d-Amino Acids in Plants: Sources, Metabolism, and Functions
Abstract
1. Introduction
2. d-AA Transport in Plants: They Get In and They Get Out, but How?
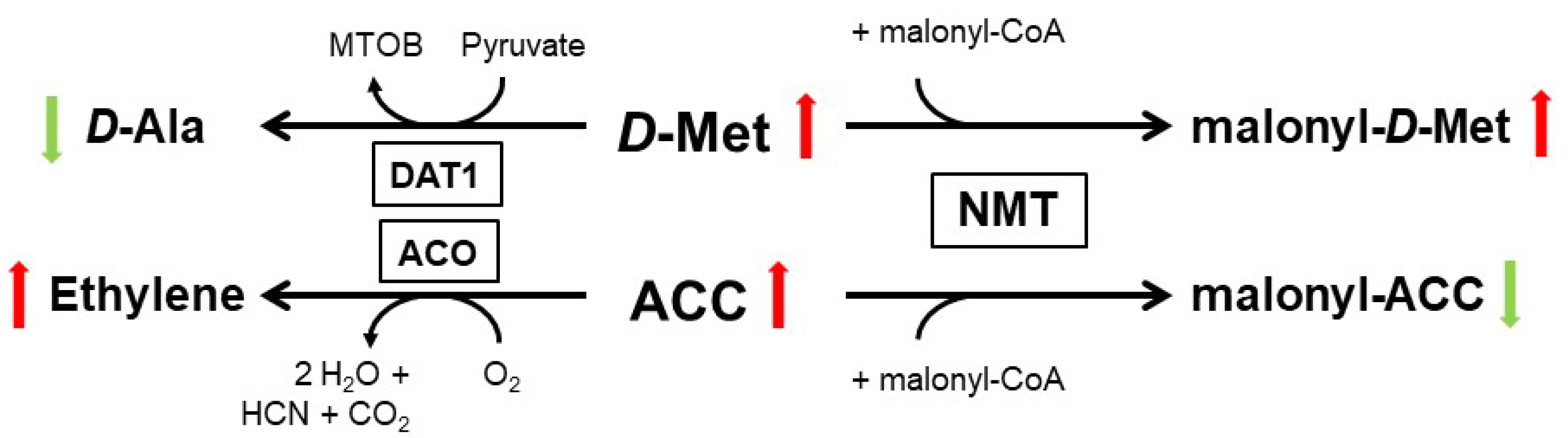
3. D-AA Metabolism in Plants: Many Ways to Handle
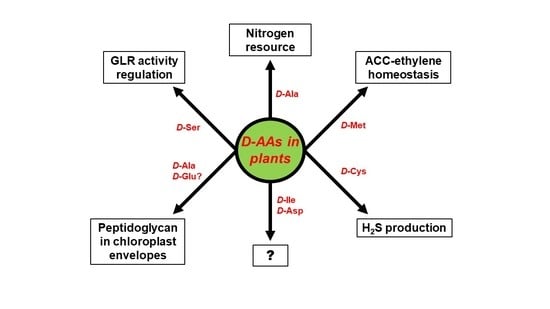
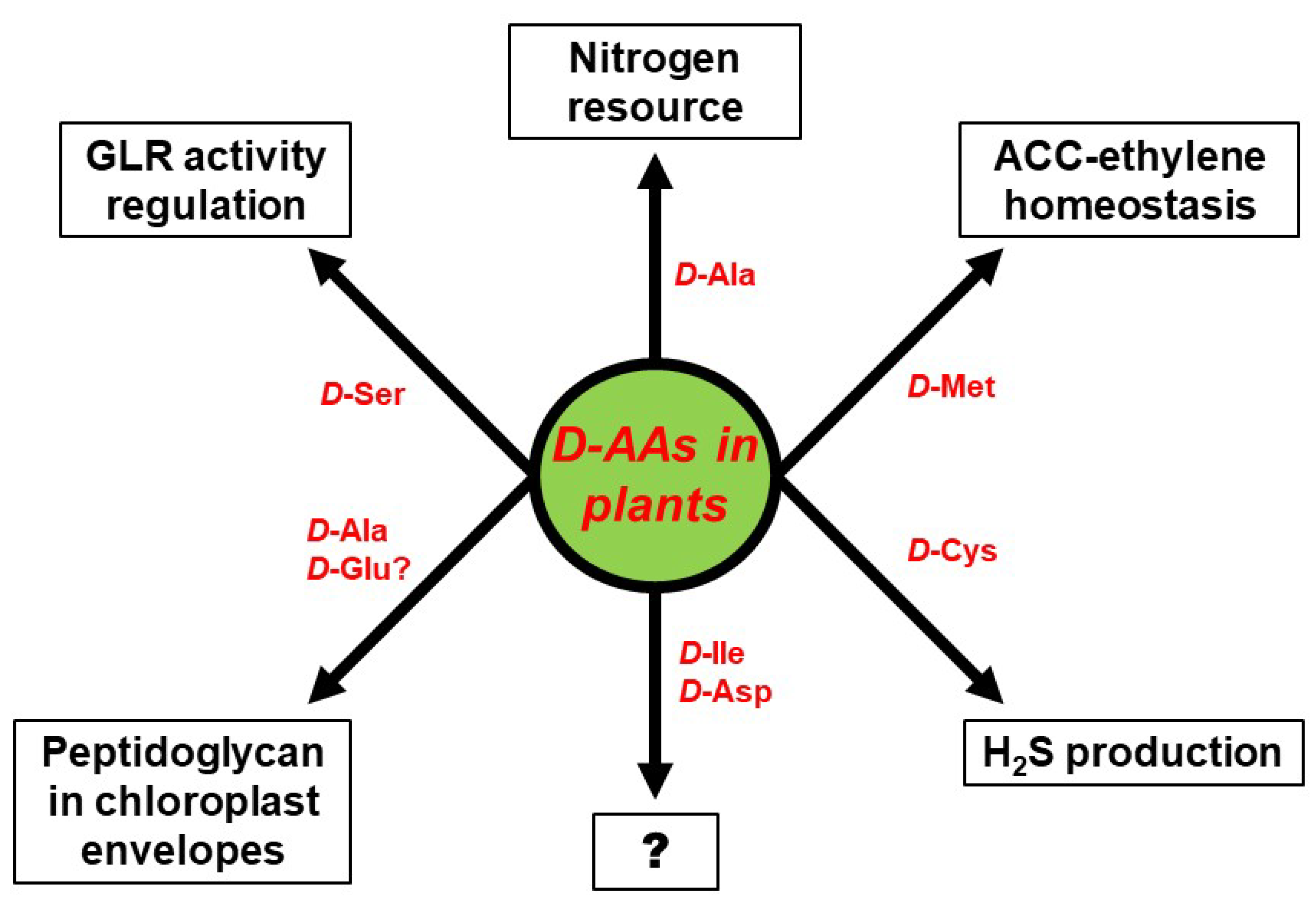
4. Physiological Functions of d-AAs in Plants: Are D-AAs Just Another Source of Nitrogen, or More?
5. Conclusions: Open Questions Galore
Funding
Acknowledgments
Conflicts of Interest
References
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2011, 10, 123–136. [Google Scholar] [CrossRef]
- Irazoki, O.; Hernandez, S.B.; Cava, F. Peptidoglycan muropeptides: Release, perception, and functions as signaling molecules. Front. Microbiol. 2019, 10, 500. [Google Scholar] [CrossRef] [PubMed]
- Ollivaux, C.; Soyez, D.; Toullec, J.Y. Biogenesis of d-amino acid containing peptides/proteins: Where, when and how? J. Pept. Sci. 2014, 20, 595–612. [Google Scholar] [CrossRef] [PubMed]
- Grishin, D.V.; Zhdanov, D.D.; Pokrovskaya, M.V.; Sokolov, N.N. d-amino acids in nature, agriculture and biomedicine. All Life 2019, 13, 11–22. [Google Scholar] [CrossRef]
- Genchi, G. An overview on d-amino acids. Amino Acids 2017, 49, 1521–1533. [Google Scholar] [CrossRef]
- Sasabe, J.; Suzuki, M. Emerging role of d-Amino acid metabolism in the innate defense. Front. Microbiol. 2018, 9, 933. [Google Scholar] [CrossRef]
- Martinez-Rodriguez, S.; Martinez-Gomez, A.I.; Rodriguez-Vico, F.; Clemente-Jimenez, J.M.; Las Heras-Vazquez, F.J. Natural occurrence and industrial applications of d-amino acids: An overview. Chem. Biodivers. 2010, 7, 1531–1548. [Google Scholar] [CrossRef]
- Matsumoto, M.; Kunisawa, A.; Hattori, T.; Kawana, S.; Kitada, Y.; Tamada, H.; Kawano, S.; Hayakawa, Y.; Iida, J.; Fukusaki, E. Free d-amino acids produced by commensal bacteria in the colonic lumen. Sci. Rep. 2018, 8, 17915. [Google Scholar] [CrossRef]
- Inselsbacher, E.; Nasholm, T. The below-ground perspective of forest plants: Soil provides mainly organic nitrogen for plants and mycorrhizal fungi. New Phytol. 2012, 195, 329–334. [Google Scholar] [CrossRef]
- Vranova, V.; Zahradnickova, H.; Janous, D.; Skene, K.R.; Matharu, A.S.; Rejsek, K.; Formanek, P. The significance of d-amino acids in soil, fate and utilization by microbes and plants: Review and identification of knowledge gaps. Plant Soil 2012, 354, 21–39. [Google Scholar] [CrossRef]
- Serralta-Interian, A.A.; Miranda-Ham, M.D.L.; Echevarría-Machado, I. Stimulation of root growth and enhanced nitrogenous metabolite content in habanero pepper (Capsicum chinense Jacq.) treated with a d-amino acid mixture. Theor. Exp. Plant Physiol. 2020, 32, 31–47. [Google Scholar] [CrossRef]
- Erikson, O.; Hertzberg, M.; Nasholm, T. A conditional marker gene allowing both positive and negative selection in plants. Nat. Biotechnol. 2004, 22, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Erikson, O.; Hertzberg, M.; Nasholm, T. The dsdA gene from Escherichia coli provides a novel selectable marker for plant transformation. Plant Mol. Biol. 2005, 57, 425–433. [Google Scholar] [CrossRef]
- Gordes, D.; Kolukisaoglu, U.; Thurow, K. Uptake and conversion of d-amino acids in Arabidopsis thaliana. Amino Acids 2011, 40, 553–563. [Google Scholar] [CrossRef]
- Gordes, D.; Koch, G.; Thurow, K.; Kolukisaoglu, U. Analyses of Arabidopsis ecotypes reveal metabolic diversity to convert d-amino acids. Springerplus 2013, 2, 559. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hill, P.W.; Quilliam, R.S.; DeLuca, T.H.; Farrar, J.; Farrell, M.; Roberts, P.; Newsham, K.K.; Hopkins, D.W.; Bardgett, R.D.; Jones, D.L. Acquisition and assimilation of nitrogen as peptide-bound and d-enantiomers of amino acids by wheat. PLoS ONE 2011, 6, e19220. [Google Scholar] [CrossRef]
- Forsum, O.; Svennerstam, H.; Ganeteg, U.; Nasholm, T. Capacities and constraints of amino acid utilization in Arabidopsis. New Phytol. 2008, 179, 1058–1069. [Google Scholar]
- Hirner, A.; Ladwig, F.; Stransky, H.; Okumoto, S.; Keinath, M.; Harms, A.; Frommer, W.B.; Koch, W. Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 2006, 18, 1931–1946. [Google Scholar] [CrossRef]
- Svennerstam, H.; Ganeteg, U.; Bellini, C.; Nasholm, T. Comprehensive screening of Arabidopsis mutants suggests the lysine histidine transporter 1 to be involved in plant uptake of amino acids. Plant Physiol. 2007, 143, 1853–1860. [Google Scholar] [CrossRef]
- Choi, J.; Eom, S.; Shin, K.; Lee, R.A.; Choi, S.; Lee, J.H.; Lee, S.; Soh, M.S. Identification of lysine histidine transporter 2 as an 1-aminocyclopropane carboxylic acid transporter in Arabidopsis thaliana by transgenic complementation approach. Front. Plant Sci. 2019, 10, 1092. [Google Scholar] [CrossRef]
- Lee, Y.H.; Foster, J.; Chen, J.; Voll, L.M.; Weber, A.P.; Tegeder, M. AAP1 transports uncharged amino acids into roots of Arabidopsis. Plant J. 2007, 50, 305–319. [Google Scholar] [CrossRef] [PubMed]
- Kolukisaoglu, Ü.; Suarez, J. d-Amino acids in plants: New insights and aspects, but also more open questions. In Amino Acid-New Insights and Roles in Plant and Animal; Asao, T., Asaduzzaman, M., Eds.; IntechOpen: London, UK, 2017; pp. 155–164. [Google Scholar]
- Lehmann, S.; Gumy, C.; Blatter, E.; Boeffel, S.; Fricke, W.; Rentsch, D. In planta function of compatible solute transporters of the AtProT family. J. Exp. Bot. 2011, 62, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Boll, M.; Foltz, M.; Anderson, C.M.; Oechsler, C.; Kottra, G.; Thwaites, D.T.; Daniel, H. Substrate recognition by the mammalian proton-dependent amino acid transporter PAT1. Mol. Membr. Biol. 2003, 20, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Foster, A.C.; Farnsworth, J.; Lind, G.E.; Li, Y.X.; Yang, J.Y.; Dang, V.; Penjwini, M.; Viswanath, V.; Staubli, U.; Kavanaugh, M.P. d-Serine is a substrate for neutral amino acid transporters ASCT1/SLC1A4 and ASCT2/SLC1A5, and is transported by both subtypes in rat hippocampal astrocyte cultures. PLoS ONE 2016, 11, e0156551. [Google Scholar] [CrossRef]
- Vollero, A.; Imperiali, F.G.; Cinquetti, R.; Margheritis, E.; Peres, A.; Bossi, E. The d-amino acid transport by the invertebrate SLC6 transporters KAAT1 and CAATCH1 from Manduca sexta. Physiol. Rep. 2016, 4. [Google Scholar] [CrossRef]
- Dinkeloo, K.; Boyd, S.; Pilot, G. Update on amino acid transporter functions and on possible amino acid sensing mechanisms in plants. Semin. Cell Dev. Biol. 2018, 74, 105–113. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interations with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Bunger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots shaping their microbiome: Global hotspots for microbial activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef]
- Baetz, U.; Martinoia, E. Root exudates: The hidden part of plant defense. Trends Plant Sci. 2014, 19, 90–98. [Google Scholar] [CrossRef]
- Hener, C.; Hummel, S.; Suarez, J.; Stahl, M.; Kolukisaoglu, U. d-amino acids are exuded by Arabidopsis thaliana roots to the rhizosphere. Int. J. Mol. Sci. 2018, 19, 1109. [Google Scholar] [CrossRef]
- Naveed, M.; Brown, L.K.; Raffan, A.C.; George, T.S.; Bengough, A.G.; Roose, T.; Sinclair, I.; Koebernick, N.; Cooper, L.; Hackett, C.A.; et al. Plant exudates may stabilize or weaken soil depending on species, origin and time. Eur. J. Soil Sci. 2017, 68, 806–816. [Google Scholar] [CrossRef] [PubMed]
- Allard-Massicotte, R.; Tessier, L.; Lecuyer, F.; Lakshmanan, V.; Lucier, J.F.; Garneau, D.; Caudwell, L.; Vlamakis, H.; Bais, H.P.; Beauregard, P.B. Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. mBio 2016, 7. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 2013, 8, e55731. [Google Scholar] [CrossRef]
- Zhou, D.; Huang, X.-F.; Chaparro, J.M.; Badri, D.V.; Manter, D.K.; Vivanco, J.M.; Guo, J. Root and bacterial secretions regulate the interaction between plants and PGPR leading to distinct plant growth promotion effects. Plant Soil 2016, 401, 259–272. [Google Scholar]
- Kawasaki, A.; Donn, S.; Ryan, P.R.; Mathesius, U.; Devilla, R.; Jones, A.; Watt, M. Microbiome and exudates of the root and rhizosphere of Brachypodium distachyon, a model for wheat. PLoS ONE 2016, 11, e0164533. [Google Scholar] [CrossRef] [PubMed]
- Badri, D.V.; Loyola-Vargas, V.M.; Broeckling, C.D.; De-la-Pena, C.; Jasinski, M.; Santelia, D.; Martinoia, E.; Sumner, L.W.; Banta, L.M.; Stermitz, F.; et al. Altered profile of secondary metabolites in the root exudates of Arabidopsis ATP-binding cassette transporter mutants. Plant Physiol. 2008, 146, 762–771. [Google Scholar] [CrossRef]
- Badri, D.V.; Quintana, N.; El Kassis, E.G.; Kim, H.K.; Choi, Y.H.; Sugiyama, A.; Verpoorte, R.; Martinoia, E.; Manter, D.K.; Vivanco, J.M. An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 2009, 151, 2006–2017. [Google Scholar] [CrossRef]
- Verrier, P.J.; Bird, D.; Burla, B.; Dassa, E.; Forestier, C.; Geisler, M.; Klein, M.; Kolukisaoglu, U.; Lee, Y.; Martinoia, E.; et al. Plant ABC proteins—A unified nomenclature and updated inventory. Trends Plant Sci. 2008, 13, 151–159. [Google Scholar] [CrossRef]
- Funakoshi, M.; Sekine, M.; Katane, M.; Furuchi, T.; Yohda, M.; Yoshikawa, T.; Homma, H. Cloning and functional characterization of Arabidopsis thaliana D-amino acid aminotransferase--d-aspartate behavior during germination. FEBS J. 2008, 275, 1188–1200. [Google Scholar] [CrossRef]
- Suarez, J.; Hener, C.; Lehnhardt, V.A.; Hummel, S.; Stahl, M.; Kolukisaoglu, U. AtDAT1 Is a Key Enzyme of d-Amino Acid Stimulated Ethylene Production in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1609. [Google Scholar] [CrossRef]
- Fujitani, Y.; Horiuchi, T.; Ito, K.; Sugimoto, M. Serine racemases from barley, Hordeum vulgare L., and other plant species represent a distinct eukaryotic group: Gene cloning and recombinant protein characterization. Phytochemistry 2007, 68, 1530–1536. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fujitani, Y.; Nakajima, N.; Ishihara, K.; Oikawa, T.; Ito, K.; Sugimoto, M. Molecular and biochemical characterization of a serine racemase from Arabidopsis thaliana. Phytochemistry 2006, 67, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Gogami, Y.; Ito, K.; Kamitani, Y.; Matsushima, Y.; Oikawa, T. Occurrence of d-serine in rice and characterization of rice serine racemase. Phytochemistry 2009, 70, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Song, Y.; Wang, C.; Sun, J.; Wang, L.; Cheng, B.; Fan, J. Crystal structure of maize serine racemase with pyridoxal 5’-phosphate. Acta Crystallogr. F Struct. Biol. Commun. 2016, 72, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Uda, K.; Edashige, Y.; Nishimura, R.; Shikano, Y.; Matsui, T.; Radkov, A.D.; Moe, L.A. Distribution and evolution of the serine/aspartate racemase family in plants. Phytochemistry 2020, 169, 112164. [Google Scholar] [CrossRef] [PubMed]
- Strauch, R.C.; Svedin, E.; Dilkes, B.; Chapple, C.; Li, X. Discovery of a novel amino acid racemase through exploration of natural variation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 11726–11731. [Google Scholar] [CrossRef]
- Gholizadeh, A.; Kohnehrouz, B.B. Molecular cloning and expression in Escherichia coli of an active fused Zea mays L. d-amino acid oxidase. Biochemistry (Mosc.) 2009, 74, 137–144. [Google Scholar] [CrossRef]
- Hou, Z.; Wang, L.; Liu, J.; Hou, L.; Liu, X. Hydrogen sulfide regulates ethylene-induced stomatal closure in Arabidopsis thaliana. J. Integr. Plant Biol. 2013, 55, 277–289. [Google Scholar] [CrossRef]
- Riemenschneider, A.; Wegele, R.; Schmidt, A.; Papenbrock, J. Isolation and characterization of a d-cysteine desulfhydrase protein from Arabidopsis thaliana. FEBS J. 2005, 272, 1291–1304. [Google Scholar] [CrossRef]
- Zhou, H.; Guan, W.; Zhou, M.; Shen, J.; Liu, X.; Wu, D.; Yin, X.; Xie, Y. Cloning and Characterization of a gene Encoding True d-cysteine Desulfhydrase from Oryza sativa. Plant Mol. Biol. Rep. 2019, 38, 95–113. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Y.; Li, X.; Xue, R.; Zhao, H. Identification of Wheat d-Cysteine Desulfhydrase (TaD-CDes) Required for Abscisic Acid Regulation of Seed Germination, Root Growth, and Stomatal Closure in Arabidopsis. J. Plant Growth Regul. 2018, 37, 1175–1184. [Google Scholar] [CrossRef]
- Hirano, T.; Tanidokoro, K.; Shimizu, Y.; Kawarabayasi, Y.; Ohshima, T.; Sato, M.; Tadano, S.; Ishikawa, H.; Takio, S.; Takechi, K.; et al. Moss chloroplasts are surrounded by a peptidoglycan wall containing d-amino acids. Plant Cell 2016, 28, 1521–1532. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Yanagida, K.; Oikawa, T.; Ogawa, T.; Soda, K. Alanine racemase of alfalfa seedlings (Medicago sativa L.): First evidence for the presence of an amino acid racemase in plants. Phytochemistry 2006, 67, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Tomoda, Y.; Nakamoto, Y.; Kawada, T.; Ishii, Y.; Nagata, Y. Alanine racemase from the green alga Chlamydomonas reinhardtii. Amino Acids 2007, 32, 59–62. [Google Scholar] [CrossRef]
- Pollegioni, L.; Piubelli, L.; Sacchi, S.; Pilone, M.S.; Molla, G. Physiological functions of d-amino acid oxidases: From yeast to humans. Cell Mol. Life Sci. 2007, 64, 1373–1394. [Google Scholar] [CrossRef]
- Noma, M.; Noguchi, M.; Tamaki, E. Isolation and Characterization of d-Alanyl-d-alanine from Tobacco Leaves. Agric. Biol. Chem. 1973, 37, 2439. [Google Scholar] [CrossRef]
- Manabe, H. Formation of Dipeptides Containing d-Alanine in wild rice plants. Phytochemistry 1991, 31, 527–529. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Ogawa, T.; Sasaoka, K. Occurence and some properties of a novel γ-Glutamyltransferase responsible for the synthesis of γ-L-Glutamine-d-Alanine in pea seedlings. Biochim. Biophys. Acta 1982, 716, 194–200. [Google Scholar] [CrossRef]
- Robinson, T. d-amino acids in higher plants. Life Sci. 1976, 19, 1097–1102. [Google Scholar] [CrossRef]
- Matilla, A.J.; Gomez-Jimenez, M.C. Metabolism of malonyl-ACC in highr plants. Recent Res. Devel. Phytochem. 2001, 5, 87–94. [Google Scholar]
- Bruckner, H.; Westhauser, T. Chromatographic determination of L- and d-amino acids in plants. Amino Acids 2003, 24, 43–55. [Google Scholar] [CrossRef]
- Michard, E.; Lima, P.T.; Borges, F.; Silva, A.C.; Portes, M.T.; Carvalho, J.E.; Gilliham, M.; Liu, L.H.; Obermeyer, G.; Feijo, J.A. Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil d-serine. Science 2011, 332, 434–437. [Google Scholar] [CrossRef] [PubMed]
- Balu, D.T.; Coyle, J.T. The NMDA receptor ‘glycine modulatory site’ in schizophrenia: d-serine, glycine, and beyond. Curr. Opin. Pharmacol. 2015, 20, 109–115. [Google Scholar] [CrossRef]
- Coyle, J.T.; Balu, D.; Wolosker, H. d-serine, the shape-shifting NMDA receptor co-agonist. Neurochem. Res. 2020, 45, 1344–1353. [Google Scholar] [CrossRef] [PubMed]
- Satoh, S.; Esashi, Y. d-amino-acid-stimulated ethylene production in seed tissues. Planta 1980, 149, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Satoh, S.; Esashi, Y. d-amino-acid-stimulated ethylene production: Molecular requirements for the stimulation and a possible receptor site. Phytochemistry 1981, 20, 947–949. [Google Scholar] [CrossRef]
- Rodrigues, M.A.; Bianchetti, R.E.; Freschi, L. Shedding light on ethylene metabolism in higher plants. Front. Plant Sci. 2014, 5, 665. [Google Scholar] [CrossRef]
- van de Poel, B.; van der Straeten, D. 1-aminocyclopropane-1-carboxylic acid (ACC) in plants: More than just the precursor of ethylene! Front. Plant Sci. 2014, 5, 640. [Google Scholar] [CrossRef]
- Houben, M.; van de Poel, B. 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (ACO): The enzyme that makes the plant hormone ethylene. Front. Plant Sci. 2019, 10, 695. [Google Scholar] [CrossRef]
- Li, Z.G.; Min, X.; Zhou, Z.H. Hydrogen sulfide: A signal molecule in plant cross-adaptation. Front. Plant Sci. 2016, 7, 1621. [Google Scholar] [CrossRef]
- Jin, Z.; Shen, J.; Qiao, Z.; Yang, G.; Wang, R.; Pei, Y. Hydrogen sulfide improves drought resistance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2011, 414, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Shivaraj, S.M.; Vats, S.; Bhat, J.A.; Dhakte, P.; Goyal, V.; Khatri, P.; Kumawat, S.; Singh, A.; Prasad, M.; Sonah, H.; et al. Nitric oxide and hydrogen sulfide crosstalk during heavy metal stress in plants. Physiol. Plant 2020, 168, 437–455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cai, W.; Ji, T.T.; Ye, L.; Lu, Y.T.; Yuan, T.T. WRKY13 Enhances Cadmium Tolerance by Promoting d-cysteine desulfhydrase and Hydrogen Sulfide Production. Plant Physiol. 2020, 183, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Machida, M.; Takechi, K.; Sato, H.; Chung, S.J.; Kuroiwa, H.; Takio, S.; Seki, M.; Shinozaki, K.; Fujita, T.; Hasebe, M.; et al. Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss. Proc. Natl. Acad. Sci. USA 2006, 103, 6753–6758. [Google Scholar] [CrossRef]
- van Baren, M.J.; Bachy, C.; Reistetter, E.N.; Purvine, S.O.; Grimwood, J.; Sudek, S.; Yu, H.; Poirier, C.; Deerinck, T.J.; Kuo, A.; et al. Evidence-based green algal genomics reveals marine diversity and ancestral characteristics of land plants. BMC Genom. 2016, 17, 267. [Google Scholar] [CrossRef]
- Woodward, A.W.; Bartel, B. Biology in Bloom: A Primer on the Arabidopsis thaliana Model System. Genetics 2018, 208, 1337–1349. [Google Scholar] [CrossRef]
- Bjorn, L.O. Peptidoglycan in eukaryotes: Unanswered questions. Phytochemistry 2020, 175, 112370. [Google Scholar] [CrossRef]
- Izumi, Y.; Kuroki, J.; Nagafuji, H.; Lin, X.; Takano, H. Effects of antibiotics that inhibit bacterial peptidoglycan synthesis on plastid division in pteridophytes. Cytologia 2008, 73, 393–400. [Google Scholar] [CrossRef]
- Izumi, Y.; Ono, K.; Takano, H. Inhibition of plastid division by ampicillin in the pteridophyte Selaginella nipponica Fr. et Sav. Plant Cell Physiol. 2003, 44, 183–189. [Google Scholar] [CrossRef]
- Katayama, N.; Takano, H.; Sugiyama, M.; Takio, S.; Sakai, A.; Tanaka, K.; Kuroiwa, H.; Ono, K. Effects of antibiotics that inhibit the bacterial peptidoglycan synthesis pathway on moss chloroplast division. Plant Cell Physiol. 2003, 44, 776–781. [Google Scholar] [CrossRef]
- Hillis, D.G.; Fletcher, J.; Solomon, K.R.; Sibley, P.K. Effects of ten antibiotics on seed germination and root elongation in three plant species. Arch. Environ. Contam. Toxicol. 2011, 60, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Minden, V.; Deloy, A.; Volkert, A.M.; Leonhardt, S.D.; Pufal, G. Antibiotics impact plant traits, even at small concentrations. AoB Plants 2017, 9, plx010. [Google Scholar] [CrossRef] [PubMed]
- Gudiño, M.E.; Blanco-Touriñán, N.; Arbona, V.; Gómez-Cadenas, A.; Blázquez, M.A.; Navarro-García, F. β-Lactam antibiotics modify root architecture and indole glucosinolate metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 2086–2098. [Google Scholar] [PubMed]
- Opris, O.; Copaciu, F.; Loredana Soran, M.; Ristoiu, D.; Niinemets, U.; Copolovici, L. Influence of nine antibiotics on key secondary metabolites and physiological characteristics in Triticum aestivum: Leaf volatiles as a promising new tool to assess toxicity. Ecotoxicol. Environ. Saf. 2013, 87, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Li, N.; Kudo, H.; Zhang, Z.; Li, J.; Wang, L.; Zhang, W.; Takechi, K.; Takano, H. Genes sufficient for synthesizing peptidoglycan are retained in gymnosperm genomes, and MurE from Larix gmelinii can rescue the albino phenotype of Arabidopsis MurE mutation. Plant Cell Physiol. 2017, 58, 587–597. [Google Scholar] [CrossRef]
- Radkov, A.D.; McNeill, K.; Uda, K.; Moe, L.A. d-amino acid catabolism is common among soil-dwelling bacteria. Microbes Environ. 2016, 31, 165–168. [Google Scholar] [CrossRef]
- Kolodkin-Gal, I.; Romero, D.; Cao, S.; Clardy, J.; Kolter, R.; Losick, R. d-amino acids trigger biofilm disassembly. Science 2010, 328, 627–629. [Google Scholar] [CrossRef]
- Forde, B.G.; Roberts, M.R. Glutamate receptor-like channels in plants: A role as amino acid sensors in plant defence? F1000Prime Rep. 2014, 6, 37. [Google Scholar] [CrossRef]
- Alfieri, A.; Doccula, F.G.; Pederzoli, R.; Grenzi, M.; Bonza, M.C.; Luoni, L.; Candeo, A.; Romano Armada, N.; Barbiroli, A.; Valentini, G.; et al. The structural bases for agonist diversity in an Arabidopsis thaliana glutamate receptor-like channel. Proc. Natl. Acad. Sci. USA 2020, 117, 752–760. [Google Scholar] [CrossRef]
- Wudick, M.M.; Michard, E.; Oliveira Nunes, C.; Feijo, J.A. Comparing plant and animal glutamate receptors: Common traits but different fates? J. Exp. Bot. 2018, 69, 4151–4163. [Google Scholar] [CrossRef]

| Proteins | EC No. | Reactions | Substrates | References |
|---|---|---|---|---|
| AtDAT1 | 2.6.1.21 | d-AA + pyruvate/2-OG → Keto acid + d-Ala/d-Glu (d-AA transamination) | d-Met (preferred) and several other d-AAs | [40,41] |
| Ser racemases (SerR) 1 | 5.1.1.10 | L-Ser → d-Ser/d-Ser → L-Ser (Ser racemization) d-Ser/L-Ser → Pyruvate + NH3 (Ser dehydration) | d- and L-Ser | [42,43,44,45] |
| Asp racemases (AspR) 2 | 5.1.1.10 | L-Asp → d- Asp/d- Asp → L- Asp (Asp racemization) | d- and L-Asp | [46] |
| AtDAAR1 + AtDAAR2 | 5.1.1.10 | L-Ile → d-Ile (Ile racemization) | L-Ile | [47] |
| ZmDAAO | 1.4.3.3 | d-AA + H2O + O2 → Keto acid + NH3 + H2O2 (d-AA oxidation) | d-Ala, d-Asp | [48] |
| d-CDes 3 | 4.4.1.15 | d-Cys + H2O → Pyruvate + H2S + NH3 (d-Cys desulfhydration) | d-Cys | [49,50,51,52] |
| d-Ala-d-Ala ligase 4 | 6.3.2.4 | 2 d-Ala + ATP → d-Ala-d-Ala + ADP + Pi | d-Ala | [53] |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolukisaoglu, Ü. d-Amino Acids in Plants: Sources, Metabolism, and Functions. Int. J. Mol. Sci. 2020, 21, 5421. https://doi.org/10.3390/ijms21155421
Kolukisaoglu Ü. d-Amino Acids in Plants: Sources, Metabolism, and Functions. International Journal of Molecular Sciences. 2020; 21(15):5421. https://doi.org/10.3390/ijms21155421
Chicago/Turabian StyleKolukisaoglu, Üner. 2020. "d-Amino Acids in Plants: Sources, Metabolism, and Functions" International Journal of Molecular Sciences 21, no. 15: 5421. https://doi.org/10.3390/ijms21155421
APA StyleKolukisaoglu, Ü. (2020). d-Amino Acids in Plants: Sources, Metabolism, and Functions. International Journal of Molecular Sciences, 21(15), 5421. https://doi.org/10.3390/ijms21155421