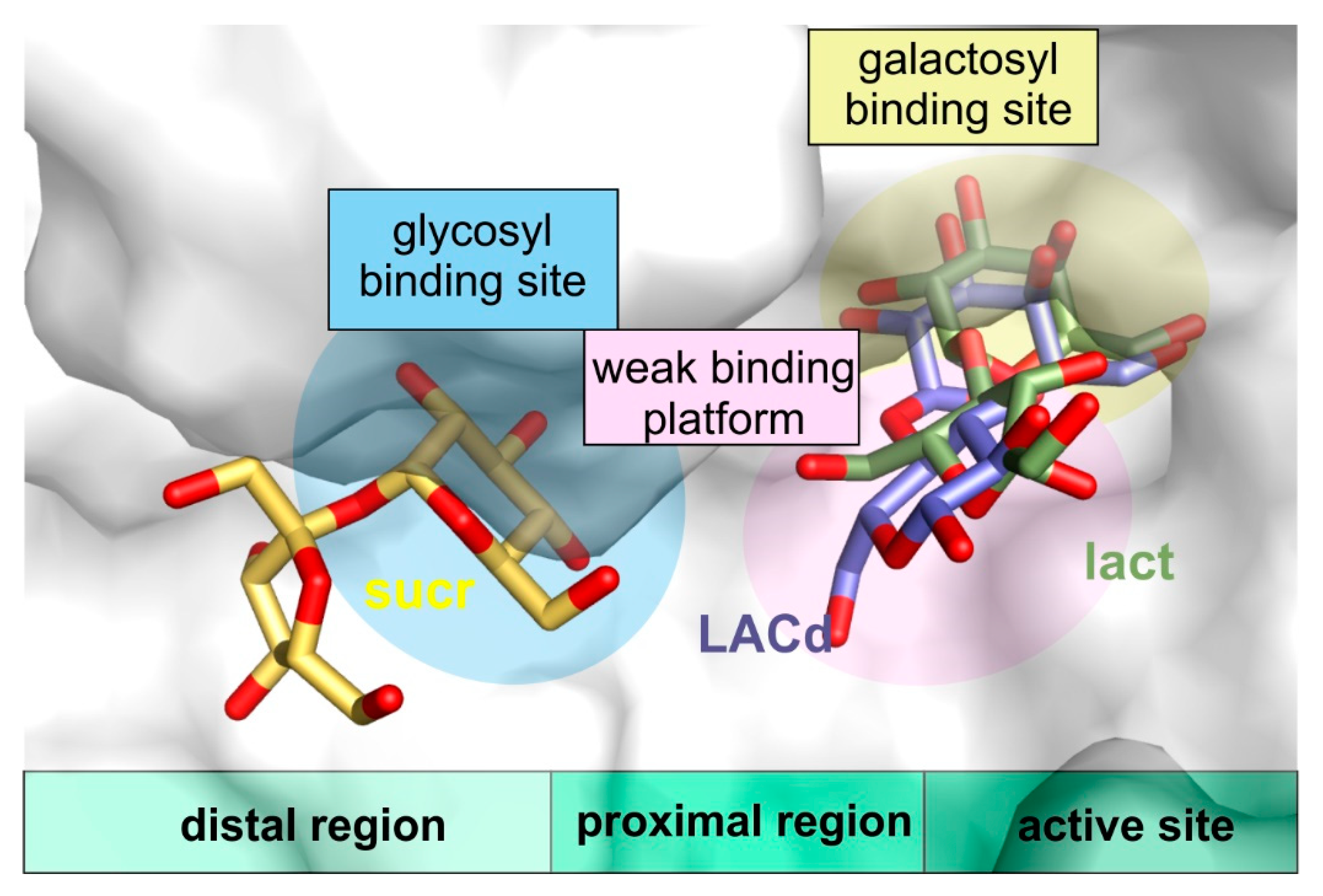
Furthermore, the structures of ArthβDG_D207A and ArthβDG_E517Q in complex with lactulose, which can be produced by ArthβDG in course of transglycosylation reaction, were, to best of our knowledge, the first crystal structures of galactosidase with this heterooligosaccharide, thus, providing a sought for insight in how a galactosyl group acceptor (larger than water) may approach and be accommodated (Figure 8). Such a detailed structural knowledge on the binding of lactulose pinpoints the residues that play role in the production of lactulose by ArthβDG, especially: N110. It also shows the region, where some additional hydrogen bonds, stabilizing fructosyl moiety, can be introduced to shift the reaction toward formation of lactulose instead of simple hydrolysis of glucose.
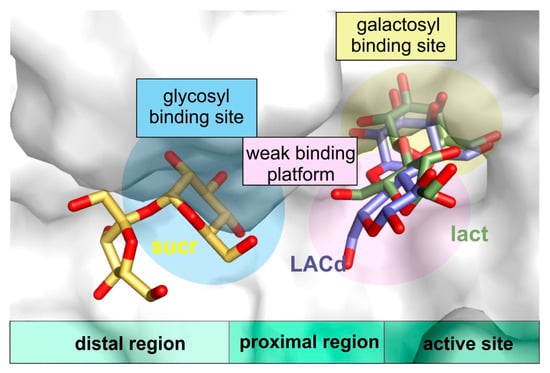
Figure 8.
The visualization of proximal region (weak binding platform) and distal region of active site (with glycosyl binding site) in superposed complex structures of ArthβDG_E441Q/sucr, ArthβDG_E441Q/LACd, and ArthβDG_E517Q/lact.
More valuable structural information that concerns transglycosylation activity of the enzyme was obtained with a solution of the complex structures with sucrose, ArthβDG_D207A/sucr and ArthβDG_E441Q/sucr. They revealed a glucopyranose binding site in the distal region of active site, showing how a galactosyl group acceptor that comprises of glucosyl moiety can approach the active site. Once again, these structures elucidate the region, mutation in which can lead to larger galactosyl group acceptor being available instead of water residue.
The results of this work can serve as a basis for knowledge-based enzyme engineering of this cold-adapted enzyme. Especially, to shift its activity toward transglycosylation reaction and improving product homogeneity. It was shown that the retention of the shape and volume of the galactosyl binding site is necessary for the enzyme’s activity. Hence, mutations, if any introduced in this region should be designed in such a way not to change the size of deep binding site, as its enlargement not only does not enhance the enzyme’s transglycosylation activity but is has a negative effect on it. It is the authors’ believe that mutations should rather be introduced in the proximal in distal regions of the active site. Especially, promoting binding of galactosyl group acceptor on the weak binding platform may prove successful in the future attempts.
Furthermore, since the architecture of the active site of galactosidases from GH2 family is highly conserved, the knowledge about the mode of binding of potential galactosyl group acceptors (larger than water) within ArthβDG and its mutant structures, can be to a certain degree transferred onto other enzymes belonging to this group, among others commonly used in the food industry β-d-galactosidase from Kluyveromyces lactis.
4. Materials and Methods
4.1. Site-Directed Mutagenesis of Gene Encoding ArthβDG
The gene encoding the ArthβDG enzyme within the pBAD-Bgal32cB plasmid [50] has been mutated using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s protocol. For this purpose, two pairs of mutagenic primers were designed and synthesized (Genomed, Warsaw, Poland). Primers For207AspAla 5′ GTGGAGGACCAGGCCATGTGGTGGCTT 3′ and Rev207AspAla 5′ GTAGCTGGCGGCCGAGAACTGGGCGA 3′ were used to introduce a point mutation A/C at 620 nucleotide position in the gene resulting in the D207A substitution in the ArthβDG amino acid sequence. Primers F32cBmut517 5′ TGGGTAACGGCCCCGGTGGAATGAGCGAAT 3′ and R32cBmut517 5′ TGGCATGCACATATTGGCAGAGGACAAAGGGCA 3′ were used to introduce a point mutation G/C at 1549 nucleotide position in the gene resulting in the E517Q substitution in the ArthβDG amino acid sequence.
Thermocycling conditions for PCRs were as follows: initial denaturation of pBAD-Bgal32cB plasmid at 98 °C for 30 s; then 25 cycles of PCR products amplification consisting of 10 s of DNA denaturation at 98 °C, 15 s of mutagenic primers annealing at 66 °C for For207AspAla and Rev207AspAla, or 69 °C for F32cBmut517 and R32cBmut517, and 3 min 20 s of PCR product extension at 72 °C; and the final extension at 72 °C for 7 min.
The obtained PCR products were treated with KLD Enzyme Mix (Kinase, Ligase, and DpnI) at 22 °C for 5 min, and then used to transform NEB 5-alpha chemically competent E.coli cells (the lacZ deletion mutant, D (lacZ) M15). Transformants were spread onto selection Luria-Bertani agar plates (10 g/L of peptone K, 5 g/L of yeast extract, 10 g/L of NaCl, and 15 g/L of agar) supplemented with ampicillin (100 μg/mL), X-gal (40 μg/mL) and l-arabinose (200 μg/mL), and incubated overnight at 37 °C and then for a few hours at room temperature.
Six colonies of E. coli + pBAD-Bgal32cB_D207A (light blue colonies with weak β-d-galactosidase activity) and E. coli + pBAD-Bgal32cB_E517Q (white colonies without enzymatic activity) were selected for further studies. Recombinant pBAD-Bgal32cB_D207A and pBAD-Bgal32cB_E517Q plasmids were isolated using the Plasmid Mini Kit (A&A Biotechnology, Gdynia, Poland) and sequenced (Genomed, Warsaw, Poland).
4.2. Production of ArthβDG, ArthβDG_D207A and ArthβDG_E517Q Proteins
Expression of recombinant ArthβDG, ArthβDG_D207A, and ArthβDG_E517Q mutants were performed in the E. coli LMG 194 cells (Invitrogen, Carlsbad, CA, USA) transformed with pBAD-Bgal32cB, pBAD-Bgal32cB_D207A and pBAD-Bgal32cB_E517Q plasmids, respectively, as previously described for wild-type protein [50] and ArthβDG_E441Q mutant [51].
Recombinant proteins were purified by a three-step chromatography comprising of ion-exchange and size-exclusion stages according to protocol developed for wild-type protein, ArthβDG [7]. The chromatography buffer was change to storage buffer (0.05 M HEPES pH 7.0) and the samples were concentrated to 20 mg/mL using 50 kDa cutoff membrane Vivaspin filters (Sartorius, Göttingen, Germany) prior to crystallization.
4.3. Determination of ArthβDG, ArthβDG_D207A and ArthβDG_E517Q Activity
The activity of ArthβDG as well as ArthβDG_D207A and ArthβDG_E517Q mutants was determined using ONPG as a substrate. 0.8 mL of ONPG solution (1 mg/mL in 20 mM potassium phosphate buffer pH 8.0) was preincubated at 28 °C for 5 min, then 0.2 mL of protein sample was added, and the reaction mixture was incubated for 1 or 10 min at 28 °C for the substrate hydrolysis. The reaction was terminated by the addition of 0.3 mL of 1 M Na2CO3 and the absorbance was measured at 410 nm. One unit of β-d-galactosidase activity was defined as the quantity of enzyme releasing of 1 μmol 2-nitrophenol per min under reaction conditions.
4.4. Thermofluor Shift Assay
Samples of ArthβDG_D207A and ArthβDG_E517Q were prepared at concentration of 0.3 mg/mL each and premixed with SYPRO. The CFX96 TouchTM (BioRad, Hercules, CA, USA) thermal cycles was used for TSA experiment. The applied temperature increment was 1 °C per 30 s, and the tested temperature ranged from 4 °C to 95 °C. The assay was performed for 24 buffers covering pH range from 4.0 to 10.0 and 24 corresponding buffers with addition of 250 mM NaCl [54].
4.5. Crystallization of ArthβDG Mutants and Obtaining Their Complexes’
Crystallization of ArthβDG_D207A, ArthβDG_E441Q and ArthβDG_E517Q was performed by diffusion hanging-drop method using TacsimateTM Hampton Research (Aliso Viejo, CA, USA) at concentration range from 27% to 45% and pH range from 4.0 to 9.0 [7]. This crystallization matrix was previously used for growing crystals of wild-type enzyme. Furthermore, the cross-seeding using seeds made of ArthβDG crystals was applied to speed up the process of obtaining diffraction quality crystals. The seed stock of crushed microcrystals diluted 10,000 times in 35% TacsimateTM pH 7.0 was premixed in 1:40 (v/v) ratio with 20 mg/mL purified protein according to previously established protocol [51]. The monocrystals suitable for diffraction experiment were obtained in a pH range of 6.4–8.8 and TacsimateTM concentration varying from 24% to 40%.
The complex ArthβDG_E517Q/gal was obtained after 24 h soaking of protein crystals with addition of globotriose and ArthβDG_D207A/gal by soaking with addition of mixture of galactose and fructose. A 24h-soaking was successful in the case of complexes ArthβDG_D207A/lact and ArthβDG_E517Q/lact, where the crystals were soaked with addition of a mixture of lactulose and galactose. The complexes of ArthβDG_E441Q and ArthβDG_D207A with saccharose were obtained by same time soaking with addition of saccharose alone.
All the crystals were cryo-protected with 60% TacsimateTM of pH corresponding to crystallization conditions prior to being flash-frozen before diffraction experiment.
4.6. Data Collection, Structure Solving, and Refinement
High-resolution diffraction data were collected using state-of-the-art BESSY II beamlines 14.1 and 14.2, Berlin, Germany [55]. The diffraction images were collected with fine slicing 0.1°. The diffraction data were processed using XDSapp [56], to avoid alternative indexing, possible in space group P3121, the log file for processing ArthβDG data was used as an input. Pair refinement was performed to determine optimal cutoff resolution for each data set. Crystal structures were solved by isomorphous replacement using rigid body refinement procedure, where the structure of ArthβDG (PDB ID: 6ETZ) was used as a model. Structure solving and further refinement was performed using the PHENIX.REFINE program [57,58] data reduction and refinement statistics are collected in Table 2 for ArthβDG_E517Q, Table 3 for ArthβDG_E441Q, and Table 4 for ArthβDG_D207A.

Table 2.
Data reduction and refinement statistics of ArthβDG_E517Q mutant’s crystal structures.

Table 3.
Data reduction and refinement statistics of ArthβDG_E441Q mutant’s crystal structures.

Table 4.
Data reduction and refinement statistics of ArthβDG_D207A mutant’s crystal structures.
4.7. Databases
The here reported crystal structures and their associated structure factor amplitudes were deposited with the Protein Data Bank under the accession codes: 6ZJP, 6ZJQ, 6ZJR, 6ZJS, 6ZJT, 6ZJU, 6ZJV, 6ZJW AND 6ZJX respectively.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/15/5354/s1, Figure S1: Selected TSA data and analysis for ArthβDG_D207A mutant, Figure S2: Selected TSA data and analysis for ArthβDG_E517Q mutant.
Author Contributions
M.R. purified enzymes, performed crystallization, synchrotron diffraction data collection, data processing, structure solving and refinement. M.R. and A.B. carried out structural analysis and mostly wrote the paper. M.W. designed and performed site-direct mutagenesis experiments resulting in genes encoding mutants, performed protein expression in E. coli, determined the purification protocol, and determined the activity of enzymes. A.B. coordinated the project. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by National Science Centre of Poland: 2016/21/B/ST5/00555 Opus 11 grant (A.B.) and 2018/28/T/ST5/00233 Etiuda 6 scholarship (M.R.).
Acknowledgments
We thank Helmholtz-Zentrum Berlin für Materialien und Energie for the allocation of synchrotron radiation beamtime at BL 14.1 and BL 14.2, where the X-ray data were collected. We gratefully acknowledge the support of the Joint Berlin MX-Laboratory, especially Manfred S. Weiss, which made this work possible.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| ArthβDG | β-d-galactosidase from Arthrobacter sp. 32cB |
| ArthβDG_D207A | β-d-galactosidase from Arthrobacter sp. 32cB mutant D207A |
| ArthβDG_E441Q | β-d-galactosidase from Arthrobacter sp. 32cB mutant E441Q |
| ArthβDG_E517Q | β-d-galactosidase from Arthrobacter sp. 32cB mutant E517Q |
| Gal | galactose |
| GOS | galactooligosaccharides |
| GH2 | glycosyl hydrolase 2 family |
| HOS | heterooligosaccharides |
| Lacd | lactose bound in deep mode |
| Lact | lactulose |
| ONPG | o-nitrophenyl-β-d-galactopyranoside |
| Sucr | sucrose |
| TSA | thermofluor shift assay |
References
- Talens-Perales, D.; Górska, A.; Huson, D.H.; Polaina, J.; Marín-Navarro, J. Analysis of Domain Architecture and Phylogenetics of Family 2 Glycoside Hydrolases (GH2). PLoS ONE 2016, 11, e0168035. [Google Scholar] [CrossRef] [PubMed]
- Białkowska, A.M.; Cieśliński, H.; Nowakowska, K.M.; Kur, J.; Turkiewicz, M. A new β-galactosidase with a low temperature optimum isolated from the Antarctic Arthrobacter sp. 20B: Gene cloning, purification and characterization. Arch. Microbiol. 2009, 191, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Juers, D.H.; Matthews, B.W.; Huber, R.E. LacZ β-galactosidase: Structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 2012, 21, 1792–1807. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.P.; Fernandez-Leiro, R.; González-Siso, M.-I.; Cerdán, M.E.; Becerra, M.; Sanz-Aparicio, J. Structural basis of specificity in tetrameric Kluyveromyces lactis β-galactosidase. J. Struct. Biol. 2012, 177, 392–401. [Google Scholar] [CrossRef]
- Skalova, T.; Dohnálek, J.; Spiwok, V.; Lipovová, P.; Vondráčková, E.; Petroková, H.; Dušková, J.; Strnad, H.; Králová, B.; Hašek, J. Cold-active β-Galactosidase from Arthrobacter sp. C2-2 Forms Compact 660kDa Hexamers: Crystal Structure at 1.9Å Resolution. J. Mol. Biol. 2005, 353, 282–294. [Google Scholar] [CrossRef]
- Rutkiewicz-Krotewicz, M.; Pietrzyk-Brzezinska, A.J.; Sekula, B.; Cieśliński, H.; Wierzbicka-Woś, A.; Kur, J.; Bujacz, A. Structural studies of a cold-adapted dimeric β-D-galactosidase from Paracoccus sp. 32d. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 1049–1061. [Google Scholar] [CrossRef]
- Rutkiewicz, M.; Pietrzyk, A.J.; Wanarska, M.; Cieśliński, H.; Bujacz, A. In Situ Random Microseeding and Streak Seeding Used for Growth of Crystals of Cold-Adapted β-d-Galactosidases: Crystal Structure of βDG from Arthrobacter sp. 32cB. Crystals 2018, 8, 13. [Google Scholar] [CrossRef]
- Brás, N.F.; Fernandes, P.A.; Ramos, M.J. QM/MM Studies on the β-Galactosidase Catalytic Mechanism: Hydrolysis and Transglycosylation Reactions. J. Chem. Theory Comput. 2010, 6, 421–433. [Google Scholar] [CrossRef]
- Juers, D.H.; Rob, B.; Dugdale, M.L.; Rahimzadeh, N.; Giang, C.; Lee, M.; Matthews, B.W.; Huber, R.E. Direct and indirect roles of His-418 in metal binding and in the activity of β-galactosidase (E. coli). Protein Sci. 2009, 18, 1281–1292. [Google Scholar] [CrossRef]
- Juers, D.H.; Heightman, T.D.; Vasella, A.; McCarter, J.D.; MacKenzie, L.; Withers, S.G.; Matthews, B.W. A structural view of the action of Escherichia coli (lacZ) beta-galactosidase. Biochemistry 2001, 40, 14781–14794. [Google Scholar] [CrossRef]
- Roberfroid, M.B. Prebiotics and probiotics: Are they functional foods? Am. J. Clin. Nutr. 2000, 71, 1682S–1687S. [Google Scholar] [CrossRef] [PubMed]
- Swennen, K.; Courtin, C.; Delcour, J. Non-digestible Oligosaccharides with Prebiotic Properties. Crit. Rev. Food Sci. Nutr. 2006, 46, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Torres, D.; Gonçalves, M.; Teixeira, J.; Rodrigues, L.R. Galacto-Oligosaccharides: Production, Properties, Applications, and Significance as Prebiotics. Compr. Rev. Food Sci. Food Saf. 2010, 9, 438–454. [Google Scholar] [CrossRef]
- Gänzle, M.G. Enzymatic synthesis of galacto-oligosaccharides and other lactose derivatives (hetero-oligosaccharides) from lactose. Int. Dairy J. 2012, 22, 116–122. [Google Scholar] [CrossRef]
- Roberfroid, M.; Gibson, G.R.; Hoyles, L.; McCartney, A.L.; Rastall, R.; Rowland, I.; Wolvers, D.; Watzl, B.; Szajewska, H.; Stahl, B.; et al. Prebiotic effects: Metabolic and health benefits. Br. J. Nutr. 2010, 104 (Suppl. S2), S1–S63. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.; Steed, H.; Macfarlane, S. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J. Appl. Microbiol. 2007, 104, 305–344. [Google Scholar] [CrossRef]
- Bujacz, A.; Jędrzejczak-Krzepkowska, M.; Bielecki, S.; Redzynia, I.; Bujacz, G.D. Crystal structures of the apo form of β-fructofuranosidase from Bifidobacterium longum and its complex with fructose. FEBS J. 2011, 278, 1728–1744. [Google Scholar] [CrossRef]
- Oliveira, L.D.; Wilbey, R.A.; Grandison, A.S.; Roseiro, L.B.; Roseiro, M.L.D.B.W. Milk oligosaccharides: A review. Int. J. Dairy Technol. 2015, 68, 305–321. [Google Scholar] [CrossRef]
- Lee, L.Y.; Bharani, R.; Biswas, A.; Lee, J.; Tran, L.-A.; Pecquet, S.; Steenhout, P. Normal growth of infants receiving an infant formula containing Lactobacillus reuteri, galacto-oligosaccharides, and fructo-oligosaccharide: A randomized controlled trial. Matern. Health Neonatol. Perinatol. 2015, 1, 9–19. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Bhandari, B.; Cichero, J.; Prakash, S. A comprehensive review on in vitro digestion of infant formula. Food Res. Int. 2015, 76, 373–386. [Google Scholar] [CrossRef]
- Musilova, S.; Rada, V.; Vlkova, E.; Bunešová, V. Beneficial effects of human milk oligosaccharides on gut microbiota. Benef. Microbes 2014, 5, 273–283. [Google Scholar] [CrossRef]
- Chierici, R.; Fanaro, S.; Saccomandi, D.; Vigi, V. Advances in the modulation of the microbial ecology of the gut in early infancy. Acta Paediatr. 2003, 92, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Orrhage, C.N.K.; Orrhage, K.; Nord, C. Factors controlling the bacterial colonization of the intestine in breastfed infants. Acta Paediatr. 1999, 88, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Kunz, C.; Rudloff, S. Biological functions of oligosaccharides in human milk. Acta Paediatr. 1993, 82, 903–912. [Google Scholar] [CrossRef]
- Li, M.; Monaco, M.H.; Wang, M.; Comstock, S.S.; Kuhlenschmidt, T.B.; Fahey, G.C., Jr.; Miller, M.J.; Kuhlenschmidt, M.S.; Donovan, S.M. Human milk oligosaccharides shorten rotavirus-induced diarrhea and modulate piglet mucosal immunity and colonic microbiota. ISME J. 2014, 8, 1609–1620. [Google Scholar] [CrossRef]
- McVeagh, P.; Brand-Miller, J. Human milk oligosaccharides: Only the breast. J. Paediatr. Child Heal. 1997, 33, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Bruggencate, S.J.T.; Bovee-Oudenhoven, I.M.; Feitsma, A.L.; Van Hoffen, E.; Schoterman, M.H. Functional role and mechanisms of sialyllactose and other sialylated milk oligosaccharides. Nutr. Rev. 2014, 72, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Wallace, T.C.; Marzorati, M.; Spence, L.; Weaver, C.M.; Williamson, P.S. New Frontiers in Fibers: Innovative and Emerging Research on the Gut Microbiome and Bone Health. J. Am. Coll. Nutr. 2017, 36, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Whisner, C.M.; Martin, B.R.; Schoterman, M.H.C.; Nakatsu, C.H.; McCabe, L.D.; McCabe, G.P.; Wastney, M.E.; Heuvel, E.V.D.; Weaver, C.M. Galacto-oligosaccharides increase calcium absorption and gut bifidobacteria in young girls: A double-blind cross-over trial. Br. J. Nutr. 2013, 110, 1292–1303. [Google Scholar] [CrossRef]
- Weaver, C.M.; Martin, B.R.; Nakatsu, C.H.; Armstrong, A.P.; Clavijo, A.; McCabe, L.D.; McCabe, G.P.; Duignan, S.; Schoterman, M.H.C.; Heuvel, E.V.D. Galactooligosaccharides Improve Mineral Absorption and Bone Properties in Growing Rats through Gut Fermentation. J. Agric. Food Chem. 2011, 59, 6501–6510. [Google Scholar] [CrossRef]
- Hughes, C.; Davoodi-Semiromi, Y.; Colee, J.C.; Culpepper, T.; Dahl, W.J.; Mai, V.; Christman, M.; Langkamp-Henken, B. Galactooligosaccharide supplementation reduces stress-induced gastrointestinal dysfunction and days of cold or flu: A randomized, double-blind, controlled trial in healthy university students. Am. J. Clin. Nutr. 2011, 93, 1305–1311. [Google Scholar] [CrossRef] [PubMed]
- Mussatto, S.I.; Mancilha, I.M. Non-digestible oligosaccharides: A review. Carbohydr. Polym. 2007, 68, 587–597. [Google Scholar] [CrossRef]
- Kim, C.S.; Ji, E.-S.; Oh, D.-K. Expression and characterization of Kluyveromyces lactis β-galactosidase in Escherichia coli. Biotechnol. Lett. 2003, 25, 1769–1774. [Google Scholar] [CrossRef] [PubMed]
- Colinas, B.R.; De Abreu, M.A.; Fernandez-Arrojo, L.; De Beer, R.; Poveda, A.; Jiménez-Barbero, J.; Haltrich, D.; Olmo, A.O.B.; Lobato, M.F.; Plou, F.J. Production of Galacto-oligosaccharides by the β-Galactosidase from Kluyveromyces lactis: Comparative Analysis of Permeabilized Cells versus Soluble Enzyme. J. Agric. Food Chem. 2011, 59, 10477–10484. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowska, A.; Klewicki, R.; Sójka, M.; Klewicka, E. Synthesis of the Galactosyl Derivative of Gluconic Acid with the Transglycosylation Activity of β-Galactosidase. Food Technol. Biotechnol. 2017, 55, 258–265. [Google Scholar] [CrossRef]
- Yin, H.; Bultema, J.B.; Dijkhuizen, L.; Van Van Leeuwen, S. Reaction kinetics and galactooligosaccharide product profiles of the β-galactosidases from Bacillus circulans, Kluyveromyces lactis and Aspergillus oryzae. Food Chem. 2017, 225, 230–238. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Siddiqui, K.S.; Andrews, D.; Sowers, K.R. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 2002, 13, 253–261. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Charlton, T.; Ertan, H.; Omar, S.M.; Siddiqui, K.S.; Williams, T.J. Biotechnological uses of enzymes from psychrophiles. Microb. Biotechnol. 2011, 4, 449–460. [Google Scholar] [CrossRef]
- Gerday, C. Psychrophily and Catalysis. Biology 2013, 2, 719–741. [Google Scholar] [CrossRef]
- Turkiewicz, M. Drobnoustroje psychrofilne i ich biotechnologiczny potencjał. Kosm. Probl. Nauk Biotechnol. 2006, 55, 307–320. [Google Scholar]
- Aghajari, N.; Haser, R.; Feller, G.; Gerday, C. Crystallization and preliminary X-ray diffraction studies of α-amylase from the antarctic psychrophile Alteromonas haloplanctis A23. Protein Sci. 1996, 5, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Arimori, T.; Ito, A.; Nakazawa, M.; Ueda, M.; Tamada, T. Crystal structure of endo-1,4-β-glucanase fromEisenia fetida. J. Synchrotron Radiat. 2013, 20, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Li, Y.; Liu, W.; Chen, C.-C.; Ko, T.-P.; He, M.; Xu, Z.; Liu, M.; Luo, H.; Guo, R.-T.; et al. Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-β-1,4-xylanase. J. Struct. Biol. 2016, 193, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-K.; An, Y.J.; Song, J.M.; Jeong, C.-S.; Kang, M.H.; Kwon, K.K.; Lee, Y.-H.; Cha, S.-S. Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-β-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus. Proteins: Struct. Funct. Bioinform. 2014, 82, 3217–3223. [Google Scholar] [CrossRef]
- Malecki, P.; Raczynska, J.E.; Vorgias, C.E.; Rypniewski, W.R. Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium. Acta Crystallogr. Sect. D Biol. Crystallogr 2013, 69, 821–829. [Google Scholar] [CrossRef]
- Zanphorlin, L.M.; De Giuseppe, P.O.; Honorato, R.; Tonoli, C.C.C.; Fattori, J.; Crespim, E.; De Oliveira, P.S.L.; Ruller, R.; Murakami, M. Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 β-glucosidase from Exiguobacterium antarcticum B7. Sci. Rep. 2016, 6, 23776. [Google Scholar] [CrossRef]
- Van Petegem, F.; Collins, T.; Meuwis, M.-A.; Gerday, C.; Feller, G.; Van Beeumen, J. The Structure of a Cold-adapted Family 8 Xylanase at 1.3 Å Resolution. J. Biol. Chem. 2002, 278, 7531–7539. [Google Scholar] [CrossRef]
- Fan, Y.; Yi, J.; Hua, X.; Feng, Y.; Yang, R.; Zhang, Y. Structure analysis of a glycosides hydrolase family 42 cold-adapted β-galactosidase from Rahnella sp. R3. RSC Adv. 2016, 6, 37362–37369. [Google Scholar] [CrossRef]
- Manas, N.H.A.; Illias, R.M.; Mahadi, N.M. Strategy in manipulating transglycosylation activity of glycosyl hydrolase for oligosaccharide production. Crit. Rev. Biotechnol. 2017, 38, 1–22. [Google Scholar]
- Pawlak-Szukalska, A.; Wanarska, M.; Popinigis, A.T.; Kur, J. A novel cold-active β-d-galactosidase with transglycosylation activity from the Antarctic Arthrobacter sp. 32cB—Gene cloning, purification and characterization. Process. Biochem. 2014, 49, 2122–2133. [Google Scholar] [CrossRef]
- Rutkiewicz, M.; Bujacz, A.; Wanarska, M.; Wierzbicka-Wos, A.; Cieśliński, H. Active Site Architecture and Reaction Mechanism Determination of Cold Adapted β-d-galactosidase from Arthrobacter sp. 32cB. Int. J. Mol. Sci. 2019, 20, 4301. [Google Scholar] [CrossRef] [PubMed]
- Rutkiewicz, M.; Bujacz, A.; Bujacz, G. Structural features of cold-adapted dimeric GH2 β-D-galactosidase from Arthrobacter sp. 32cB. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2019, 1867, 776–786. [Google Scholar] [CrossRef] [PubMed]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2017, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, L.; Mayerhofer, H.; Geerlof, A.; Mueller-Dieckmann, J.; Weiss, M.S. Optimization of protein buffer cocktails using Thermofluor. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 209–214. [Google Scholar] [CrossRef]
- Mueller, U.; Forster, R.; Hellmig, M.; Huschmann, F.U.; Kastner, A.; Malecki, P.; Pühringer, S.; Röwer, M.; Sparta, K.; Steffien, M.; et al. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. Eur. Phys. J. Plus 2015, 130, 141. [Google Scholar] [CrossRef]
- Sparta, K.; Krug, M.; Heinemann, U.; Mueller, U.; Weiss, M.S. XDSAPP2.0. J. Appl. Crystallogr. 2016, 49, 1085–1092. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–2211. [Google Scholar] [CrossRef]
- Moriarty, N.W.; Tronrud, D.E.; Adams, P.D.; Karplus, P.A. A new default restraint library for the protein backbone inPhenix: A conformation-dependent geometry goes mainstream. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 176–179. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).