High-Throughput Sequencing and Expression Analysis Suggest the Involvement of Pseudomonas putida RA-Responsive microRNAs in Growth and Development of Arabidopsis
Abstract
1. Introduction
2. Result
2.1. P. putida RA Improves Plant Growth by Modifications of Lateral Branches and Root Architecture
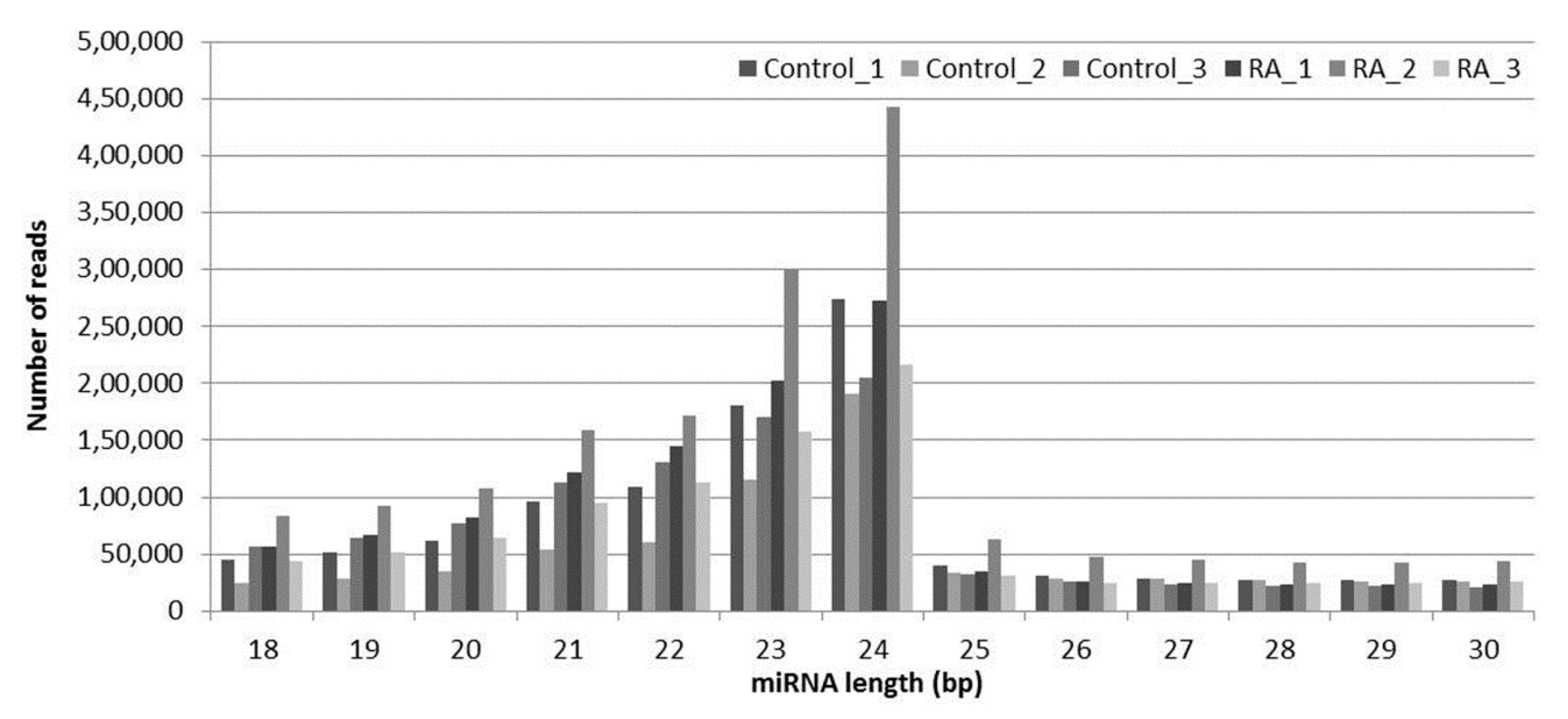
2.2. Summary of sRNA Sequencing
2.3. Identification and Analysis of Known and Novel miRNAs
2.4. miRNA Targets and Their GO Assessment
2.5. Differential Expression Analysis of miRNAs and Their Validation
2.6. Expression Analysis of miRNAs Targets
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Preparation of sRNA Libraries for Sequencing and Data Analysis
4.3. Identification and Expression Analysis of Arabidopsis miRNAs
4.4. Target Prediction and Their Functional Annotation
4.5. Validation and Expression Analysis of miRNAs and Their Targets
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.L.; Touraine, B.; Moenne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dye, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef] [PubMed]
- Asari, S.; Tarkowska, D.; Rolcik, J.; Novak, O.; Palmero, D.V.; Bejai, S.; Meijer, J. Analysis of plant growth-promoting properties of Bacillus amyloliquefaciens UCMB5113 using Arabidopsis thaliana as host plant. Planta 2017, 245, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.P.; Hartmann, A.; Gao, X.W.; Borriss, R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42-a review. Front. Microbiol. 2015, 6, 780. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Prasad, V.; Chauhan, P.S.; Lata, C. Bacillus amyloliquefaciens Confers Tolerance to Various Abiotic Stresses and Modulates Plant Response to Phytohormones through Osmoprotection and Gene Expression Regulation in Rice. Front. Plant Sci. 2017, 8, 1510. [Google Scholar] [CrossRef]
- Jatan, R.; Lata, C. Role of MicroRNAs in Abiotic and Biotic Stress Resistance in Plants. Proc. Indian Natn. Sci. Acad. 2019, 85, 553–567. [Google Scholar]
- Wang, Q.; Li, Y.; Li, J.; Wang, Y.; Wang, C.; Wang, P. Experimental and kinetic study on the cometabolic biodegradation of phenol and 4-chlorophenol by psychrotrophic Pseudomonas putida LY1. Environ. Sci. Pollut. Res. Int. 2015, 22, 565–573. [Google Scholar] [CrossRef]
- Arrebola, E.; Tienda, S.; Vida, C.; de Vicente, A.; Cazorla, F.M. Fitness Features Involved in the Biocontrol Interaction of Pseudomonas chlororaphis With Host Plants: The Case Study of PcPCL1606. Front. Microbiol. 2019, 10, 719. [Google Scholar] [CrossRef]
- Dangi, A.K.; Sharma, B.; Hill, R.T.; Shukla, P. Bioremediation through microbes: Systems biology and metabolic engineering approach. Crit. Rev. Biotechnol. 2019, 39, 79–98. [Google Scholar] [CrossRef]
- Mardiyono; Sajidan; Masykuri, M.; Setyono, P. Bioremediation of Nickel Heavy Metals in Electroplating Industrial Liquid Waste with Bacillus Subtilis. In Proceedings of the AIP Conference Proceedings, Surakarta, Indonesia, 20 July 2019. [Google Scholar]
- Gouda, S.; Kerry, R.G.; Das, G.; Paramithiotis, S.; Shin, H.-S.; Patra, J.K. Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 2018, 206, 131–140. [Google Scholar] [CrossRef]
- Itelima, J.; Bang, W.; Onyimba, I.; Oj, E. A review: Biofertilizer; a key player in enhancing soil fertility and crop productivity. J. Microbiol. Biotechnol. Rep. 2018, 2, 22–28. [Google Scholar]
- Katiyar, D.; Hemantaranjan, A.; Singh, B. Plant growth promoting Rhizobacteria-an efficient tool for agriculture promotion. Adv. Plants Agric. Res. 2016, 4, 426–434. [Google Scholar] [CrossRef]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Nasrulhaq Boyce, A. Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability-A Review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, P.S.; Lata, C.; Tiwari, S.; Chauhan, A.S.; Mishra, S.K.; Agrawal, L.; Chakrabarty, D.; Nautiya, C.S. Transcriptional alterations reveal Bacillus amyloliquefaciens-rice cooperation under salt stress. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Jatan, R.; Chauhan, P.S.; Lata, C. Pseudomonas putida modulates the expression of miRNAs and their target genes in response to drought and salt stresses in chickpea (Cicer arietinum L.). Genomics 2019, 111, 509–519. [Google Scholar] [CrossRef]
- Jatan, R.; Tiwari, S.; Asif, M.H.; Lata, C. Genome-wide profiling reveals extensive alterations in Pseudomonas putida-mediated miRNAs expression during drought stress in chickpea (Cicer arietinum L.). Environ. Exp. Bot. 2019, 157, 217–227. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Sunkar, R.; Li, Y.F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Yu, Y.; Jia, T.R.; Chen, X.M. The “how” and “where” of plant microRNAs. New Phytol. 2017, 216, 1002–1017. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Zhang, L.; Wu, G.; Fu, C.; Long, Y.; Xiang, J.; Gan, J.; Zhou, Y.; Yu, L.; Li, M. Genome-Wide Identification and Characterization of MicroRNAs and Target Genes in Lonicera japonica. PLoS ONE 2016, 11, e0164140. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, W.; Sun, X.; Liang, Z.; Wang, F. Conserved and novel heat stress-responsive microRNAs were identified by deep sequencing in Saccharina japonica (Laminariales, Phaeophyta). Plant Cell Environ. 2015, 38, 1357–1367. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yu, H.; Zhao, G.; Huang, Q.; Lu, Y.; Ouyang, B. Profiling of drought-responsive microRNA and mRNA in tomato using high-throughput sequencing. BMC Genom. 2017, 18, 481. [Google Scholar] [CrossRef]
- Yang, X.; Liu, F.; Zhang, Y.; Wang, L.; Cheng, Y.F. Cold-responsive miRNAs and their target genes in the wild eggplant species Solanum aculeatissimum. BMC Genom. 2017, 18, 1000. [Google Scholar] [CrossRef]
- Yin, Z.; Han, X.; Li, Y.; Wang, J.; Wang, D.; Wang, S.; Fu, X.; Ye, W. Comparative Analysis of Cotton Small RNAs and Their Target Genes in Response to Salt Stress. Genes (Basel) 2017, 8, 369. [Google Scholar] [CrossRef]
- Khraiwesh, B.; Zhu, J.K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 2012, 1819, 137–148. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Fan, L.; Liu, Y.; Xu, H.; Llewellyn, D.; Wilson, I. miR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS ONE 2013, 8, e84390. [Google Scholar] [CrossRef]
- Combier, J.P.; Frugier, F.; de Billy, F.; Boualem, A.; El-Yahyaoui, F.; Moreau, S.; Vernie, T.; Ott, T.; Gamas, P.; Crespi, M.; et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006, 20, 3084–3088. [Google Scholar] [CrossRef]
- Boualem, A.; Laporte, P.; Jovanovic, M.; Laffont, C.; Plet, J.; Combier, J.P.; Niebel, A.; Crespi, M.; Frugier, F. MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. 2008, 54, 876–887. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Zou, Y.; Chen, L.; Cai, Z.; Zhang, S.; Zhao, F.; Tian, Y.; Jiang, Q.; Ferguson, B.J.; et al. Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 2014, 26, 4782–4801. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Hossain, M.S.; Valdes-Lopez, O.; Hoang, N.T.; Zhai, J.; Wang, J.; Libault, M.; Brechenmacher, L.; Findley, S.; Joshi, T.; et al. Identification and functional characterization of soybean root hair microRNAs expressed in response to Bradyrhizobium japonicum infection. Plant Biotechnol. J. 2016, 14, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Tsikou, D.; Yan, Z.; Holt, D.B.; Abel, N.B.; Reid, D.E.; Madsen, L.H.; Bhasin, H.; Sexauer, M.; Stougaard, J.; Markmann, K. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 2018, 362, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Yu, H.; Li, E.; Wang, Y.; Liu, J.; Jiang, H. Identification of miRNAs Involved in Bacillus velezensis FZB42-Activated Induced Systemic Resistance in Maize. Int. J. Mol. Sci. 2019, 20, 5057. [Google Scholar] [CrossRef]
- Xie, S.; Jiang, H.; Xu, Z.; Xu, Q.; Cheng, B. Small RNA profiling reveals important roles for miRNAs in Arabidopsis response to Bacillus velezensis FZB42. Gene 2017, 629, 9–15. [Google Scholar] [CrossRef]
- Kumar, M.; Mishra, S.; Dixit, V.; Agarwal, L.; Chauhan, P.S.; Nautiyal, C.S. Synergistic effect of Pseudomonas putida and Bacillus amyloliquefaciens ameliorates drought stress in chickpea (Cicer arietinum L.). Plant Signal. Behav. 2016, 11, e1071004. [Google Scholar] [CrossRef]
- Srivastava, S.; Chaudhry, V.; Mishra, A.; Chauhan, P.S.; Rehman, A.; Yadav, A.; Tuteja, N.; Nautiyal, C.S. Gene expression profiling through microarray analysis in Arabidopsis thaliana colonized by Pseudomonas putida MTCC5279, a plant growth promoting rhizobacterium. Plant Signal. Behav. 2012, 7, 235–245. [Google Scholar] [CrossRef]
- Tiwari, S.; Lata, C.; Chauhan, P.S.; Nautiyal, C.S. Pseudomonas putida attunes morphophysiological, biochemical and molecular responses in Cicer arietinum L. during drought stress and recovery. Plant Physiol. Biochem. 2016, 99, 108–117. [Google Scholar] [CrossRef]
- He, Y.; Pantigoso, H.A.; Wu, Z.; Vivanco, J.M. Co-inoculation of Bacillus sp. and Pseudomonas putida at different development stages acts as a biostimulant to promote growth, yield and nutrient uptake of tomato. J. Appl. Microbiol. 2019, 127, 196–207. [Google Scholar] [CrossRef]
- Srivastava, S.; Bist, V.; Awasthi, S.; Chauhan, R.; Chaudhry, V.; Singh, P.C.; Dwivedi, S.; Niranjan, A.; Agrawal, L.; Chauhan, P.S.; et al. Chlorella vulgaris and Pseudomonas putida interaction modulates phosphate trafficking for reduced arsenic uptake in rice (Oryza sativa L.). J. Hazard. Mater. 2018, 351, 177–187. [Google Scholar] [CrossRef]
- Srivastava, S. Prescience of endogenous regulation in Arabidopsis thaliana by Pseudomonas putida MTCC 5279 under phosphate starved salinity stress condition. Sci. Rep. 2020, 10, 5855. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Bossuyt, S.; Engelen, K.; Marchal, K.; Vanderleyden, J. Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 2014, 201, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Bhogale, S.; Mahajan, A.S.; Natarajan, B.; Rajabhoj, M.; Thulasiram, H.V.; Banerjee, A.K. MicroRNA156: A potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 2014, 164, 1011–1027. [Google Scholar] [CrossRef] [PubMed]
- Buhtz, A.; Pieritz, J.; Springer, F.; Kehr, J. Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant. Biol. 2010, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Adam, H.; Diaz-Mendoza, M.; Zurczak, M.; Gonzalez-Schain, N.D.; Suarez-Lopez, P. Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 2009, 136, 2873–2881. [Google Scholar] [CrossRef]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and functional diversification of MIRNA genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef]
- Fahlgren, N.; Howell, M.D.; Kasschau, K.D.; Chapman, E.J.; Sullivan, C.M.; Cumbie, J.S.; Givan, S.A.; Law, T.F.; Grant, S.R.; Dangl, J.L.; et al. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE 2007, 2, e219. [Google Scholar] [CrossRef]
- Song, C.; Wang, C.; Zhang, C.; Korir, N.K.; Yu, H.; Ma, Z.; Fang, J. Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genom. 2010, 11, 431. [Google Scholar] [CrossRef]
- Breakfield, N.W.; Corcoran, D.L.; Petricka, J.J.; Shen, J.; Sae-Seaw, J.; Rubio-Somoza, I.; Weigel, D.; Ohler, U.; Benfey, P.N. High-resolution experimental and computational profiling of tissue-specific known and novel miRNAs in Arabidopsis. Genome Res. 2012, 22, 163–176. [Google Scholar] [CrossRef]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.G.; Park, J.H.; Lim, J.Y.; Kim, D.; Choi, Y.; Kim, S.; Reeves, G.; Yeom, S.I.; Lee, J.S.; Park, M.; et al. The hot pepper (Capsicum annuum) microRNA transcriptome reveals novel and conserved targets: A foundation for understanding MicroRNA functional roles in hot pepper. PLoS ONE 2013, 8, e64238. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Shao, C.; Wang, H.; Chen, M. The regulatory activities of plant microRNAs: A more dynamic perspective. Plant Physiol. 2011, 157, 1583–1595. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Raman, S.; Greb, T.; Peaucelle, A.; Blein, T.; Laufs, P.; Theres, K. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008, 55, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, F.D.R.; Suarez, Y.R.; Rangel, R.M.C.; Garcia, V.L.; Aguilera, K.L.G.; Martinez, N.M.; de Folter, S. Effect of Constitutive miR164 Expression on Plant Morphology and Fruit Development in Arabidopsis and Tomato. Agronomy (Basel) 2017, 7, 48. [Google Scholar] [CrossRef]
- Yu, H.; Huang, T. Molecular Mechanisms of Floral Boundary Formation in Arabidopsis. Int. J. Mol. Sci. 2016, 17, 317. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.C.; Dugas, D.V.; Bartel, D.P.; Bartel, B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 2004, 14, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.C.; Sieber, P.; Wellmer, F.; Meyerowitz, E.M. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 2005, 15, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Wei, W.; Li, Y.; Kan, L.; Wang, F.; Zhang, X.; Li, F.; Liu, Z.; Kang, C. Conserved and novel roles of miR164-CUC2 regulatory module in specifying leaf and floral organ morphology in strawberry. New Phytol. 2019, 224, 480–492. [Google Scholar] [CrossRef]
- Meng, J.G.; Zhang, X.D.; Tan, S.K.; Zhao, K.X.; Yang, Z.M. Genome-wide identification of Cd-responsive NRAMP transporter genes and analyzing expression of NRAMP 1 mediated by miR167 in Brassica napus. Biometals 2017, 30, 917–931. [Google Scholar] [CrossRef] [PubMed]
- Tejada-Jimenez, M.; Castro-Rodriguez, R.; Kryvoruchko, I.; Lucas, M.M.; Udvardi, M.; Imperial, J.; Gonzalez-Guerrero, M. Medicago truncatula natural resistance-associated macrophage Protein1 is required for iron uptake by rhizobia-infected nodule cells. Plant Physiol. 2015, 168, 258–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Qiu, W.; Dai, J.; Guo, X.; Lu, Q.; Wang, T.; Li, S.; Liu, T.; Zuo, Y. AhNRAMP1 Enhances Manganese and Zinc Uptake in Plants. Front. Plant Sci. 2019, 10, 415. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Kobayashi, T.; Kakei, Y.; Senoura, T.; Nakazono, M.; Takahashi, H.; Nakanishi, H.; Shen, H.; Duan, P.; Guo, X.; et al. AhNRAMP1 iron transporter is involved in iron acquisition in peanut. J. Exp. Bot. 2012, 63, 4437–4446. [Google Scholar] [CrossRef] [PubMed]
- Bian, H.; Xie, Y.; Guo, F.; Han, N.; Ma, S.; Zeng, Z.; Wang, J.; Yang, Y.; Zhu, M. Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 2012, 196, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Parry, G.; Calderon-Villalobos, L.I.; Prigge, M.; Peret, B.; Dharmasiri, S.; Itoh, H.; Lechner, E.; Gray, W.M.; Bennett, M.; Estelle, M. Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. USA 2009, 106, 22540–22545. [Google Scholar] [CrossRef]
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef]
- Chen, Z.H.; Bao, M.L.; Sun, Y.Z.; Yang, Y.J.; Xu, X.H.; Wang, J.H.; Han, N.; Bian, H.W.; Zhu, M.Y. Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol. Biol. 2011, 77, 619–629. [Google Scholar] [CrossRef]
- Iglesias, M.J.; Terrile, M.C.; Windels, D.; Lombardo, M.C.; Bartoli, C.G.; Vazquez, F.; Estelle, M.; Casalongue, C.A. MiR393 regulation of auxin signaling and redox-related components during acclimation to salinity in Arabidopsis. PLoS ONE 2014, 9, e107678. [Google Scholar] [CrossRef]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef]
- Wojcik, A.M.; Gaj, M.D. miR393 contributes to the embryogenic transition induced in vitro in Arabidopsis via the modification of the tissue sensitivity to auxin treatment. Planta 2016, 244, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Bian, H.; Zeng, Z.; Hou, N.; Shi, B.; Wang, J.; Zhu, M.; Han, N. miR393-Mediated Auxin Signaling Regulation is Involved in Root Elongation Inhibition in Response to Toxic Aluminum Stress in Barley. Plant Cell Physiol. 2017, 58, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Hu, L.; Han, N.; Hu, J.; Yang, Y.; Xiang, T.; Zhang, X.; Wang, L. Overexpression of a miR393-resistant form of transport inhibitor response protein 1 (mTIR1) enhances salt tolerance by increased osmoregulation and Na+ exclusion in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Araus, V.; Lu, C.; Parry, G.; Green, P.J.; Coruzzi, G.M.; Gutierrez, R.A. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 4477–4482. [Google Scholar] [CrossRef]
- Li, X.; Xia, K.; Liang, Z.; Chen, K.; Gao, C.; Zhang, M. MicroRNA393 is involved in nitrogen-promoted rice tillering through regulation of auxin signal transduction in axillary buds. Sci. Rep. 2016, 6, 32158. [Google Scholar] [CrossRef]
- Xu, J.; Li, J.; Cui, L.; Zhang, T.; Wu, Z.; Zhu, P.Y.; Meng, Y.J.; Zhang, K.J.; Yu, X.Q.; Lou, Q.F.; et al. New insights into the roles of cucumber TIR1 homologs and miR393 in regulating fruit/seed set development and leaf morphogenesis. BMC Plant Biol. 2017, 17, 130. [Google Scholar] [CrossRef]
- Zeng, W.; Sun, Z.; Lai, Z.; Yang, S.; Chen, H.; Yang, X.; Tao, J.; Tang, X. Determination of the MiRNAs Related to Bean Pyralid Larvae Resistance in Soybean Using Small RNA and Transcriptome Sequencing. Int. J. Mol. Sci. 2019, 20, 2966. [Google Scholar] [CrossRef]
- Hewezi, T.; Maier, T.R.; Nettleton, D.; Baum, T.J. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol. 2012, 159, 321–335. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A Small Transcription Factor Family with Important Functions in Plant Biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, B.H.; Jung, J.H.; Park, S.K.; Song, J.T.; Kim, J.H. GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR Specify Meristematic Cells of Gynoecia and Anthers. Plant Physiol. 2018, 176, 717–729. [Google Scholar] [CrossRef]
- Choi, S.; Cho, Y.H.; Kim, K.; Matsui, M.; Son, S.H.; Kim, S.K.; Fujioka, S.; Hwang, I. BAT1, a putative acyltransferase, modulates brassinosteroid levels in Arabidopsis. Plant J. 2013, 73, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Breuer, C.; Kawamura, A.; Jikumaru, Y.; Hanada, A.; Fujioka, S.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Kamiya, Y.; et al. Arabidopsis PIZZA has the capacity to acylate brassinosteroids. PLoS ONE 2012, 7, e46805. [Google Scholar] [CrossRef]
- Han, Y.J.; Kim, Y.S.; Hwang, O.J.; Roh, J.; Ganguly, K.; Kim, S.K.; Hwang, I.; Kim, J.I. Overexpression of Arabidopsis thaliana brassinosteroid-related acyltransferase 1 gene induces brassinosteroid-deficient phenotypes in creeping bentgrass. PLoS ONE 2017, 12, e0187378. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.; Fujioka, S.; Noguchi, T.; Takatsuto, S.; Yoshida, S.; Feldmann, K.A. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001, 26, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Trieu, A.; Radhakrishnan, P.; Kwok, S.F.; Harris, S.; Zhang, K.; Wang, J.; Wan, J.; Zhai, H.; Takatsuto, S.; et al. Brassinosteroids regulate grain filling in rice. Plant Cell 2008, 20, 2130–2145. [Google Scholar] [CrossRef] [PubMed]
- Kang, N.Y.; Lee, H.W.; Kim, J. The AP2/EREBP gene PUCHI Co-Acts with LBD16/ASL18 and LBD18/ASL20 downstream of ARF7 and ARF19 to regulate lateral root development in Arabidopsis. Plant Cell Physiol. 2013, 54, 1326–1334. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, M.; Lee, M.R.; Park, S.K.; Kim, J. LATERAL ORGAN BOUNDARIES DOMAIN (LBD)10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. Plant J. 2015, 81, 794–809. [Google Scholar] [CrossRef]
- Oh, S.A.; Park, K.S.; Twell, D.; Park, S.K. The SIDECAR POLLEN gene encodes a microspore-specific LOB/AS2 domain protein required for the correct timing and orientation of asymmetric cell division. Plant J. 2010, 64, 839–850. [Google Scholar] [CrossRef]
- Fan, M.; Xu, C.; Xu, K.; Hu, Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012, 22, 1169–1180. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, J.; Jia, H.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B.; White, F.F.; Wang, N.; Jones, J.B. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, E521–E529. [Google Scholar] [CrossRef]
- Mangeon, A.; Bell, E.M.; Lin, W.C.; Jablonska, B.; Springer, P.S. Misregulation of the LOB domain gene DDA1 suggests possible functions in auxin signalling and photomorphogenesis. J. Exp. Bot. 2011, 62, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, L.F.; Powell, J.J.; Aitken, E.A.B.; Kazan, K.; Manners, J.M. The Lateral Organ Boundaries Domain Transcription Factor LBD20 Functions in Fusarium Wilt Susceptibility and Jasmonate Signaling in Arabidopsis. Plant Physiol. 2012, 160, 407–418. [Google Scholar] [CrossRef]
- Xu, C.; Luo, F.; Hochholdinger, F. LOB Domain Proteins: Beyond Lateral Organ Boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Goh, T.; Joi, S.; Mimura, T.; Fukaki, H. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 2012, 139, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Goh, T.; Toyokura, K.; Yamaguchi, N.; Okamoto, Y.; Uehara, T.; Kaneko, S.; Takebayashi, Y.; Kasahara, H.; Ikeyama, Y.; Okushima, Y.; et al. Lateral root initiation requires the sequential induction of transcription factors LBD16 and PUCHI in Arabidopsis thaliana. New Phytol. 2019, 224, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Somerville, C. Isolation of photorespiration mutants in Arabidopsis thaliana. Methods Chloroplast Mol. Biol. 1982, 129–138. [Google Scholar]
- Stocks, M.B.; Moxon, S.; Mapleson, D.; Woolfenden, H.C.; Mohorianu, I.; Folkes, L.; Schwach, F.; Dalmay, T.; Moulton, V. The UEA sRNA workbench: A suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 2012, 28, 2059–2061. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Bateman, A.; Marshall, M.; Khanna, A.; Eddy, S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003, 31, 439–441. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef] [PubMed]
- Qibin, L.; Jiang, W. MIREAP: microRNA Discovery by Deep Sequencing. Available online: http://sourceforge.net/project/mireap/ (accessed on 8 April 2018).
- Lorenz, R.; Bernhart, S.H.; Zu Siederdissen, C.H.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




| Control_1 | Control_2 | Control_3 | RA_1 | RA_2 | RA_3 | |
|---|---|---|---|---|---|---|
| Total Reads | 14,686,655 | 12,373,692 | 14,639,734 | 14,202,847 | 29,7684,85 | 13,162,721 |
| Trimmed unique reads | 1,319,211 | 986,556 | 1,235,562 | 1,404,413 | 2,156,182 | 1,215,179 |
| Reads aligned to genome | 1,174,700 | 846,192 | 1,114,548 | 1,270,093 | 1,888,067 | 1,100,485 |
| rRNA | 172,731 | 172,194 | 128,009 | 135,128 | 248,281 | 145,704 |
| snoRNA | 6578 | 7353 | 6214 | 7699 | 9254 | 8057 |
| snRNA | 4184 | 3679 | 4371 | 4609 | 6912 | 4566 |
| tRNA | 23 | 13 | 25 | 29 | 60 | 27 |
| Clustered reads | 584,288 | 408,865 | 542,203 | 614,695 | 863,035 | 526,561 |
| Reads aligned to miRBase | 4448 | 3467 | 4574 | 4943 | 7029 | 4108 |
| Known miRNA unique | 238 | 216 | 235 | 234 | 256 | 233 |
| Reads utilized for Novel miRNA | 567,702 | 397,267 | 524,730 | 597,830 | 832,030 | 513,748 |
| Novel miRNA predicted | 13 | 11 | 27 | 16 | 18 | 15 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jatan, R.; Chauhan, P.S.; Lata, C. High-Throughput Sequencing and Expression Analysis Suggest the Involvement of Pseudomonas putida RA-Responsive microRNAs in Growth and Development of Arabidopsis. Int. J. Mol. Sci. 2020, 21, 5468. https://doi.org/10.3390/ijms21155468
Jatan R, Chauhan PS, Lata C. High-Throughput Sequencing and Expression Analysis Suggest the Involvement of Pseudomonas putida RA-Responsive microRNAs in Growth and Development of Arabidopsis. International Journal of Molecular Sciences. 2020; 21(15):5468. https://doi.org/10.3390/ijms21155468
Chicago/Turabian StyleJatan, Ram, Puneet Singh Chauhan, and Charu Lata. 2020. "High-Throughput Sequencing and Expression Analysis Suggest the Involvement of Pseudomonas putida RA-Responsive microRNAs in Growth and Development of Arabidopsis" International Journal of Molecular Sciences 21, no. 15: 5468. https://doi.org/10.3390/ijms21155468
APA StyleJatan, R., Chauhan, P. S., & Lata, C. (2020). High-Throughput Sequencing and Expression Analysis Suggest the Involvement of Pseudomonas putida RA-Responsive microRNAs in Growth and Development of Arabidopsis. International Journal of Molecular Sciences, 21(15), 5468. https://doi.org/10.3390/ijms21155468





