Abstract
Acinetobacter baumannii is considered one of the most persistent pathogens responsible for nosocomial infections. Due to the emergence of multidrug resistant strains, as well as high morbidity and mortality caused by this pathogen, A. baumannii was placed on the World Health Organization (WHO) drug-resistant bacteria and antimicrobial resistance research priority list. This review summarizes current studies on mechanisms that protect A. baumannii against multiple stresses caused by the host immune response, outside host environment, and antibiotic treatment. We particularly focus on the ability of A. baumannii to survive long-term desiccation on abiotic surfaces and the population heterogeneity in A. baumannii biofilms. Insight into these protective mechanisms may provide clues for the development of new strategies to fight multidrug resistant strains of A. baumannii.
1. Introduction
Gram-negative coccobacillus Acinetobacter baumannii belongs to a group of ESKAPE pathogens. ESKAPE is the acronym for the group of bacteria that include Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter spp. Due to their ability to effectively escape antibiotic treatments, these multidrug-resistant (MDR) pathogens are common causes of life-threatening infections affecting mainly immunocompromised and critically ill patients in intensive care units (ICUs) [1]. In recent years, the overall number of antibiotics that act on the ESKAPE pathogens decreased significantly [1,2]. In 2017, the World Health Organization (WHO) published a list of 12 “priority” pathogens encompassing the ESKAPE group, for which new antibiotics are urgently needed. The WHO classifies carbapenem-resistant A. baumannii as the number one critical pathogen. Major risk factors for the acquisition of A. baumannii include antibiotic usage, especially β-lactams—the most commonly used drugs to treat infections caused by important pathogens which cause a variety of diseases in humans and animals. The second most common risk factor is mechanical ventilation, while other risks include surgical wound infections and invasive procedures such as central venous or urinary catheters [3]. It was demonstrated that approximately 1000,000 people globally are infected with A. baumannii every year, while the emergence of MDR strains is reported worldwide [4,5,6]. Antimicrobial treatment of MDR A. baumannii infections include colistin, sulbactam, and tigecycline, used in combination with other antibiotics [1,7,8]. A recently published global study, the Tigecycline Evaluation and Surveillance Trial (TEST), revealed that the percentage of MDR A. baumannii isolates was the highest among all analyzed Gram-negative bacteria, and it increased from 23% in 2004 to 63% in 2014 [4]. A. baumannii causes a range of infections, including ventilator-associated pneumonia, bacteremia, meningitis, urinary tract, wound, and bone infections [2,9]. The risk of mortality is high and often reaches 40–50% in ICU [10,11]. A. baumannii is a life-threatening problem not only because of multidrug resistance but also its ability to evade the host immune response and survive under harsh environmental conditions. In this review, we present various mechanisms that protect A. baumannii against the innate host immune response and stresses caused by the outside host environment. We focus on (1) the ability of A. baumannii to survive long-term desiccation, (2) factors involved in maintaining proteome homeostasis in A. baumannii cells, (3) the population heterogeneity in A. baumannii biofilms, and (4) the mechanism underlying A. baumannii antibiotic resistance.
2. A. baumannii and the Host Innate Immune Response
2.1. The First Line of Host Defense against A. baumannii
Neutrophils, macrophages, antimicrobial peptides (AMPs), and complement system components are the first line of innate immune defense that A. baumannii encounters during infection. Neutrophils can kill bacteria via phagocytosis, degranulation, or NETosis—a specific type of cell death pathway resulting in the release of the neutrophil extracellular traps (NETs). In NETs, chromatin forms a web-like structure decorated with antibacterial factors, including neutrophil elastase and AMPs [9]. Multiple studies suggest that neutrophils play a crucial role in the control of A. baumannii infection [12,13,14,15]; however, contradictory results showing that neutrophils do not kill A. baumannii were also reported [16,17]. After phagocytosis, neutrophil clearance of A. baumannii is mainly dependent on reduced nicotinamide adenine dinucleotide phosphate (NADPH) oxidase which generates reactive oxygen species (ROS) to kill the pathogen [12]. The stimulation of H2O2 production in the lung of a mouse model in response to A. baumannii infection confirmed this observation [18]. It was also found that A. baumannii inhibits NETosis, in contrast to other Gram-negative bacteria that trigger NET formation [17,19,20]. The role of macrophages in eliminating A. baumannii remains controversial. Most studies showed that macrophages play a minor role during A. baumannii infection [14,15,21]. However, Qiu et al. demonstrated that macrophages could be the first line of defense against respiratory A. baumannii infections; the depletion of alveolar macrophages significantly enhanced the susceptibility of mice to A. baumannii [22]. In addition, phagocytosis and killing of A. baumannii were observed in vitro, and the macrophages produced nitric oxide and secreted proinflammatory cytokines and chemokines [22].
AMPs are expressed constitutively or induced in different types of cells and tissues. Most of the AMPs are cationic; therefore, they can easily target the negatively charged surface of bacteria [23,24]. To date, 139 human host defense peptides were identified [25]. Cathelicidin-derived LL-37 is the best-studied AMP that kills A. baumannii cells through binding to the outer membrane protein A (OmpA) [24,26]. The LL-37 precursor, human cationic antibacterial peptide (hCAP-18), is produced by epithelial cells and neutrophils [13]. Recently, it was demonstrated that LL-37 forms a dimer of two anti-parallel amphipathic α-helices without supercoiling [27]. LL-37 helices target and extract lipopolysaccharides (LPS) to form holes in the outer membrane (OM). After diffusion into the periplasmic space, LL-37 may extract lipids from the inner membrane, forming a fibril-like structure [27]. Two other AMPs that belong to the human beta defensins, hBD-2 and hBD-3, were shown to kill A. baumannii in a concentration-dependent manner [28].
Another element of the immune system directed against A. baumannii involves the complement system components. The complement system consists of more than 30 plasma proteins that collaborate as a cascade triggering either bacterial cell lysis or opsonization and phagocytosis [29]. There are three pathways of complement activation: classical, lectin, and alternative pathways. The classical pathway is initiated by immune complexes, whereas the lectin pathway is triggered by pathogen-specific carbohydrates. The alternative pathway, which is responsible for the killing of A. baumannii [30,31], is permanently active due to spontaneous hydrolysis of the central complement system component, C3, enabling fast detection of pathogens [29,32]. Several studies reported that the depletion of the complement results in an increase of A. baumannii viability in human serum or a mouse model of A. baumannii infection [15,33,34].
2.2. Mechanisms Protecting A. baumannii against the Innate Immune Response
A. baumannii uses different virulence factors or mechanisms to evade the innate immune response. Surface glycoconjugates play key roles, but other strategies, including secreted proteins and metabolic pathways, also participate in the defense against the immune system [35,36].
The first barrier that protects A. baumannii against the immune host response is an exopolysaccharide capsule. The capsule is formed from long-chain polysaccharides composed of repeated carbohydrate units (K units). The synthesis of capsular polysaccharides (CPS) is dependent on a K locus (KL), which contains genes for synthesis of activated sugar precursors, glycosyl transfer, glycan modification, and oligosaccharide repeat-unit processing [37]. To date, 128 KL gene clusters were identified in A. baumannii [38]. Therefore, the monosaccharide composition and CPS structure are highly variable in A. baumannii strains [37,39,40,41,42,43]. K units vary in length and may consist of two to six residues. Common neutral sugars such as d-glucose, d-galactose, N-acetyl-d-glucosamine, and N-acetyl-d-galactosamine or rare sugars, including derivatives of pseudaminic, legionaminic, or acinetaminic acid, can be incorporated into CPS. Interestingly, acinetaminic acid was never found in nature before its detection in A. baumanni isolates with K12 and K13 gene clusters [42]. The CPS of different isolates are linear or branched and may possess non-carbohydrate substituents, including the most frequent O- and N-acyl groups [44]. The Wzy pathway encoded by the K locus is responsible for the export and extracellular assembly of CPS [45]. The K locus is regulated by the two-component signal transduction system BfmRS. The global BfmR regulator, along with histidine kinase BfmS, controls a variety of processes, including biogenesis of A. baumannii envelope elements, formation of biofilms, desiccation tolerance, and multiple stress responses [46,47,48,49]. Several studies demonstrated that the production of capsules affects A. baumannii virulence and persistence in the host [50,51]. Capsule-enriched strains cause more severe disease or higher lethality [35,45,50,52,53]. These observations may be partly explained by the high hydrophilicity of CPS and negative charges of CPS monosaccharides that prevent phagocytosis by hindering interactions with the negatively charged surfaces of neutrophils and macrophages, preventing phagocytosis [35]. However, it should be noted that the highly variable structure of the CPS may affect its capacity as a protective barrier against the immune response and other stresses, including complement-mediated killing, lysozyme degradation, and ROS [45,50,51,52,53,54].
Another barrier protecting A. baumannii against the host response is the OM. LPS, the main component of the outer layer of the OM, contains three domains: (1) lipid A, the membrane anchor glycosylated with (2) a core oligosaccharide which may provide an attachment site for (3) a long-chain O-antigenic polysaccharide [35]. In A. baumannii, the primary component of the outer layer of the OM is lipooligosaccharide (LOS) which, in contrast to the typical LPS, lacks the O-antigen [37]. A. baumannii LOS belongs to the group of pathogen-associated molecular patterns (PAMPs) recognized by Toll-like receptor 4 (TLR4) [13,55]. TLR4 is one of the pattern recognition receptors (PRRs) which function as cell-surface sensors of bacterial infection. A. baumannii LOS triggers a TLR4-mediated release of tumor necrosis factor (TNF) and interleukin 8 (IL-8) from macrophages [56]. This inflammation response is beneficial for the host; however, if it is upregulated, it can result in a cytokine storm and septic shock [57]. In the case of highly virulent A. baumannii, enhanced TLR4 activation is correlated to increased shedding of LOS into growth medium [58]. A. baumannii constitutively synthesizes hepta-acylated lipid A, under standard growth conditions, in contrast to other Gram-negative bacteria that upregulate its synthesis only under stress conditions [59]. Constitutive hepta-acylation of lipid A fortifies the OM to protect A. baumannii from cationic AMPs, lysozyme, and colistin, which is the last-resort antibiotic to treat MDR A. baumannii infections [59]. Colistin resistance may result from a complete lack of LOS or the addition of galactosamine to LOS. LOS deficiency significantly alters the interaction of A. baumannii with the host innate immune system. The overall pro-inflammatory response to LOS-deficient A. baumannii is reduced due to the lack of TLR-4 mediated stimulation. Instead of the TLR4-dependent mechanism, the TLR2-dependent mechanism is activated [60]. This alternative response is probably a consequence of increased expression of specific lipoproteins, transporters, and other surface components that compensate for the lack of LOS [61]. Although LOS-deficient strains are colistin-resistant, they exhibit decreased virulence and increased susceptibility to LL-37 and lysozyme [60,62,63,64].
Apart from the surface glycoconjugates, essential virulence factors of A. baumannii are OM proteins. The best-characterized A. baumannii OM protein, OmpA porin, is responsible for adhesion and invasion of A. baumannii into human epithelial cells [65,66,67,68]. It was found that the overproduction of OmpA is a risk factor for the development of A. baumannii pneumonia and bacteraemia, as well as for an increased mortality rate [69]. Consistently, the deletion of the ompA gene reduced A. baumannii bacterial dissemination and development of secondary pneumonia in the murine peritoneal sepsis model [69]. OmpA is secreted and enters epithelial cells via outer membrane vesicles (OMVs) [70,71,72]. OMVs are used by Gram-negative bacteria to deliver toxins, virulence factors, and other effector molecules to host cells. After entering the epithelial cell, OmpA migrates to mitochondria and stimulates the release of cytochrome c, finally promoting apoptosis [65]. In addition, OmpA triggers cell death by inducing the expression of TLR2 and production of nitrogen oxide. Lee et al. found that high concentrations of OmpA induces ROS production, leading to early-onset apoptosis and delayed-onset necrosis in dendritic cells (DCs) [73]. These results demonstrate that OmpA can cause the death of DSc, thereby impairing T-cell responses against A. baumannii. Other studies revealed that the interaction of OmpA with the fluid-phase complement regulator factor H (FH) enables A. baumannii to escape complement and to survive in human serum [31]. Two other A. baumannii proteins, CipA and the protein killing factor (PKF) serine protease, also contribute to serum resistance. The recently identified CipA is a plasminogen-binding protein exposed on the OM [32]. Plasminogen, in the complex with CipA, is cleaved and converted to the active serine protease plasmin, which degrades fibrinogen and the complement component C3b. Thus, CipA can prevent entrapment of A. baumannii in fibrin thrombi, and it facilitates dissemination of the pathogen. Regardless of plasminogen binding, CipA can also inhibit the alternative complement pathway [32]. The PKF serine protease is involved in serum resistance possibly through degradation of yet unidentified complement components [74]. It was suggested that both CipA and PKF are secreted via a type II secretion system (T2SS) [75]. The T2SS enables secretion of effector proteins including multiple enzymes critical for A. baumannii virulence [9]. The hypothesis that the T2SS participates in the secretion of CipA and PKF is based on results showing that deletion of the T2SS gene gspD resulted in significantly decreased resistance to human serum [75].
Recent studies revealed that, in addition to the aforementioned proteins and structures, A. baumannii possesses numerous protective mechanisms against complement-mediated killing. Sanchez-Larrayoz et al. identified 50 genes essential for the survival of A. baumannii in human serum, including the Mla system, which encodes proteins required for the maintenance of OM lipid asymmetry [54]. The Mla proteins prevent the accumulation of phospholipids in the outer leaflet of the OM by transporting them to the inner membrane. This study suggests that the accumulation of surface-exposed phospholipids in mla-deficient strains can activate the alternative pathway of the complement system [54].
An example of a metabolic adaptation that enables A. baumannii to evade neutrophil chemotaxis is the phenylacetic acid catabolism pathway encoded by the paa system [14]. The paa genes were identified in 16% of the sequenced bacterial species [76,77]. The paa operon is involved in degradation of aromatic compounds including phenylacetate to form acetyl-coenzyme A (CoA) and succinyl-CoA [76]. Bhuiyan et al. demonstrated that the loss of function of this catabolic pathway resulted in the accumulation of phenylacetate, which acted as an attractant of neutrophils, leading finally to bacterial clearance [14].
An interesting example of the A. baummanii virulence strategy is its ability to adhere to neutrophils without being eliminated by phagocytosis. Since neutrophils can transmigrate from the infection site to vasculature, it was proposed that the reverse migration of neutrophils can disseminate the infection to other organs [17]. Moreover, Sato et al. demonstrated that MDR A. baumannii isolates can survive in macrophages after phagocytosis. The MDR strains induced ROS production in macrophages, and they concomitantly exhibited upregulated catalase activity which allowed them to resist oxidative stress [78]. These results indicate that A. baumannii can spread in the infected organism using both neutrophils and macrophages.
Most of the strategies described above (Figure 1) allow A. baumannii not only to evade the innate immune response, but also to survive in the external environment.
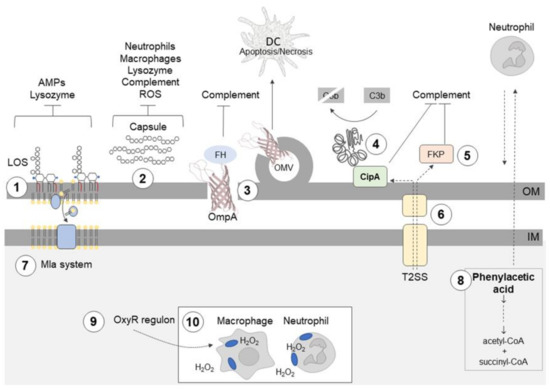
Figure 1.
Acinetobacter baumannii uses different mechanisms to evade the innate immune response. ① Hepta-acylation of lipid A in lipooligosaccharide (LOS) fortifies the outer membrane (OM) and protects A. baumannii from cationic antimicrobial peptides (AMPs), colistin, and lysozyme. ② Highly hydrophilic and negatively charged capsular polysaccharides (CPS) hinder interactions with negatively charged surfaces of neutrophils and macrophages; the capsule is also a barrier which protects against complement-mediated killing, lysozyme degradation, and reactive oxygen species (ROS). ③ Outer membrane protein A (OmpA) interacts with factor H (FH), thereby inhibiting the complement-mediated killing; OmpA induces ROS production and the death of dendritic cells (DCs). ④ CipA forms a complex with plasminogen/plasmin, which degrades the complement component C3b; CipA and ⑤ the protein killing factor (PKF) serine protease inhibit the alternative complement pathway. ⑥ The type II secretion system (T2SS) contributes to serum resistance, and it probably participates in CipA and PKF serine protease secretion. ⑦ Surface-exposed phospholipids are potential activators of the alternative complement pathway. The Mla system prevents the accumulation of phospholipids in the outer leaflet of the OM. ⑧ Phenylacetate, a derivative of phenylalanine and neutrophil attractant, is removed from the bacterial cell by conversion to acetyl-coenzyme A (CoA) and succinyl-CoA. ⑨ Enhanced catalase activity enables A. baumannii to survive in macrophages in the presence of ROS. ⑩ A. baumannii can spread during infection using neutrophils and macrophages.
3. Mechanisms Protecting A. baumannii against Desiccation
Desiccation, as a common environmental stressor, poses challenges to bacterial cells. Water molecules, as the only nonvolatile solvent in cells, are critical in reaction mechanisms; they also confer stability to lipids, DNA, and proteins, as well as contributing structural order. Loss of membrane integrity during desiccation disrupts the respiratory chain leading to the accumulation of superoxide ions. Furthermore, the malfunction of transport proteins and destabilization of proteins with iron-sulfur clusters cause an increase in the level of intracellular iron. Superoxide radicals can participate in Fenton and Haber-Weiss reactions with ferrous or ferric ions leading to the production of highly toxic hydroxyl radicals [79]. Therefore, oxidative damage of DNA, lipids, and proteins is one of the effects of water loss. Acinetobacter spp., compared with other Gram-negative rods, are more resistant to dry conditions [80]. A. baumannii uniquely survives on inanimate objects and fingertips for extended periods, which explains its potency in cross-infection breakouts [81].
The main structures that facilitate bacteria to enhance water retention are CPS and LPS/LOS [36]. CPS were shown to contribute to desiccation tolerance in A. baumannii [51]. However, other studies demonstrated that there is no simple correlation between the capsule thickness and survival rate under desiccation [49]. Therefore, it seems that the type and structure of CPS or other mechanisms, including hepta-acetylated lipid A, must contribute to the outstanding desiccation tolerance of A. baumannii. Boll et al. demonstrated that the A. baumannii mutant lacking the LpxM acylsynthetaze produces penta-acylated lipid A, instead of hepta-acylated lipid A, and it exhibits decreased desiccation tolerance, probably due to increased membrane fluidity [59].
The accumulation of organic osmolytes, generally designated as compatible solutes, is a prerequisite for the adaptation of bacteria to osmotic stress imposed by water loss. A crucial role in desiccation resistance in various microorganisms is played by the non-reducing disaccharide, trehalose. Trehalose acts as an osmolyte, chemical chaperone, and metabolite that can directly or indirectly stabilize proteins and membranes [82,83,84,85]. It seems that endogenous trehalose is not involved in desiccation tolerance in A. baumannii, but exogenous trehalose was found to efficiently protect A. baumannii on dry surfaces [68]. In response to osmotic stress, A. baumannii also accumulates mannitol and glutamate; however, their contribution to desiccation resistance remains mostly unexplored [81]. To counteract the effects of oxidative stress, the expression of anti-oxidant enzymes, such as catalases KatE and KatG, superoxide dismutase, and glutathione peroxidase, is induced in desiccation-stressed A. baumannii [49,86,87]. A recent study by Farrow et al. proved that the global BfmR regulator contributes to that desiccation tolerance [49].
Gayoso et al. found that desiccation stress affects the composition of the OM. The overproduction of OMPs (Omp25, DcaP-like, and CarO) was observed, indicating a shift in membrane permeability. This study also revealed that several genes encoding proteins involved in transcription and translation, including RNA polymerase subunits RpoA and RpoC, ribosome-associated proteins, and the elongation factor Tu, are downregulated in A. baumannii during desiccation. Proteins whose expression was upregulated included ribosomal recycling factor (RRF), integration host factor (IHF), and the histone-like protein HU. RRF facilitates disassembly of the ribosome at the end of translation. IHF and HU are involved in transcription regulation, and they are essential for maintaining DNA supercoiling and compaction. Consistent with this finding, the presence of an electron-dense region inside desiccation-stressed A. baumannii cells was detected [87]. All these observations led to the conclusion that A. baumannii cells exposed to desiccation stress enter a dormant state [87]. Under favourable conditions, dormant bacteria can recover and resume growth.
The protection of proteins is crucial for the survival of bacteria during desiccation stress and subsequent rehydration. In the next section, we discuss mechanisms that counteract protein damage caused by water loss and other stresses. Then, we present the current knowledge of biofilm formation, which is one of the main strategies used by bacterial populations to survive desiccation stress [88].
4. Protein Homeostasis in A. baumannii
Upon oxidative stress, proteins, which are the main target of ROS, are damaged by metal-catalyzed oxidation and non-enzymatic glycation [79,82,89]. During desiccation, these detrimental reactions are facilitated by the reduction of the hydration shell around proteins and protein condensation, which in turn may lead to misfolding and aggregation of proteins. Apart from anti-oxidant enzymes, bacteria evolved additional mechanisms that protect proteins, including molecular chaperones and proteases. The main role of molecular chaperones is maintaining protein homeostasis (proteostasis), i.e., a proper balance of protein synthesis, folding, transport, and degradation [90]. Molecular chaperones are highly conserved among prokaryotes and, under stress conditions, they prevent aggregation of unfolded proteins, facilitate degradation of irreversibly misfolded proteins by proteases, and enable solubilization of protein aggregates for subsequent refolding or degradation [91,92]. The key molecular chaperones in bacteria include the heat-shock protein 70 (Hsp70) family chaperone DnaK, its DnaJ (Hsp40) co-chaperone, and the nucleotide exchange factor GrpE, as well as the chaperonin GroEL (Hsp60) and its co-chaperone GroES (Hsp10). The efficient solubilization of aggregated proteins requires the cooperation of the DnaK–DnaJ–GrpE system with ClpB (Hsp100), IbpA/B (the small Hsp family), or Hsp33, which is the primary chaperone redox-activated upon oxidative stress. Most of these chaperones were found to be upregulated in A. baumannii submitted to stresses that impair homeostasis: heat shock (DnaK, GroEL), oxidative stress (GrpE, DnaK, GroES, GroEL), antibiotic exposure (DnaK, GroEL), and desiccation (TF, GroES, GrpE, DnaJ, DnaK, ClpX, ClpB) [18,86,87,93,94]. Wang et al. reported that the expression of more than 50 genes encoding proteins related to proteostasis, including chaperones and the Lon protease, was increased during desiccation [86]. The induction of proteins involved in the proteostasis system was accompanied by protein aggregation. Surprisingly, the accumulation of protein aggregates correlated positively with the ability of A. baumannii to survive desiccation. The survival rate was also increased when protein aggregation was induced prior to desiccation by a subinhibitory concentration of streptomycin, or it was enhanced by the Δlon mutation. It was also demonstrated that the model proteins sequestered in the aggregates, β-lactamase and GFP, retained their activities [86]. These results are in agreement with previous studies showing that bacterial inclusion bodies contain functional proteins, and they confirm that aggregates may serve as compartments that protect proteins against inactivation [95,96]. Upon desiccation, the sequestration of native molecules into aggregates may be favored due to the gradual concentration of proteins. In contrast, high temperatures or other stressors that cause fast and abundant protein misfolding may lead to decreased survival or cell death due to the formation of aggregates enriched in non-functional proteins.
Recent studies showed that the induction of protein aggregation and disturbance of proteostasis may be an efficient strategy to kill pathogenic bacteria. Khodaparast et al. identified several peptides that induced bactericidal protein aggregation in Escherichia coli and A. baumannii [97]. The peptides contained aggregation-prone sequences (APRs) that naturally occur in hydrophobic cores of globular proteins or on protein–protein interaction surfaces. When aggregation was nucleated in bacteria by the peptides containing APRs, it led to the lethal formation of inclusion bodies containing hundreds of proteins. The quaternary amine compounds (QACs), including benzalkonium chloride (BZK), can also trigger protein aggregation in A. baumannii when used at low concentrations [98]. QACs are commonly used biocides that, at high concentrations, disrupt membranes. The exact mechanism of BZK action on proteostasis remains unclear, although it was found that resistance to BZK was acquired through ribosomal protein mutations that protected A. baumannii against BZK-induced protein aggregation.
5. Biofilm and Heterogeneity of A. baumannii Populations
5.1. Formation of A. baumannii Biofilms
Biofilms are multicellular consortia of single or multiple bacterial species enclosed in extracellular polymeric substances (EPS) which comprise polysaccharides, proteins, and nucleic acids secreted by bacteria. The structure of mature biofilms is often very complex with clusters of bacterial cells separated by fluid-filled channels. Diffusion of nutrients and oxygen is limited in biofilms; therefore, the environmental conditions are not homogeneous throughout a biofilm. This leads to the formation of heterogeneous cell subpopulations adapted to local microenvironments. Biofilm-dwelling bacteria are more resistant to antibiotics and other stressors than planktonic cells [99,100]. A number of factors are known to lead to the enhanced antibiotic resistance of biofilms, e.g., impaired drug diffusion due to microbial aggregations and overexpression of the extracellular polymeric substance (EPS) matrix, biofilm-specific efflux pumps, alterations in microbial phenotypic and genotypic features due to stress responses, and specific microenvironment conditions that inactivate antibiotics and the presence of persister cells (see below) [101]. Antibiotics administered at concentrations below the minimum inhibitory concentration (MIC) often induce biofilm formation in a variety of bacterial species [100,102]. A. baumannii forms biofilms on both biotic and abiotic surfaces which contributes to its remarkable ability to survive in hospital environments. While extrinsic factors such as surface hydrophobicity, temperature, and oxygen concentration are reported to influence A. baumannii biofilms, numerous physicochemical and microbial features (e.g., capsular polysaccharides, surface appendages, adhesins, and virulence and resistance determinants) facilitate the formation and maintenance of A. baumannii biofilms (Figure 2A) [103]. In addition to biofilms on solid surfaces, A. baumannii also forms “pellicles” at the air-liquid interface (Figure 2B) [104,105]. The formation of these floating biofilms is a rare trait in clinical A. baumannii isolates, and it is associated with surface-associated motility [104,106]. The relationship between motility and pellicles or surface-attached biofilms is complex. Although a motile state seems to be the opposite to a sedentary lifestyle in biofilms, motility may be required for the formation of microcolonies at the early stages of biofilm development and during the reorganisation of mature three-dimensional biofilm structures [107].
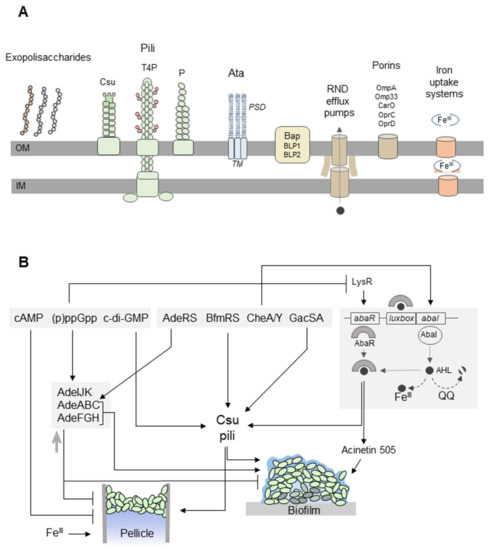
Figure 2.
A. baumannii forms biofilms on solid surfaces and pellicles at the air-liquid interface. (A) Extracellular appendages involved in biofilm/pellicle formation include exopolysaccharides (capsular polysaccharides, poly-β-(1–6)-N-acetylglucosamine (PNAG), alginate), and pili [108]. Csu pili are assembled via the chaperone-usher pathway. The CsuE adhesin, which is located at the pilus tip, exposes three hydrophobic finger-like loops that may insert into cavities in abiotic surfaces [109]. Type IV pili (T4P) are composed of PilA subunits with variable amino-acid sequences in different A. baumannii strains. Depending on the PilA structure, the pili promote twitching motility or biofilm formation [110]. The chaperone–usher P pili are overproduced in A. baumannii pellicles [105]. The homotrimeric Ata autotransporter binds to extracellular matrix components and abiotic surfaces. The transmembrane anchor domain (TM) facilitates the export of the passenger domain (PSD) to the cell surface through a pore formed by the TM. Flexible PSDs allow interactions with different surfaces [111]. Bap and Bap-like proteins (BLP1, BLP2) stabilize the three-dimensional biofilm structure on abiotic surfaces and play a role in the adhesion of A. baumannii to the host cell [112,113]. Three resistance-nodulation-division (RND) efflux pumps (AdeABC, AdeFGH, and AdeIJK) affect A. baumannii biofilm development [114,115,116]. The AdeFGH efflux pump participates in the transport of quorum-sensing (QS) molecules [116]. OmpA is responsible for the attachment of A. baumannii to plastic surfaces and epithelial cells [67]. The CarO, OprC, and OprD porins may be involved in the uptake of metabolites required for the synthesis of siderophores in pellicles [105]. The iron uptake systems, including acinetobactin and enterobactin receptors, are upregulated during pellicle formation [105,117]. (B) The formation of the A. baumannii biofilm and pellicle is regulated by the nucleotide second messengers, two-component signal transduction systems, and QS. cAMP inhibits pellicle formation [104]. The synthesis of Csu pili depends on cyclic di-GMP (c-di-GMP) [118] and the BfmRS and GacSA systems [48,119,120]. The hybrid two-component regulator CheA/Y controls the expression of Csu pili and acinetin-505 via QS [106,121]. The QS system of A. baumannii consists of an AbaI inducer and its cognate receptor AbaR. AbaI is an autoinducer synthase producing N-acyl homoserine lactone (AHL) molecules bound by AbaR. The AbaR–AHL complexes activate the synthesis of AbaI and the expression of QS-dependent genes, which in turn triggers the production of acinetin-505 and Csu pili [121,122]. Biofilm formation may be inhibited by quorum-quenching (QQ) enzymes, which degrade AHLs [123,124], as well as high concentrations of FeIII ions that bind AHLs [125]. On the other hand, FeIII ions are required for pellicle development [105,117]. The AdeABC (controlled by the two-component signal transduction AdeRS system) and AdeFGH efflux pumps participate in biofilm formation [114,116]. However, other studies revealed that the overproduction of efflux pumps may result in decreased biofilm/pellicle growth [115]. ppGpp regulates the expression of genes encoding the efflux pump’s components [126] and inhibits the production of AbaR and acinetin-505 [127].
A. baumannii does not produce flagella; however, it can move via surface-associated motility or twitching motility [104,128]. Multiple genes required for surface-associated motility, including genes associated with purine and pyrimidine biosynthesis or natural competence, were recently identified [128], but its mechanism remains poorly understood. Twitching motility is mediated by the extension and retraction of type IV pili, which are composed of helically assembled PilA subunits. PilA produced by various A. baumannii isolates differ in amino-acid sequence and O-linked glycosylation. It was proposed that, when negatively charged residues dominate on the surface of the headgroup domain of PilA, the pili retract from each other due to electrostatic repulsion and promote twitching motility. The opposite effect, i.e., pili bundling, cell–cell attachment, and biofilm formation, may occur in the case of PilA variants without negatively charged headgroup domains [110].
Several studies showed that one of the main structures required for cell attachment and biofilm development is CPS [129]. For example, it was shown that, in the case of the A. baumannii Δwza, capsule-deficient strain biofilm growth and adhesion to epithelial cells were reduced [45]. However, under certain conditions, enhanced production of the capsule may be associated with biofilm reduction. It was recently reported that the Lon protease affects biofilm formation in A. baumannii [130]. Although the Lon-deficient mutant produced thicker capsule compared to wild-type (WT) cells, it displayed lowered adherence to polystyrene surfaces, decreased motility, and formed a weak pellicle biofilm, but strongly upregulated a surface antigen, encoded by surA1. The exact mechanism of biofilm regulation by Lon remains to be elucidated. A. baumannii produces an additional surface exopolysaccharide, poly-β-(1–6)-N-acetylglucosamine (PNAG). Proteins involved in the polymerization and secretion of PNAG are encoded by the pgaABCD operon widely distributed among A. baumannii clinical isolates [131].
The ability of A. baumannii to form biofilms on abiotic surfaces depends on the production of pili assembled via the CsuA/BABCDE chaperone–usher secretion system which is controlled by the BfmR global regulator [109,119]. OmpA also participates in the development of biofilms on plastic surfaces. In contrast to the CsuA/BABCDE pili system, OmpA is required during the attachment to Candida albicans filaments and human alveolar epithelial cells. After the attachment, OmpA triggers apoptosis of the eukaryotic cells [67]. The giant Bap protein, consisting of 8621 amino acids, is involved in the formation and stabilization of the complex three-dimensional biofilm architecture on abiotic surfaces, and it plays a role in adhesion of A. baumannii to the host cell [112,113]. Bap possess immunoglobulin-like (Ig-like) repeats that seem to be typical for proteins involved in biofilm development, for example, Bap-like proteins, BLP1 and BLP2, produced by some A. baumannii strains [112,132]. Another surface adhesin in A. baumannii, the trimeric Ata autotransporter, is involved in biofilm production by binding various extracellular matrix/basal membrane (ECM/BM) components, including the basement protein laminin and collagen types I, III, IV, and V. During tissue damage, ECM/BM proteins become exposed serving as docking sites for A. baumannii and a niche supporting biofilm growth. Ata is also responsible for self-adhesion of A. baumannii cells and biofilm formation on various abiotic materials [111,133,134]. Other proteins and structures contributing to A. baumannii biofilm development comprise CarO, Omp33, the resistance–nodulation–division (RND) efflux pumps, Pap pilus, and alginate [102,103,108,114,116,135,136]. Depending on the experimental conditions, various structures and mechanisms responsible for pellicle formation were identified [104,105,106,117,137]. Two independent studies demonstrated that the iron uptake systems, as well as CarO, OprD, and OprC porins, are required to develop pellicles [105,117] In addition, the overexpression of multiple pili systems, including Fil and Csu pili, was also observed [105,117,137].
At least three two-component systems regulate surface motility and biofilm/pellicle formation: the aforementioned BmfRS pathway, GacSA, and CheA/Y [48,106,120]. CheA/Y is a hybrid sensor histidine kinase/response regulator that controls the csuA/ABCDE operon and the AbaI-dependent quorum-sensing (QS) pathway [106]. The QS system of A. baumannii consists of AbaI autoinducer synthase and the AbaR receptor protein for the autoinducer, N-acyl homoserine lactone (AHL) (for more details, see Figure 2B) [138]. Different types of AHLs were detected in A. baumannii [139,140,141]. Interestingly, C8-AHL and 3-oxo-C8-AHL were produced by both soil and nosocomial A. baumannii strains, whereas long-chain AHLs with C10, C12, C14, and C16 acyl chains were detected only in the nosocomial isolates [140]. During biofilm formation, the AdeFGH efflux pump participates in the transport of AHLs [116]. It was suggested that QS signals may initiate twitching motility and the attachment of A. baumannii to abiotic surfaces via the CsuA/BABCDE secretion system [122]. Other studies reported that AbaI is required for the later stages of biofilm development [139,141]. Since QS signaling molecules in some bacteria are strong iron chelators, ferric iron (Fe3+) limitation increases the AHL level in A. baumannii in a dose-dependent manner, leading to a stress response and biofilm formation [125].
Nucleotide second messengers such as cAMP, cyclic di-GMP (c-di-GMP), and penta/tetra-guanosine phosphate ((p)ppGpp) are key regulators of numerous bacterial traits including adaptation to harsh environments, as well as transition from biofilm to motility, mutualism to commensalism, acute to chronic virulence characteristics, and cell division to cell differentiation [142,143]. The exact functions of these messengers in A. baumannii were only partially examined. It was reported that enhanced cAMP levels, caused by the lack of cAMP phosphodiesterase, lead to the inhibition of surface-associated motility and pellicle formation [104]. C-di-GMP is synthesized by diguanylate cyclase activity of GGDEF domain-containing proteins, while degradation of c-di-GMP into two GMP molecules is catalyzed by the phosphodiesterase activity of EAL domain-containing proteins [144,145]. Most c-di-GMP-dependent signalling pathways regulate the bacteria ability to interact with abiotic surfaces or with other bacterial and eukaryotic cells. Eleven genes for GGDEF/EAL proteins in the genome of the A. baumannii 17,978 strain were identified, and most of the predicted proteins were enzymatically active [118]. It was demonstrated that distinct panels of these enzymes promote biofilm formation, macro-colony growth, and surface-associated motility [118].
The (p)ppGpp alarmone is produced by the RelA/SpoT proteins in response to amino-acid starvation and other stresses. (p)ppGpp triggers the stringent response resulting in the downregulated transcription of most metabolic genes and the upregulation of genes responsible for amino-acid biosynthesis [146,147,148]. The stringent response was linked to biofilm formation in a range of pathogens, including Acinetobacter spp. [142,149,150]. The formation of biofilms is impaired [149,151] or enhanced [152,153] in (p)ppGpp-deficient bacteria. Recent studies revealed the interplay among the stringent response, QS, motility, and biofilm/pellicle formation in A. baumannii, but the exact mechanisms remain unclear (Figure 2B). It was reported that the formation of A. baumannii biofilms can be inhibited by a synthetic dodecapeptide 1081, which triggers degradation of (p)ppGpp [149]. However, the activity of peptide 1081 and its link with the stringent response were recently questioned [154]. The ΔrelA mutation in A. baumannii results in a hypermotile phenotype, as well as in the overproduction of AbaR and acinetin-505. Acinetin-505 is a 505-Da lipopeptide that may act as a surfactant promoting surface-associated motility [127], biofilm formation, and virulence [121]. Numerous studies indicated that (p)ppGpp is involved in the formation of dormant persister bacteria, which are implicated in biofilm tolerance to antibiotics [127,155].
5.2. Persisters and Heterogeneity of A. baumannii Populations
Persisters are able to survive exposure to a bactericidal drug concentration, and they usually constitute a small fraction of bacterial populations [156]. Antibiotic persistence is a transient state, and, when persisters resume growth after drug treatment, their progeny become antibiotic susceptible. There is an ongoing debate about mechanisms underlying persister formation [157,158,159,160]. It is well known that biofilms provide conducive niches that favor the formation of persisters [161]. Persisters can arise spontaneously or in response to stress caused by antibiotics, the host immune response, ROS, high osmolarity, pH changes, diauxic shift, desiccation, or nutrient starvation. In addition to the stringent response mentioned above, toxin–antitoxin modules, quorum signaling, efflux pumps, the SOS, and oxidative stress responses can be activated during persister formation. These pathways and stimuli may lead to decreased metabolism, depletion of ATP, protein aggregation, and inhibition of translation [132,162,163]. It was recently demonstrated that, in E. coli, (p)ppGpp induces production of the ribosome modulation factor (RMF), the hibernation-promoting factor (Hpf), and the ribosome-associated inhibitor (RaiA), which convert active 70S ribosomes into inactive 70S, 90S, and 100S ribosomes, leading to translation inhibition [164]. It should be noted that, due to diverse conditions in biofilm structures, multiple mechanisms triggering antibiotic tolerance, and stochastic effects, persister subpopulations should be considered as a heterogeneous group of cells.
The formation of A. baumannii persisters induced by polymixin B, meropenem, and ceftazidime was reported [165,166,167,168,169]. It was also found that the ppGpp deficiency in the A. baumannii ΔrelA strain reduced formation of persister cells tolerant to colistin and rifampicin [127]. The analysis of the transcriptome of persisters tolerant to ceftazidime revealed upregulation of two toxin–antitoxin systems HigB/HigA and DUF1044/RelB, as well as downregulation of certain metabolic pathways, including the electron transport chain and citrate cycle [168]. Interestingly, the expression of genes associated with biodegradation pathways of aromatic compounds was detected in persistent cells. It was suggested that the degradation of aromatic rings in antibiotics could be utilized by A. baumannii persisters during nutrient starvation [168]. Zou et al. found that the major fraction of A. baumannii persisters that survive β-lactam antibiotic treatment contains spherical non-walled, but metabolically active cells [170]. In contrast to wall-deficient, so-called L-forms of other Gram-negative bacteria, A. baumannii non-walled cells were able to survive without any osmoprotective agent. This type of A. baumannii persister cell was also formed during antibiotic therapy in vivo in Galleria melonella larvae which were used as the infection model.
Biofilms provide a conducive environment facilitating not only phenotypic heterogeneity (e.g., persister formation) but also genetic diversification. It was demonstrated that evolution within A. baumannii biofilms can generate greater genetic diversity than in planktonic, well-mixed populations [171]. Planktonic cells exposed to ciprofloxacin shared the same limited number of mutations in topoisomerase (the primary drug target), whereas biofilm-adapted populations acquired different types of mutations in the regulators of the efflux pumps. The emergence of a certain trade-off between fitness and resistance level was detected; biofilm-adapted clones were less drug-resistant than planktonic cells, but more fit in the absence of the drug [171]. Other studies demonstrated that the exposure of A. baumannii biofilms to sub-inhibitory concentrations of ciprofloxacin or tetracycline led to the generation of genetic and phenotypic diversity among biofilm dispersal isolates [172]. Dispersed cells accumulate a wide diversity of mutations that enhance biofilm formation and antibiotic resistance. For example, the efflux transport system AdeABC was upregulated in the presence of both ciprofloxacin and tetracycline, whereas the expression of RecA and UmuD, which are involved in DNA repair and mutagenesis, was increased during ciprofloxacin treatment.
Phenotypic alteration between opaque (VIR-O) and translucent (AV-T) colonies is another example of A. baumannii population heterogeneity. It was shown that both phenotypes exhibited significant differences in cell morphology, biofilm formation, surface motility, antibiotic resistance, and virulence [173]. VIR-O cells were covered with a thicker coating of the extracellular capsule, and they were more resistant to disinfectants, ROS, antibiotics, lysozyme, and the cathelin-related antimicrobial peptide [53]. This highly virulent subpopulation dominated in the bloodstream of human patients. The AV-T cells produced more dense biofilms and a larger quantity of OMVs in comparison with the VIR-O variant [174]. The analysis of VIR-O and AV-T transcriptomes suggested that the AV-T subpopulation is better adapted for natural environments outside the host than VIR-O cells. The phenotype switching between VIR-O and AV-T subpopulations depends on several factors, including a TetR-type transcriptional regulator ABUW_1645, the ArpAB efflux system, the EnvZ/OmpR two-component system [174], and ppGpp [127].
Antibiotic heteroresistance is another common phenotype that may contribute to the heterogeneity of bacterial populations. We describe this phenomenon in the next section, discussing the broader problem of multidrug resistance of A. baumannii.
6. Multidrug Resistance of A. baumannii
Multidrug-resistant pathogens pose serious threats in healthcare settings worldwide. For the past number of years, antimicrobial discovery and resistance development to new antimicrobials occurred almost at the same time. Not surprisingly, A. baumannii, similarly to other bacteria, also acquired resistance to newly developed antimicrobial agents [175]. To characterize the various patterns of resistance, the following terms are used: MDR, extensively drug-resistant (XDR), and pandrug-resistant (PDR) bacteria. According to the definition proposed by Magiorakos et al. [176], MDR is defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories, XDR is defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories, and PDR is defined as non-susceptibility to all agents in all antimicrobial categories. In the case of Acinetobacter spp., 22 antimicrobial agents belong to nine categories: aminoglycosides, antipseudomonal carbapenems, antipseudomonal fluoroquinolones, (antipseudomonal) penicillins + β lactamase inhibitors, extended spectrum cephalosporins, folate pathway inhibitors, polymixins, and tetracyclines. A. baumannii can become resistant to a variety of antibiotics via intrinsic and acquisition mechanisms. Its ability to acquire drug resistance genes from other human pathogens is not well understood. However, considering the capability of the A. baumannii genome to exchange genetic material both within and between species, it is quite likely that these bacteria may have evolved toward enhanced pathogenicity.
6.1. Mechanisms Responsible for A. baumannii Multidrug Resistance
The main mechanisms conferring resistance to different classes of antibiotics include the presence of β-lactamases, modifying enzymes, permeability defects, alteration of target sites, and multidrug efflux pumps [177]. Severe hospital-acquired infections caused by A. baumannii involve the use of carbapenems as highly effective drugs of choice used for the treatment of such infections [178]. Because of their broad spectrum, carbapenems are often active against microorganisms resistant to other antimicrobial compounds, and they are frequently used to treat complicated bacterial infections. Over the last few years, A. baumannii MDR strains became increasingly resistant to carbapenems, the drug of choice to treat severe infections caused by these bacteria. The main cause of carbapenem resistance is the presence of oxacillinases (OXA), which belong to the Ambler class D β-lactamases. Over 400 OXA enzymes encoded by chromosome- or plasmid-located genes were characterized. Other classes of β-lactamases: class A, class B (metallo-β-lactamases, MBL), and class C (AmpC) were also identified in A. baumannii strains [6,179,180,181,182,183,184,185]. The most frequent MBLs in A. baumannii are imipenemases (IMPs), Verona integron-encoded MBL (VIM), and MBL from New Delhi (NDM). Class C β-lactamases are encoded by the ampC gene. Overexpression of ampC, regulated by an upstream insertion sequence (IS) element known as ISAba1, is the main mechanism of resistance to third-generation cephalosporins in A. baumannii [186]. Overexpression of the OXA and AmpC enzymes due to the presence of IS elements (see below), as well as the emergence of new OXA and AmpC variants, contributes to the increasing problem of A. baumannii resistance [187,188].
Another mechanism of A. baumannii resistance is associated with enzymatic modification of the antimicrobial molecule. One of the best examples of resistance via modification of the drug is the presence of a large group of aminoglycoside-modifying enzymes (AMEs). These enzymes possess unique substrate specificity and modify amino- or hydroxyl- groups of the aminoglycosides. There are three different types of AMEs, acetyltransferases, nucleotidyl transferases, and phosphotransferases, while all of them were identified in A. baumannii isolates [6].
Proteomic analysis of A. baumannii MDR strains shows protein variability that could be correlated with the appearance of resistance phenotypes, especially OMPs, which are involved in cellular drug uptake or efflux. The emergence of an antibiotic resistance level is often related to diverse variations in the expression of OMPs. It was found that, in A. baumannii, OmpA is strongly associated not only with adhesion to epithelial cells and biofilm formation, as mentioned earlier, but also with the modulation of cellular permeability and antibiotic resistance [103]. Importantly, changes in permeability frequently result in low-level resistance; therefore, the combination with other mechanisms, such as increased expression of efflux pumps, to confer a high-level antibiotic resistance phenotype is required [189].
Another common mechanism of antibiotic resistance in A. baumannii is alteration of the target site or cellular functions [2]. This often results from spontaneous mutation of a bacterial gene on the chromosome. Modification of the target site results in decreased affinity for the drug molecule. One of the most known examples of target changes is enzymatic alteration of the binding site. In an alternative pathway, bacteria produce new proteins that protect the target against an antibiotic. Examples of drugs affected by this mechanism include fluoroquinolones and tetracyclines [190]. In A. baumannii, point mutations in the gyrA/parC topoisomerases result in fluoroquinolones resistance, whereas a mutation in rpsJ, the gene that encodes the ribosomal S10 protein, is responsible for tigecycline resistance [191]. Another interesting example is the mechanism responsible for colistin resistance. Positively charged colistin kills bacteria by interacting with the negatively charged lipid A and destabilization of the OM. Mutations in the lipid A biosynthesis genes, lpxA, lpxC, and lpxD, result in the complete loss of lipooligosaccharides, which in turn abolish interactions with colistin. The second mechanism depends on the PmrAB two-component system. Mutations in pmrA or pmrB lead to the activation of the pmrC gene located upstream of pmrAB, encoding phosphoetanolamine transferase. Phosphoethanolamine transferred to lipid A decreases the negative charge of LOS, preventing colistin binding [62,63].
Among several types of efflux pumps that confer multidrug resistance, the RND efflux systems (AdeABC, AdeFGH, AdeIJK) are the most prevalent in A. baumannii. The AdeABC pump, found in 80% of A. baumannii isolates, extrudes a wide range of antibiotics, including β-lactams, aminoglicosides, fluoroquinolones, tetracyclines-tigecycline, macrolides–lincosamides, and chloramphenicol [103,177,192].The expression of AdeABC is tightly regulated by the AdeRS two-component system which was found to control almost 600 other genes [192,193]. Point mutations in the adeR–adeS genes or the presence of an ISAba1 insertion sequence upstream from the adeABC operon result in the overexpression of the AdeABC pump and multidrug resistance [194]. The AdeFGH pump, when overexpressed, confers enhanced resistance to fluoroquinolones, tetracycline–tigecycline, chloramphenicol, clindamycin, trimethoprim, sulfamethoxazole, sodium dodecyl sulfate, and dyes such as ethidium bromide, safranin O, and acridine orange [195]. It was found that overexpression of AdeFGH is caused by mutation in the adeL gene located upstream from the adeFGH operon, as well as the encoding of an a-Lys-type transcriptional regulator. The AdeIJK pump is produced in A. baumannii constitutively, and it is responsible for resistance to the same major drug classes as AdeABC, as well as antifolates and fusidic acid [115].
6.2. Genetic Elements Responsible for A. baumannii Multidrug Resistance
Members of the genus Acinetobacter quickly develop resistance to antimicrobial compounds. Antibiotic resistance genes can be disseminated through various mechanisms of horizontal gene transfer such as transformation, conjugation, and transduction. A. baumannii appears to use all the mechanisms; however, recent studies point to natural transformation as the mechanism playing an important role in the acquisition of the multidrug resistance phenotype [196]. In this process, bacteria appeared to be capable of uptake, integration, and functional expression of naked fragments of extracellular DNA from the environment. Bacteria could use transformation to avoid being targeted by antibiotics by accepting the genetic variation present in their neighborhood, including drug resistance genes [197]. Multidrug resistance of A. baumannii is mainly due to the horizontal acquisition of resistance genes, although recent studies showed that increased expression of chromosomal genes for the efflux system plays a major role in MDR [177].
Often, A. baumannii resistance to more than one class of antibiotics occurs when genes encoding resistance to antimicrobial agents are physically located in close proximity to each other on mobile genetic elements such as plasmids, transposons, and integrons. Plasmid profiling revealed the presence of multiple plasmids of varying molecular sizes in more than 80% of Acinetobacter isolates [198]. They constitute a reservoir of genes important not only for the dissemination of antibiotic resistance but also essential for bacteria adaptation and evolution. Recent analysis of 173 complete plasmid sequences from A. baumannii isolates originated from 17 countries revealed that 34.6% of the plasmids pose antibiotic resistance genes [199]. Bertini et al. constructed a classification system for the A. baumannii plasmids based on the sequence of replicase genes, and they identified 19 homology groups (GR1–GR19) [200]. Fourteen additional groups of plasmids were recently proposed [199,201,202]. GR6 was the most prevalent group detected in antibiotic-resistant A. baumannii isolates from Europe [203]. The GR6 plasmids harbor class D β-lactamase genes, including blaOXA-23, blaOXA-58, and blaOXA-40, aminoglycoside-resistant genes (aph(3’)-Via, aadB, aadA2, strA, strB, aacA4, aph(3’)-Via), and sulfonamide (sul2) and streptomycin (strAB) resistance genes [199,200,203].
IS and transposons (Tn) are able to move from one genomic location to another in the chromosome or plasmid DNA within a single cell [204]. IS may include a strong promoter that initiates the expression of a downstream gene, e.g., ISAba1 located upstream of blaOXA-51 genes or the adeABC operon leading to intrinsic A. baumannii carbapenem resistance or multidrug resistance, respectively [194,205]. A diverse range of composite Tn, which harbor antibiotic resistance genes flanked by IS, was identified in A. baumannii isolates. These transposons encode AmpC cephalosporinases, OXA carbapenemases, aminoglycosidases, and NDM or VIM metallo-carbapenemases [179,186,206,207,208,209,210]. For example, a chromosomally located Tn125-like transposon containing the blaNDM-1 gene was identified in NDM-1-producing A. baumannii from European countries [179]. The blaOXA-23 genes with adjacent ISAba1 were detected in globally disseminated transposons Tn2006 and Tn2008, as well as in Tn2009 from A. baumannii strains isolated in China [206,208]. The largest antibiotic resistance gene clusters in various A. baumannii isolates are resistance islands designated AbaR1–R30. These complex transposons are located in the chromosomal comM gene (encoding the ATP-ase), and they carry heavy-metal resistance determinants apart from antibiotic resistance genes [125,211]. Most AbaRs from A. baumannii strains of international clone I share a backbone transposon Tn6019 interrupted by a large compound transposon that contains a variable-resistance region flanked by two copies of Tn6018 [212]. AbaR1 contains genes conferring resistance to ampicillin (veb1 and oxa10), sulfonamides (three copies of the sul1 gene), streptomycin (two copies of aadA1; strA, strB), aminoglycoside (aadB, aacC1, aphA1b, aacA), chloramphenicol (cmlA1, cmlA5, cmlA9, and catA1), rifampin (arr2), trimethoprim (dfrA1 and dfrA10), and tetracycline (tetA-A and tetA-G) [211].
Integrons are responsible for the dissemination of antibiotic resistance, especially among Gram-negative bacteria [213]. They can integrate into chromosomes or plasmid via site-specific recombination. These genetic elements are able to acquire, integrate, and express gene cassettes which can carry antibiotic resistance. Class 1 integrons are commonly found in A. baumannii and typically encode genes for aminoglycoside resistance, Ambler class A β-lactamases, metallo-beta-lactamases, and oxacillinases, as well resistance to antiseptics and sulfonamides [196]. They were also reported in clinical A. baumannii strains. Many reports showed that clinical A. baumannii strains carrying integrons were significantly more resistant to all tested antibiotics than strains lacking integrons [214]. It should be noted that some mobile genetic elements and resistance genes are disseminated worldwide (e.g., ISAbaI, blaOXA-23, blaOXA-51), whereas others are distributed locally across different countries or regions (e.g., integrons and class B carbapenemases are more frequently found in Asia) [215,216].
6.3. Cross-Resistance, Co-Regulatory Resistance, and Stress-Induced Resistance to Antibiotics in A. baumannii
Antibiotics can induce selective pressure on bacterial populations, leading to antimicrobial resistance through a mechanism called cross-resistance. This mechanism confers resistance to an entire class of antibiotic and is mainly achieved via multidrug efflux pumps. In A. baumannii, efflux pumps can be specific for a single substrate or can confer resistance to multiple antimicrobials by facilitating the extrusion of a broad range of compounds including antibiotics, heavy metals, and biocides from the bacterial cell [177]. Other studies demonstrated that treatment of A. baumannii infection with cationic microbial peptide colistin can induce not only increased resistance to antibiotics but also resistance to host cationic antimicrobials typically found at sites of inflammation. These findings showed that understanding the molecular basis of cross-resistance is important for the development of more effective therapeutic schemes [217].
Another mechanism involved in bacterial resistance is called co-regulatory resistance. This occurs when resistant genes to antimicrobial agents are controlled by regulatory proteins [218]. Commonality of target sites between different class of antibiotics leads to the selection of mutants and the emergence of cross-resistance, as well as the co-selection and persistence of antibiotic-resistant strains. The presence of class 1 integrons in A. baumannii strains confers several phenotypes, including resistance, to a broad range of antibiotic classes, in addition to heavy metals and biocides [213]. In integrons, antibiotic resistance genes are under the control of a single promoter. As a result, these genes are expressed in a coordinated manner.
Some microorganisms readily acquire antibiotic resistance mechanisms in response to environmental stresses. It was shown that different physiological conditions influenced antimicrobial susceptibility and porin expression in A. baumannii. For example, putative efflux transporters were induced by the physiological concentrations of NaCl, contributing to increased tolerance of A. baumannii to aminoglycosides, carbapenems, quinolones, and colistin. Moreover, the physiological level of some cations present within the host promotes the upregulation of genes coding for multidrug efflux pumps [219]. Such regulated changes in efflux pump expression may increase the ability of this pathogen to survive antibiotic challenge.
As mentioned, many clinical A. baumannii strains can survive drying for a prolonged period of time. However, when rehydration happens, DNA damage may occur such as various DNA lesions, cross-linking, base removal, and strand breaks. To repair some of the DNA damage, A. baumannii developed an inducible DNA damage response in which RecA plays a major regulatory role in mechanisms involved in stress survival [220]. The RecA protein is involved in DNA damage repair and, consequently, in cellular protection against stresses induced by DNA damaging agents, several classes of antibiotics, and oxidative agents. This response increases mutagenesis and is one of the mechanisms used by A. baumannii to acquire antibiotic resistance, particularly in hospitals under clinically relevant DNA-damaging conditions [221].
In another study, it was shown that A. baumannii cells pretreated at 45 °C for 30 min were better able to survive a subsequent streptomycin exposure than cells pretreated at 37 °C. This phenomenon may be explained by the synthesis of misfolded proteins, produced by the streptomycin-disrupted ribosome and inserted into the bacterial membrane. Treating A. baumannii cells with the aminoglycoside antibiotic streptomycin induces not only ribosomal mistranslation but also expression of the heat-shock proteins DnaK and GroEL, responsible for elimination of aberrant polypeptides, thereby reducing their toxicity to bacterial cells [222].
The emergence of antibiotic-resistant A. baumannii strains may be preceded by the formation of persister cells. The evolution of antibiotic resistance promoted by persistence or tolerance was observed in vitro or in patients in the case of other bacterial species [223,224,225,226,227]. Persisters, as non-dividing cells, can accumulate de novo mutations via mechanisms independent of DNA replication or via horizontal gene transfer.
6.4. Heteroresistance to Antibiotics in A. baumannii
The term “heteroresistance” is defined as the presence of subpopulations of cells that have a higher MIC than the dominant population [228,229]. In contrast to persisters which are dormant cells, heteroresistant subpopulations can proliferate in the presence of antibiotics.
A. baumannii heteroresistance to colistin in “colistin-susceptible” clinical isolates was described for the first time by Li et al. [230]. Colistin heteroresistance was caused by mutations and the insertional inactivation of the lipid A biosynthesis genes, leading to the complete loss of lipooligosacchrides [63,231]. Heteroresistance to carbapenems, aminoglycosides, and trimethoprim/sulfamethoxazole in A. baumannii was also reported [167,232,233]. For example, it was found that increased tobramycin resistance was an unstable phenotype that emerged due to the extensive RecA-dependent amplification of the aadB gene encoding an aminoglycoside adenylyltransferase. The aadB gene was carried on a plasmid, in the region containing four other resistance genes [233]. Gene amplification seems to be the main mechanism conferring heteroresistance in various pathogens. Analysis of the prevalence and mechanisms of heteroresistance in A. baumannii, E. coli, Salmonella enterica, and Klebsiella pneumoniae revealed that almost 28% of clinical isolates were heteroresistant to various antibiotics. The majority of heteroresistance cases were unstable, and they resulted from tandem amplification of resistance genes [234].
7. Concluding Remarks
A. baumannii is the primary species detected and isolated from hospital environments including intensive care units. The ease with which A. baumannii colonizes patients makes it problematic as these patients might transmit or become infected when the immune system is stressed. Most clinical A. baumannii isolates are naturally competent; thus, they can rapidly acquire resistance genes. The success of A. baumannii as a human pathogen is also associated with its outstanding ability to survive long-term desiccation in nosocomial environments. The formation of biofilms and antibiotic-tolerant persisters contributes to the heterogeneity of A. baumannii populations, facilitating their adaptation to fluctuating environments. It was proposed that, in response to desiccation stress, A. baumannii follows the “bust-and-boom” strategy [87,235]. The “bust-and-boom” strategy implies the death of the main stressed population (e.g., in the biofilm), where a few viable surviving bacteria can resume growth and restore the original population, once the environmental conditions are suitable. Persister cells and antibiotic heteroresistance are the main causes of recurrent and difficult-to-eradicate infections. Huge progress was made in the last decade toward understanding the mechanisms underlying these processes. However, there are still several issues that remain to be elucidated: (1) the controversy about the definition and metabolic state of persister cells still exists, and multiple definitions of heteroresistance used in the literature may often lead to confusing and inconsistent conclusions [228]; (2) furthermore, different types of heteroresistant or persister cells, including viable but non-culturable bacteria and L-forms, may coexist in the same population; (3) persisters and heteroresistance are difficult to detect or diagnose with standard procedures; (4) it should also be kept in mind that most data regarding antibiotic persistence and heteroresistance originate from experiments performed in laboratory settings and animal models; therefore, they may not reflect the fate of pathogen cells in the human host; (5) inappropriate use of drugs may cause rapid development of persistence and resistance. These problems are associated with most pathogenic infections, but they are particularly important in the case of A. baumannii, due to its nosocomial origin and dramatically increasing prevalence of MDR isolates. Antibiotic persistence, population heterogeneity, and biofilm-related resistance should be considered as significant risk factors in the course of choosing an appropriate therapy. In the past few years, several strategies that eliminate A. baumannii biofilms and kill persister cells were reported. These approaches include the combination of antibiotics, natural or synthetic AMPs, stringent response inhibitors, QS antagonists, and biofilm disruptors [149,236,237,238,239,240,241,242,243,244,245]. Although these results are promising, further studies are needed to implement novel therapeutic strategies or design drug candidates that will effectively combat A. baumannii infections.
Author Contributions
Conceptualization, S.M., B.F.-B. and E.L.; writing—original draft preparation, S.M., B.F.-B., A.Ł., D.K.-W., K.S.-S. and E.L.; writing—review and editing, S.M., B.F.-B. and E.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Center, Poland, grant number 2018/29/Z/NZ6/01040 (project funded under the Joint Program Initiative on Antimicrobial Resistance JPI EC AMR Call 2018).
Acknowledgments
We would like to thank Martin Blaszk for his assistance with the preparation of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AHL | N-Acyl homoserine lactone |
| AMEs | Aminoglycoside-modifying enzymes |
| AMP | Antimicrobial peptide |
| APRs | Aggregation-prone sequences |
| BZK | Benzalkonium chloride |
| CPS | Capsular polysaccharides |
| DCs | Dendritic cells |
| EPS | Extracellular polymeric substances |
| ICU | Intensive care units |
| IHF | Integration host factor |
| IM | Inner membrane |
| LOS | Lipooligosaccharides |
| LPS | Lipopolysaccharides |
| MDR | Multidrug-resistant |
| NETs | Neutrophil extracellular traps |
| OM | Outer membrane |
| OMP | Outer membrane protein |
| OMVs | Outer membrane vesicles |
| PAMPs | Pathogen-associated molecular patterns |
| PDR | Pan-drug resistant |
| PNAG | Poly-β-(1–6)-N-acetylglucosamine |
| PRRs | Pattern recognition receptors |
| QACs | Quaternary amine compounds |
| QS | Quorum-sensing |
| Quorum-quenching | |
| ROS | Reactive oxygen species |
| RRF | Ribosomal recycling factor |
| T2SS | Type II secretion system |
| TLR4 | Toll-like receptor 4 |
| TNF | Tumor necrosis factor |
| XDR | Extensively drug-resistant |
References
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Manchanda, V.; Sinha, S.; Singh, N. Multidrug resistant Acinetobacter. J. Glob. Infect. Dis. 2010, 2, 291. [Google Scholar] [CrossRef] [PubMed]
- Blanco, N.; Harris, A.D.; Rock, C.; Johnson, J.K.; Pineles, L.; Bonomo, R.A.; Srinivasan, A.; Pettigrew, M.M.; Thom, K.A. Risk factors and outcomes associated with multidrug-resistant Acinetobacter baumannii upon intensive care unit admission. Antimicrob. Agents Chemother. 2018, 62, e01631-17. [Google Scholar] [CrossRef] [PubMed]
- Giammanco, A.; Calà, C.; Fasciana, T.; Dowzicky, M.J. Global Assessment of the Activity of Tigecycline against Multidrug-Resistant Gram-Negative Pathogens between 2004 and 2014 as Part of the Tigecycline Evaluation and Surveillance Trial. mSphere 2017, 2, e00310–e00316. [Google Scholar] [CrossRef]
- Spellberg, B.; Rex, J.H. The value of single-pathogen antibacterial agents. Nat. Rev. Drug Discov. 2013, 12, 963–964. [Google Scholar] [CrossRef]
- Vrancianu, C.O.; Gheorghe, I.; Czobor, I.B.; Chifiriuc, M.C. Antibiotic Resistance Profiles, Molecular Mechanisms and Innovative Treatment Strategies of Acinetobacter baumannii. Microorganisms 2020, 8, 935. [Google Scholar] [CrossRef]
- Piperaki, E.T.; Tzouvelekis, L.S.; Miriagou, V.; Daikos, G.L. Carbapenem-resistant Acinetobacter baumannii: In pursuit of an effective treatment. Clin. Microbiol. Infect. 2019, 25, 951–957. [Google Scholar] [CrossRef]
- Assimakopoulos, S.F.; Karamouzos, V.; Lefkaditi, A.; Sklavou, C.; Kolonitsiou, F.; Christofidou, M.; Fligou, F.; Gogos, C.; Marangos, S. Triple combination therapy with high-dose ampicillin/sulbactam, high-dose tigecycline and colistin in the treatment of ventilator-associated pneumonia caused by pan-drug resistant Acinetobacter baumannii: A case series study. Infez. Med. 2019, 11, 11–16. [Google Scholar]
- Morris, F.C.; Dexter, C.; Kostoulias, X.; Uddin, M.I.; Peleg, A.Y. The Mechanisms of Disease Caused by Acinetobacter baumannii. Front. Microbiol. 2019, 10, 1601. [Google Scholar] [CrossRef]
- Zhou, H.; Yao, Y.; Zhu, B.; Ren, D.; Yang, Q.; Fu, Y.; Yu, Y.; Zhou, J. Risk factors for acquisition and mortality of multidrug-resistant Acinetobacter baumannii bacteremia: A retrospective study from a Chinese hospital. Medicine 2019, 98, e14937. [Google Scholar] [CrossRef]
- Greene, C.; Vadlamudi, G.; Newton, D.; Foxman, B.; Xi, C. The influence of biofilm formation and multidrug resistance on environmental survival of clinical and environmental isolates of Acinetobacter baumannii. Am. J. Infect. Control 2016, 44, e65–e71. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Kuolee, R.; Harris, G.; Chen, W. Role of NADPH Phagocyte oxidase in host defense against acute respiratory Acinetobacter baumannii infection in mice. Infect. Immun. 2009, 77, 1015–1021. [Google Scholar] [CrossRef] [PubMed]
- García-Patiño, M.G.; García-Contreras, R.; Licona-Limón, P. The immune response against Acinetobacter baumannii, an emerging pathogen in nosocomial infections. Front. Immunol. 2017, 8, 441. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, M.S.; Ellett, F.; Murray, G.L.; Kostoulias, X.; Cerqueira, G.M.; Schulze, K.E.; Maifiah, M.H.M.; Li, J.; Creek, D.J.; Lieschke, G.J.; et al. Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc. Natl. Acad. Sci. USA 2016, 113, 9599–9604. [Google Scholar] [CrossRef]
- Bruhn, K.W.; Pantapalangkoor, P.; Nielsen, T.; Tan, B.; Junus, J.; Hujer, K.M.; Wright, M.S.; Bonomo, R.A.; Adams, M.D.; Chen, W.; et al. Host fate is rapidly determined by innate effector-microbial interactions during Acinetobacter baumannii bacteremia. J. Infect. Dis. 2015, 211, 1296–1305. [Google Scholar] [CrossRef]
- Kamoshida, G.; Kikuchi-Ueda, T.; Tansho-Nagakawa, S.; Nakano, R.; Nakano, A.; Kikuchi, H.; Ubagai, T.; Ono, Y. Acinetobacter baumannii escape from neutrophil extracellular traps (NETs). J. Infect. Chemother. 2014, 21, 43–49. [Google Scholar] [CrossRef]
- Kamoshida, G.; Tansho-Nagakawa, S.; Kikuchi-Ueda, T.; Nakano, R.; Hikosaka, K.; Nishida, S.; Ubagai, T.; Higashi, S.; Ono, Y. A novel bacterial transport mechanism of Acinetobacter baumannii via activated human neutrophils through interleukin-8. J. Leukoc. Biol. 2016, 100, 1405–1412. [Google Scholar] [CrossRef]
- Juttukonda, L.J.; Green, E.R.; Lonergan, Z.R.; Heffern, M.C.; Chang, C.J.; Skaara, E.P. Acinetobacter baumannii OxyR Regulates the Transcriptional Response to Hydrogen Peroxide. Infect. Immun. 2019, 87, e00413–e00418. [Google Scholar]
- Konstantinidis, T.; Kambas, K.; Mitsios, A.; Panopoulou, M.; Tsironidou, V.; Dellaporta, E.; Kouklakis, G.; Arampatzioglou, A.; Angelidou, I.; Mitroulis, I.; et al. Immunomodulatory role of clarithromycin in Acinetobacter baumannii infection via formation of neutrophil extracellular traps. Antimicrob. Agents Chemother. 2016, 60, 1040–1048. [Google Scholar] [CrossRef]
- Kamoshida, G.; Kikuchi-Ueda, T.; Nishida, S.; Tansho-Nagakawa, S.; Ubagai, T.; Ono, Y. Pathogenic bacterium Acinetobacter baumannii inhibits the formation of neutrophil extracellular traps by suppressing neutrophil adhesion. Front. Immunol. 2018, 9, 178. [Google Scholar] [CrossRef]
- Lázaro-Díez, M.; Chapartegui-González, I.; Redondo-Salvo, S.; Leigh, C.; Merino, D.; Segundo, D.S.; Navas, J.; Icardo, J.M.; Acosta, F.; Ocampo-Sosa, A.; et al. Human neutrophils phagocytose and kill Acinetobacter baumannii and A. pittii. Sci. Rep. 2017, 7, 4571. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; KuoLee, R.; Harris, G.; Van Rooijen, N.; Patel, G.B.; Chen, W. Role of macrophages in early host resistance to respiratory Acinetobacter baumannii infection. PLoS ONE 2012, 7, e40019. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Human antimicrobial peptides and proteins. Pharmaceuticals 2014, 7, 545–594. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Tsai, P.W.; Chen, J.Y.; Lin, Y.Y.; Lan, C.Y. OmpA binding mediates the effect of antimicrobial peptide LL-37 on Acinetobacter baumannii. PLoS ONE 2015, 10, e0141107. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Sambanthamoorthy, K.; Palys, T.; Paranavitana, C. The Human Antimicrobial Peptide LL-37 and Its Fragments Possess both Antimicrobial and Antibiofilm Activities Against Multidrug-Resistant Acinetobacter Baumannii; Elsevier Inc.: Amsterdam, The Netherlands, 2013; Volume 49, ISBN 3013199172. [Google Scholar]
- Sancho-Vaello, E.; François, P.; Bonetti, E.J.; Lilie, H.; Finger, S.; Gil-Ortiz, F.; Gil-Carton, D.; Zeth, K. Structural remodeling and oligomerization of human cathelicidin on membranes suggest fibril-like structures as active species. Sci. Rep. 2017, 7, 15371. [Google Scholar] [CrossRef]
- Feng, Z.; Jia, X.; Adams, M.D.; Ghosh, S.K.; Bonomo, R.A.; Weinberg, A. Epithelial innate immune response to Acinetobacter baumannii challenge. Infect. Immun. 2014, 82, 4458–4465. [Google Scholar] [CrossRef]
- Merle, N.S.; Church, S.E.; Fremeaux-Bacchi, V.; Roumenina, L.T. Complement system part I—molecular mechanisms of activation and regulation. Front. Immunol. 2015, 6, 262. [Google Scholar] [CrossRef]
- King, L.B.; Swiatlo, E.; Swiatlo, A.; McDaniel, L.S. Serum resistance and biofilm formation in clinical isolates of Acinetobacter baumannii. FEMS Immunol. Med. Microbiol. 2009, 55, 414–421. [Google Scholar] [CrossRef]
- Kim, S.W.; Choi, C.H.; Moon, D.C.; Jin, J.S.; Lee, J.H.; Shin, J.H.; Kim, J.M.; Lee, Y.C.; Seol, S.Y.; Cho, D.T.; et al. Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol. Lett. 2009, 301, 224–231. [Google Scholar] [CrossRef]
- Koenigs, A.; Stahl, J.; Averhoff, B.; Göttig, S.; Wichelhaus, T.A.; Wallich, R.; Zipfel, P.F.; Kraiczy, P. CipA of Acinetobacter baumannii Is a Novel Plasminogen Binding and Complement Inhibitory Protein. J. Infect. Dis. 2016, 213, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Harris, G.; Lee, R.K.; Lam, C.K.; Kanzaki, G.; Patel, G.B.; Xu, H.H.; Chen, W. A Mouse model of Acinetobacter baumannii-associated pneumonia using a clinically isolated hypervirulent strain. Antimicrob. Agents Chemother. 2013, 57, 3601–3613. [Google Scholar] [CrossRef] [PubMed]
- Subashchandrabose, S.; Smith, S.; DeOrnellas, V.; Crepin, S.; Kole, M.; Zahdeh, C.; Mobley, H.L.T. Acinetobacter baumannii Genes Required for Bacterial Survival during Bloodstream Infection. mSphere 2016, 1, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Geisinger, E.; Huo, W.; Hernandez-Bird, J.; Isberg, R.R. Acinetobacter baumannii: Envelope Determinants That Control Drug Resistance, Virulence, and Surface Variability. Annu. Rev. Microbiol. 2019, 73, 481–506. [Google Scholar] [CrossRef]
- Singh, J.K.; Adams, F.G.; Brown, M.H. Diversity and function of capsular polysaccharide in Acinetobacter baumannii. Front. Microbiol. 2019, 9, 3301. [Google Scholar] [CrossRef]
- Kenyon, J.J.; Hall, R.M. Variation in the Complex Carbohydrate Biosynthesis Loci of Acinetobacter baumannii Genomes. PLoS ONE 2013, 8, e62160. [Google Scholar] [CrossRef]
- Wyres, K.L.; Cahill, S.M.; Holt, K.E.; Hall, R.M.; Kenyon, J.J. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with kaptive. Microb. Genom. 2020, 6, e000339. [Google Scholar] [CrossRef]
- Wright, M.S.; Jacobs, M.R.; Bonomo, R.A.; Adams, M.D. Transcriptome remodeling of Acinetobacter baumannii during infection and treatment. mBio 2017, 8, e02193-16. [Google Scholar] [CrossRef]
- Shashkov, A.S.; Kenyon, J.J.; Arbatsky, N.P.; Shneider, M.M.; Popova, A.V.; Miroshnikov, K.A.; Volozhantsev, N.V.; Knirel, Y.A. Structures of three different neutral polysaccharides of Acinetobacter baumannii, NIPH190, NIPH201, and NIPH615, assigned to K30, K45, and K48 capsule types, respectively, based on capsule biosynthesis gene clusters. Carbohydr. Res. 2015, 417, 81–88. [Google Scholar] [CrossRef]
- Kenyon, J.J.; Senchenkova, S.N.; Shashkov, A.S.; Shneider, M.M.; Popova, A.V.; Knirel, Y.A.; Hall, R.M. K17 capsular polysaccharide produced by Acinetobacter baumannii isolate G7 contains an amide of 2-acetamido-2-deoxy-D-galacturonic acid with D-alanine. Int. J. Biol. Macromol. 2020, 144, 857–862. [Google Scholar] [CrossRef]
- Kenyon, J.J.; Marzaioli, A.M.; De Castro, C.; Hall, R.M. 5,7-di-N-acetyl-acinetaminic acid: A novel non-2-ulosonic acid found in the capsule of an Acinetobacter baumannii isolate. Glycobiology 2014, 25, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Shashkov, A.S.; Cahill, S.M.; Arbatsky, N.P.; Westacott, A.C.; Kasimova, A.A.; Shneider, M.M.; Popova, A.V.; Shagin, D.A.; Shelenkov, A.A.; Mikhailova, Y.V.; et al. Acinetobacter baumannii K116 capsular polysaccharide structure is a hybrid of the K14 and revised K37 structures. Carbohydr. Res. 2019, 484, 107774. [Google Scholar] [CrossRef] [PubMed]
- Giguère, D. Surface polysaccharides from Acinetobacter baumannii: Structures and syntheses. Carbohydr. Res. 2015, 418, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Niu, T.; Guo, L.; Luo, Q.; Zhou, K.; Yu, W.; Chen, Y.; Huang, C.; Xiao, Y. Wza gene knockout decreases Acinetobacter baumannii virulence and affects Wzy-dependent capsular polysaccharide synthesis. Virulence 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Geisinger, E.; Mortman, N.J.; Vargas-Cuebas, G.; Tai, A.K.; Isberg, R.R. A global regulatory system links virulence and antibiotic resistance to envelope homeostasis in Acinetobacter baumannii. PLoS Pathog. 2018, 14, e1007030. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, M.H.; Il Kim, S.; Son, J.H.; Kim, S.; Lee, Y.C.; Shin, M.; Oh, M.H.; Lee, J.C. The sensor kinase BfmS controls production of outer membrane vesicles in Acinetobacter baumannii. BMC Microbiol. 2019, 19, 301. [Google Scholar] [CrossRef]
- Krasauskas, R.; Skerniškytė, J.; Armalytė, J.; Sužiedėlienė, E. The role of Acinetobacter baumannii response regulator BfmR in pellicle formation and competitiveness via contact-dependent inhibition system. BMC Microbiol. 2019, 19, 241. [Google Scholar] [CrossRef]
- Farrow, J.M.; Wells, G.; Pesci, E.C. Desiccation tolerance in Acinetobacter baumannii is mediated by the two-component response regulator BfmR. PLoS ONE 2018, 13, e0205638. [Google Scholar] [CrossRef]
- Russo, T.A.; Luke, N.R.; Beanan, J.M.; Olson, R.; Sauberan, S.L.; MacDonald, U.; Schultz, L.W.; Umland, T.C.; Campagnari, A.A. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect. Immun. 2010, 78, 3993–4000. [Google Scholar] [CrossRef]
- Tipton, K.A.; Chin, C.; Farokhyfar, M.; Weiss, D.S.; Rather, P.N. Role of Capsule in Resistance to Disinfectants, Host Antimicrobials, and Desiccation in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2018, 62, 18. [Google Scholar] [CrossRef]
- Geisinger, E.; Isberg, R.R. Antibiotic Modulation of Capsular Exopolysaccharide and Virulence in Acinetobacter baumannii. PLoS Pathog. 2015, 11, e1004691. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.Y.; Tipton, K.A.; Farokhyfar, M.; Burd, E.M.; Weiss, D.S.; Rather, P.N. A high-frequency phenotypic switch links bacterial virulence and environmental survival in Acinetobacter baumannii. Nat. Microbiol. 2018, 3, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Larrayoz, A.F.; Elhosseiny, N.M.; Chevrette, M.G.; Fu, Y.; Giunta, P.; Spallanzani, R.G.; Ravi, K.; Pier, G.B.; Lory, S.; Maira-Litrán, T. Complexity of Complement Resistance Factors Expressed by Acinetobacter baumannii Needed for Survival in Human Serum. J. Immunol. 2017, 199, 2803–2814. [Google Scholar] [CrossRef] [PubMed]
- Cochet, F.; Peri, F. The role of carbohydrates in the lipopolysaccharide (LPS)/toll-like receptor 4 (TLR4) Signalling. Int. J. Mol. Sci. 2017, 18, 2318. [Google Scholar] [CrossRef] [PubMed]
- Erridge, C.; Moncayo-Nieto, O.L.; Morgan, R.; Young, M.; Poxton, I.R. Acinetobacter baumannii lipopolysaccharides are potent stimulators of human monocyte activation via Toll-like receptor 4 signalling. J. Med. Microbiol. 2007, 56, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and pathophysiological overview of Acinetobacter infections: A century of challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Tan, B.; Pantapalangkoor, P.; Ho, T.; Baquir, B.; Tomaras, A.; Montgomery, J.I.; Reilly, U.; Barbacci, E.G.; Hujer, K.; et al. Inhibition of LpxC Protects Mice from Resistant Acinetobacter baumannii by Modulating Inflammation and Enhancing Phagocytosis. mBio 2012, 3, 23–29. [Google Scholar] [CrossRef]
- Boll, J.M.; Tucker, A.T.; Klein, D.R.; Beltran, A.M.; Brodbelt, J.S.; Davies, B.W.; Trent, M.S. Reinforcing lipid a acylation on the cell surface of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. mBio 2015, 6, e00478-15. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Mansell, A.; Crane, B.; Fitzsimons, T.C.; Nation, R.L.; Li, J.; Adler, B.; Boyce, J.D. Lipopolysaccharide-deficient Acinetobacter baumannii shows altered signaling through host toll-like receptors and increased susceptibility to the host antimicrobial peptide LL-37. Infect. Immun. 2013, 81, 684–689. [Google Scholar] [CrossRef]
- Boll, J.M.; Crofts, A.A.; Peters, K.; Cattoir, V.; Vollmer, W.; Davies, B.W.; Trent, M.S. A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proc. Natl. Acad. Sci. USA 2016, 113, E6228–E6237. [Google Scholar] [CrossRef]
- Beceiro, A.; Moreno, A.; Fernández, N.; Vallejo, J.A.; Aranda, J.; Adler, B.; Harper, M.; Boyce, J.D.; Bou, G. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2014, 58, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.F.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St. Michael, F.; Cox, A.D.; et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef] [PubMed]
- García-Quintanilla, M.; Pulido, M.R.; Moreno-Martínez, P.; Martín-Peña, R.; López-Rojas, R.; Pachón, J.; McConnell, M.J. Activity of host antimicrobials against multidrug-resistant Acinetobacter baumannii acquiring colistin resistance through loss of lipopolysaccharide. Antimicrob. Agents Chemother. 2014, 58, 2972–2975. [Google Scholar] [CrossRef] [PubMed]
- Nie, D.; Hu, Y.; Chen, Z.; Li, M.; Hou, Z.; Luo, X.; Mao, X.; Xue, X. Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. J. Biomed. Sci. 2020, 27, 26. [Google Scholar] [CrossRef]
- Choi, C.H.; Hyun, S.H.; Lee, J.Y.; Lee, J.S.; Lee, Y.S.; Kim, S.A.; Chae, J.P.; Yoo, S.M.; Lee, J.C. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell. Microbiol. 2008, 10, 309–319. [Google Scholar] [CrossRef]
- Gaddy, J.A.; Tomaras, A.P.; Actis, L.A. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 2009, 77, 3150–3160. [Google Scholar] [CrossRef]
- Schweppe, D.K.; Harding, C.; Chavez, J.D.; Wu, X.; Ramage, E.; Singh, P.K.; Manoil, C.; Bruce, J.E. Host-Microbe Protein Interactions during Bacterial Infection. Chem. Biol. 2015, 22, 1521–1530. [Google Scholar] [CrossRef]
- Sánchez-Encinales, V.; Álvarez-Marín, R.; Pachón-Ibáñez, M.E.; Fernández-Cuenca, F.; Pascual, A.; Garnacho-Montero, J.; Martínez-Martínez, L.; Vila, J.; Tomás, M.M.; Cisneros, J.M.; et al. Overproduction of Outer membrane protein a by Acinetobacter baumannii as a risk factor for nosocomial pneumonia, bacteremia, and mortality rate increase. J. Infect. Dis. 2017, 215, 966–974. [Google Scholar]
- Weber, B.S.; Kinsella, R.L.; Harding, C.M.; Feldman, M.F. The Secrets of Acinetobacter Secretion. Trends Microbiol. 2017, 25, 532–545. [Google Scholar] [CrossRef]
- Jin, J.S.; Kwon, S.O.; Moon, D.C.; Gurung, M.; Lee, J.H.; Il Kim, S.; Lee, J.C. Acinetobacter baumannii secretes cytotoxic outer membrane protein a via outer membrane vesicles. PLoS ONE 2011, 6, e17027. [Google Scholar] [CrossRef]
- Kwon, S.O.; Gho, Y.S.; Lee, J.C.; Kim, S.I. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol. Lett. 2009, 297, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Choi, C.H.; Kim, J.W.; Lee, J.C. Acinetobacter baumannii outer membrane protein a induces dendritic cell death through mitochondrial targeting. J. Microbiol. 2010, 48, 387–392. [Google Scholar] [CrossRef] [PubMed]
- King, L.B.; Pangburn, M.K.; McDaniel, L.S. Serine protease PKF of Acinetobacter baumannii results in serum resistance and suppression of biofilm formation. J. Infect. Dis. 2013, 207, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- Waack, U.; Johnson, T.L.; Chedid, K.; Xi, C.; Simmons, L.A.; Mobley, H.L.T.; Sandkvist, M. Targeting the type II secretion system: Development, optimization, and validation of a high-throughput screen for the identification of small molecule inhibitors. Front. Cell. Infect. Microbiol. 2017, 7, 380. [Google Scholar] [CrossRef]
- Teufel, R.; Mascaraque, V.; Ismail, W.; Voss, M.; Perera, J.; Eisenreich, W.; Haehnel, W.; Fuchs, G. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. USA 2010, 107, 14390–14395. [Google Scholar] [CrossRef]
- Fernández, C.; Díaz, E.; García, J.L. Insights on the regulation of the phenylacetate degradation pathway from Escherichia coli. Environ. Microbiol. Rep. 2014, 6, 239–250. [Google Scholar] [CrossRef]
- Sato, Y.; Unno, Y.; Miyazaki, C.; Ubagai, T.; Ono, Y. Multidrug-resistant Acinetobacter baumannii resists reactive oxygen species and survives in macrophages. Sci. Rep. 2019, 9, 17462. [Google Scholar] [CrossRef]
- Lebre, P.H.; De Maayer, P.; Cowan, D.A. Xerotolerant bacteria: Surviving through a dry spell. Nat. Rev. Microbiol. 2017, 15, 285–296. [Google Scholar] [CrossRef]
- Houang, E.T.S.; Sormunen, R.T.; Lai, L.; Chan, C.Y.; Leong, A.S.Y. Effect of desiccation on the ultrastructural appearances of Acinetobacter baumannii and Acinetobacter lwoffii. J. Clin. Pathol. 1998, 51, 786–788. [Google Scholar] [CrossRef]
- Zeidler, S.; Müller, V. The role of compatible solutes in desiccation resistance of Acinetobacter baumannii. Microbiologyopen 2019, 8, e00740. [Google Scholar] [CrossRef]
- Laskowska, E.; Kuczyńska-Wiśnik, D. New insight into the mechanisms protecting bacteria during desiccation. Curr. Genet. 2020, 66, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Moruno Algara, M.; Kuczyńska-Wiśnik, D.; Dębski, J.; Stojowska-Swędrzyńska, K.; Sominka, H.; Bukrejewska, M.; Laskowska, E. Trehalose protects Escherichia coli against carbon stress manifested by protein acetylation and aggregation. Mol. Microbiol. 2019, 112, 866–880. [Google Scholar] [CrossRef] [PubMed]
- Zeidler, S.; Müller, V. Coping with low water activities and osmotic stress in Acinetobacter baumannii: Significance, current status and perspectives. Environ. Microbiol. 2019, 21, 2212–2230. [Google Scholar] [CrossRef] [PubMed]
- Potts, M. Desiccation tolerance of prokaryotes. Microbiol. Rev. 1994, 58, 755–805. [Google Scholar] [CrossRef]
- Wang, X.; Cole, C.G.; Dupai, C.D.; Davies, B.W. Protein aggregation is associated with Acinetobacter baumannii desiccation tolerance. Microorganisms 2020, 8, 23–29. [Google Scholar] [CrossRef]
- Gayoso, C.M.; Mateos, J.; Méndez, J.A.; Fernández-Puente, P.; Rumbo, C.; Tomás, M.; Martínez De Ilarduya, Ó.; Bou, G. Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J. Proteome Res. 2014, 13, 460–476. [Google Scholar] [CrossRef]
- Espinal, P.; Martí, S.; Vila, J. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J. Hosp. Infect. 2012, 80, 56–60. [Google Scholar] [CrossRef]
- Boteva, E.; Mironova, R. Maillard reaction and aging: Can bacteria shed light on the link? Biotechnol. Biotechnol. Equip. 2019, 33, 481–497. [Google Scholar] [CrossRef]
- Schramm, F.D.; Schroeder, K.; Jonas, K. Protein aggregation in bacteria. FEMS Microbiol. Rev. 2019, 44, 54–72. [Google Scholar] [CrossRef]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354-1. [Google Scholar] [CrossRef]
- Mogk, A.; Huber, D.; Bukau, B. Integrating protein homeostasis strategies in prokaryotes. Cold Spring Harb. Perspect. Biol. 2011, 3, a004366. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, K.; Gandra, R.F.; Wisniewski, E.S.; Osaku, C.A.; Kadowaki, M.K.; Felipach-Neto, V.; Haus, L.F.A.Á.; Simão, R.D.C.G. DnaK and GroEL are induced in response to antibiotic and heat shock in Acinetobacter baumannii. J. Med. Microbiol. 2010, 59, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Nwugo, C.C.; Gaddy, J.A.; Zimbler, D.L.; Actis, L.A. Deciphering the iron response in Acinetobacter baumannii: A proteomics approach. J. Proteom. 2011, 74, 44–58. [Google Scholar] [CrossRef] [PubMed]
- Rinas, U.; Garcia-Fruitós, E.; Corchero, J.L.; Vázquez, E.; Seras-Franzoso, J.; Villaverde, A. Bacterial Inclusion Bodies: Discovering Their Better Half. Trends Biochem. Sci. 2017, 42, 726–737. [Google Scholar] [CrossRef]
- Kuczyńska-Wiśnik, D.; Zurawa-Janicka, D.; Narkiewicz, J.; Kwiatkowska, J.; Lipińska, B.; Laskowska, E. Escherichia coli small heat shock proteins IbpA/B enhance activity of enzymes sequestered in inclusion bodies. Acta Biochim. Pol. 2004, 51, 925–931. [Google Scholar]
- Khodaparast, L.; Khodaparast, L.; Gallardo, R.; Louros, N.N.; Michiels, E.; Ramakrishnan, R.; Ramakers, M.; Claes, F.; Young, L.; Shahrooei, M.; et al. Aggregating sequences that occur in many proteins constitute weak spots of bacterial proteostasis. Nat. Commun. 2018, 9, 866. [Google Scholar] [CrossRef]
- Knauf, G.A.; Cunningham, A.L.; Kazi, M.I.; Riddington, I.M.; Crofts, A.A.; Cattoir, V.; Trent, M.S.; Davies, B.W. Exploring the antimicrobial action of quaternary amines against Acinetobacter baumannii. mBio 2018, 9, e02394-17. [Google Scholar] [CrossRef]
- Steenackers, H.P.; Parijs, I.; Foster, K.R.; Vanderleyden, J. Experimental evolution in biofilm populations. FEMS Microbiol. Rev. 2016, 40, 373–397. [Google Scholar] [CrossRef]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef]
- Lebeaux, D.; Ghigo, J.-M.; Beloin, C. Biofilm-Related Infections: Bridging the Gap between Clinical Management and Fundamental Aspects of Recalcitrance toward Antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef]
- Chen, H.; Cao, J.; Zhou, C.; Liu, H.; Zhang, X.; Zhou, T. Biofilm formation restrained by subinhibitory concentrations of tigecyclin in Acinetobacter baumannii is associated with downregulation of efflux pumps. Chemotherapy 2017, 62, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Eze, E.C.; Chenia, H.Y.; El Zowalaty, M.E. Acinetobacter baumannii biofilms: Effects of physicochemical factors, virulence, antibiotic resistance determinants, gene regulation, and future antimicrobial treatments. Infect. Drug Resist. 2018, 11, 2277–2299. [Google Scholar] [CrossRef] [PubMed]
- Giles, S.K.; Stroeher, U.H.; Eijkelkamp, B.A.; Brown, M.H. Identification of genes essential for pellicle formation in Acinetobacter baumannii ’Microbial biochemistry, physiology and metabolism. BMC Microbiol. 2015, 15, 116. [Google Scholar] [CrossRef] [PubMed]
- Marti, S.; Chabane, Y.N.; Alexandre, S.; Coquet, L.; Vila, J.; Jouenne, T.; Dé, E. Growth of Acinetobacter baumannii in pellicle enhanced the expression of potential virulence factors. PLoS ONE 2011, 6, e26030. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Lv, R.; Xiao, L.; Wang, M.; Du, Z.; Tan, Y.; Cui, Y.; Yan, Y.; Luo, Y.; Yang, R.; et al. A1S_2811, a CheA/Y-like hybrid two-component regulator from Acinetobacter baumannii ATCC17978, is involved in surface motility and biofilm formation in this bacterium. Microbiologyopen 2017, 6, e00510. [Google Scholar] [CrossRef] [PubMed]
- Verstraeten, N.; Braeken, K.; Debkumari, B.; Fauvart, M.; Fransaer, J.; Vermant, J.; Michiels, J. Living on a surface: Swarming and biofilm formation. Trends Microbiol. 2008, 16, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.K.; Iyer, P.S.; Barage, S.H.; Sonawane, K.D.; Chopade, B.A. Characterization of the algC gene expression pattern in the multidrug resistant Acinetobacter baumannii AIIMS 7 and correlation with biofilm development on abiotic surface. Sci. World J. 2014, 2014, 593546. [Google Scholar] [CrossRef]
- Pakharukova, N.; Tuittila, M.; Paavilainen, S.; Malmi, H.; Parilova, O.; Teneberg, S.; Knight, S.D.; Zavialov, A.V. Structural basis for Acinetobacter baumannii biofilm formation. Proc. Natl. Acad. Sci. USA 2018, 115, 5558–5563. [Google Scholar] [CrossRef]
- Ronish, L.A.; Lillehoj, E.; Fields, J.K.; Sundberg, E.J.; Piepenbrink, K.H. The structure of PilA from Acinetobacter baumannii AB5075 suggests a mechanism for functional specialization in Acinetobacter type IV pili. J. Biol. Chem. 2019, 294, 218–230. [Google Scholar] [CrossRef]
- Koiwai, K.; Hartmann, M.D.; Linke, D.; Lupas, A.N.; Hori, K. Structural basis for toughness and flexibility in the C-terminal passenger domain of an Acinetobacter trimeric autotransporter adhesin. J. Biol. Chem. 2016, 291, 3705–3724. [Google Scholar] [CrossRef]
- De Gregorio, E.; Del Franco, M.; Martinucci, M.; Roscetto, E.; Zarrilli, R.; Di Nocera, P.P. Biofilm-associated proteins: News from Acinetobacter. BMC Genom. 2015, 16, 933. [Google Scholar] [CrossRef] [PubMed]
- Loehfelm, T.W.; Luke, N.R.; Campagnari, A.A. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J. Bacteriol. 2008, 190, 1036–1044. [Google Scholar] [CrossRef] [PubMed]
- Richmond, G.E.; Evans, L.P.; Anderson, M.J.; Wand, M.E.; Bonney, L.C.; Ivens, A.; Chua, L.; Webber, M.A.; Sutton, J.M.; Peterson, M.L.; et al. Regulates Genes Required for Multidrug Efflux, Biofilm Formation, and Virulence in a Strain-Specific Manner. Am. Soc. Microbiol. 2016, 7, e00430-16. [Google Scholar]
- Yoon, E.J.; Chabane, Y.N.; Goussard, S.; Snesrud, E.; Courvalin, P.; Dé, E.; Grillot-Courvalin, C. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio 2015, 6, e00309–e00315. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Lu, F.; Yuan, F.; Jiang, D.; Zhao, P.; Zhu, J.; Cheng, H.; Cao, J.; Lu, G. Biofilm formation caused by clinical Acinetobacter baumannii isolates is associated with overexpression of the AdeFGH efflux pump. Antimicrob. Agents Chemother. 2015, 59, 4817–4825. [Google Scholar] [CrossRef] [PubMed]
- Kentache, T.; Abdelkrim, A.B.; Jouenne, T.; Dé, E.; Hardouin, J. Global dynamic proteome study of a pellicle-forming Acinetobacter baumannii strain. Mol. Cell. Proteom. 2017, 16, 100–112. [Google Scholar] [CrossRef]
- Ahmad, I.; Nygren, E.; Khalid, F.; Myint, S.L.; Uhlin, B.E. A Cyclic-di-GMP signalling network regulates biofilm formation and surface associated motility of Acinetobacter baumannii 17978. Sci. Rep. 2020, 10, 1991. [Google Scholar] [CrossRef]
- Tomaras, A.P.; Flagler, M.J.; Dorsey, C.W.; Gaddy, J.A.; Actis, L.A. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 2008, 154, 3398–3409. [Google Scholar] [CrossRef]
- Cerqueira, G.M.; Kostoulias, X.; Khoo, C.; Aibinu, I.; Qu, Y.; Traven, A.; Peleg, A.Y. A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis. 2014, 210, 46–55. [Google Scholar] [CrossRef]
- Rumbo-Feal, S.; Pérez, A.; Ramelot, T.A.; Álvarez-Fraga, L.; Vallejo, J.A.; Beceiro, A.; Ohneck, E.J.; Arivett, B.A.; Merino, M.; Fiester, S.E.; et al. Contribution of the A. baumannii A1S_0114 Gene to the Interaction with Eukaryotic Cells and Virulence. Front. Cell. Infect. Microbiol. 2017, 7, 108. [Google Scholar] [CrossRef]
- Luo, L.M.; Wu, L.J.; Xiao, Y.L.; Zhao, D.; Chen, Z.X.; Kang, M.; Zhang, Q.; Xie, Y. Enhancing pili assembly and biofilm formation in Acinetobacter baumannii ATCC19606 using non-native acyl-homoserine lactones. BMC Microbiol. 2015, 15, 62. [Google Scholar] [CrossRef] [PubMed]
- López, M.; Mayer, C.; Fernández-García, L.; Blasco, L.; Muras, A.; Ruiz, F.M.; Bou, G.; Otero, A.; Tomás, M.; Rodríguez-Baño, J.; et al. Quorum sensing network in clinical strains of A. baumannii: AidA is a new quorum quenching enzyme. PLoS ONE 2017, 12, e0174454. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Muras, A.; Romero, M.; López, M.; Tomás, M.; Otero, A. Multiple quorum quenching enzymes are active in the nosocomial pathogen Acinetobacter baumannii ATCC17978. Front. Cell. Infect. Microbiol. 2018, 8, 310. [Google Scholar] [CrossRef] [PubMed]
- Modarresi, F.; Azizi, O.; Shakibaie, M.R.; Motamedifar, M.; Mosadegh, E.; Mansouri, S. Iron limitation enhances acyl homoserine lactone (AHL) production and biofilm formation in clinical isolates of Acinetobacter baumannii. Virulence 2015, 6, 152–161. [Google Scholar] [CrossRef]
- Jung, H.W.; Kim, K.; Islam, M.M.; Lee, J.C.; Shin, M. Role of ppGpp-regulated efflux genes in Acinetobacter baumannii. J. Antimicrob. Chemother. 2020, 75, 1130–1134. [Google Scholar] [CrossRef]
- Pérez-Varela, M.; Tierney, A.R.P.; Kim, J.-S.; Vazquez-Torres, A.; Rather, P. Characterization of RelA in Acinetobacter baumannii. J. Bacteriol. 2020, 202. [Google Scholar] [CrossRef]
- Blaschke, U.; Skiebe, E.; Wilharm, G. Novel genes required for surface-associated motility in Acinetobacter baumannii. bioRxiv 2020, 2251. [Google Scholar] [CrossRef]
- Lees-Miller, R.G.; Iwashkiw, J.A.; Scott, N.E.; Seper, A.; Vinogradov, E.; Schild, S.; Feldman, M.F. A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol. Microbiol. 2013, 89, 816–830. [Google Scholar] [CrossRef]
- Ching, C.; Yang, B.; Onwubueke, C.; Lazinski, D.; Camilli, A.; Godoy, V.G. Lon protease has multifaceted biological functions in Acinetobacter baumannii. J. Bacteriol. 2019, 201, e00536-1. [Google Scholar] [CrossRef]
- Choi, A.H.K.; Slamti, L.; Avci, F.Y.; Pier, G.B.; Maira-Litrán, T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 2009, 191, 5953–5963. [Google Scholar] [CrossRef]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, M.; Nakatani, H.; Hori, K. AtaA, a New Member of the Trimeric Autotransporter Adhesins from Acinetobacter sp. Tol 5 Mediating High Adhesiveness to Various Abiotic Surfaces. PLoS ONE 2012, 7, e48830. [Google Scholar] [CrossRef] [PubMed]
- Bentancor, L.V.; Camacho-Peiro, A.; Bozkurt-Guzel, C.; Pier, G.B.; Maira-Litrán, T. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J. Bacteriol. 2012, 194, 3950–3960. [Google Scholar] [CrossRef] [PubMed]
- Cabral, M.P.; Soares, N.C.; Aranda, J.; Parreira, J.R.; Rumbo, C.; Poza, M.; Valle, J.; Calamia, V.; Lasa, Í.; Bou, G. Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J. Proteome Res. 2011, 10, 3399–3417. [Google Scholar] [CrossRef] [PubMed]
- Wood, C.R.; Ohneck, E.J.; Edelmann, R.E.; Actis, L.A. A light-regulated type I pilus contributes to Acinetobacter baumannii biofilm, motility, and virulence functions. Infect. Immun. 2018, 86, e00442-18. [Google Scholar] [CrossRef] [PubMed]
- Chabane, Y.N.; Marti, S.; Rihouey, C.; Alexandre, S.; Hardouin, J.; Lesouhaitier, O.; Vila, J.; Kaplan, J.B.; Jouenne, T.; Dé, E. Characterisation of pellicles formed by Acinetobacter baumannii at the air-liquid interface. PLoS ONE 2014, 9, e111660. [Google Scholar] [CrossRef] [PubMed]
- Saipriya, K.; Swathi, C.H.; Ratnakar, K.S.; Sritharan, V. Quorum-sensing system in Acinetobacter baumannii: A potential target for new drug development. J. Appl. Microbiol. 2020, 128, 15–27. [Google Scholar] [CrossRef]
- Anbazhagan, D.; Mansor, M.; Yan, G.O.S.; Yusof, M.Y.M.; Hassan, H.; Sekaran, S.D. Detection of quorum sensing signal molecules and identification of an autoinducer synthase gene among biofilm forming clinical isolates of Acinetobacter spp. PLoS ONE 2012, 7, e36696. [Google Scholar] [CrossRef]
- Erdönmez, D.; Rad, A.Y.; Aksöz, N. Quorum sensing molecules production by nosocomial and soil isolates Acinetobacter baumannii. Arch. Microbiol. 2017, 199, 1325–1334. [Google Scholar] [CrossRef]
- Niu, C.; Clemmer, K.M.; Bonomo, R.A.; Rather, P.N. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 2008, 190, 3386–3392. [Google Scholar] [CrossRef]
- Hengge, R. Linking bacterial growth, survival, and multicellularity—Small signaling molecules as triggers and drivers. Curr. Opin. Microbiol. 2020, 55, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Jenal, U.; Reinders, A.; Lori, C. Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 2017, 15, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, T. C-di-GMP Synthesis: Structural Aspects of Evolution, Catalysis and Regulation. J. Mol. Biol. 2016, 428, 3683–3701. [Google Scholar] [CrossRef] [PubMed]
- Romling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The First 25 Years of a Universal Bacterial Second Messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef]
- Potrykus, K.; Cashel, M. (p)ppGpp: Still Magical? Annu. Rev. Microbiol. 2008, 62, 35–51. [Google Scholar] [CrossRef]
- Hobbs, J.K.; Boraston, A.B. (p)ppGpp and the Stringent Response: An Emerging Threat to Antibiotic Therapy. ACS Infect. Dis. 2019, 5, 1505–1517. [Google Scholar] [CrossRef]
- Hauryliuk, V.; Atkinson, G.C.; Murakami, K.S.; Tenson, T.; Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 2015, 13, 298–309. [Google Scholar] [CrossRef]
- De la Fuente-Núñez, C.; Reffuveille, F.; Haney, E.F.; Straus, S.K.; Hancock, R.E.W. Broad-Spectrum Anti-biofilm Peptide That Targets a Cellular Stress Response. PLoS Pathog. 2014, 10, e1004152. [Google Scholar] [CrossRef]
- Shin, B.; Park, C.; Park, W. Stress responses linked to antimicrobial resistance in Acinetobacter species. Appl. Microbiol. Biotechnol. 2020, 104, 1423–1435. [Google Scholar] [CrossRef]
- Weiss, L.A.; Stallings, C.L. Essential roles for Mycobacterium tuberculosis rel beyond the production of (p)ppGpp. J. Bacteriol. 2013, 195, 5629–5638. [Google Scholar] [CrossRef]
- Liu, H.; Xiao, Y.; Nie, H.; Huang, Q.; Chen, W. Influence of (p)ppGpp on biofilm regulation in Pseudomonas putida KT2440. Microbiol. Res. 2017, 204, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.M.; Davey, M.E. Synthesis of ppGpp impacts type IX secretion and biofilm matrix formation in Porphyromonas gingivalis. NPJ Biofilms Microb. 2020, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Andresen, L.; Tenson, T.; Hauryliuk, V. Cationic bactericidal peptide 1018 does not specifically target the stringent response alarmone (p)ppGpp. Sci. Rep. 2016, 6, 36549. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Gollan, B.; Grabe, G.; Michaux, C.; Helaine, S. Bacterial Persisters and Infection: Past, Present, and Progressing. Annu. Rev. Microbiol. 2019, 73, 359–385. [Google Scholar] [CrossRef]
- Wilmaerts, D.; Windels, E.M.; Verstraeten, N.; Michiels, J. General Mechanisms Leading to Persister Formation and Awakening. Trends Genet. 2019, 35, 401–411. [Google Scholar] [CrossRef]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Kim, J.S.; Wood, T.K. Persistent persister misperceptions. Front. Microbiol. 2016, 7, 2134. [Google Scholar] [CrossRef]
- Lewis, K.; Manuse, S. Persister Formation and Antibiotic Tolerance of Chronic Infections. In Persister Cells and Infectious Disease; Lewis, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 59–76. [Google Scholar]
- Kuczyńska-Wiśnik, D.; Stojowska, K.; Matuszewska, E.; Leszczyńska, D.; Algara, M.M.; Augustynowicz, M.; Laskowska, E. Lack of intracellular trehalose affects formation of Escherichia coli persister cells. Microbiology 2015, 161, 786–796. [Google Scholar] [CrossRef]
- Leszczynska, D.; Matuszewska, E.; Kuczynska-Wisnik, D.; Furmanek-Blaszk, B.; Laskowska, E. The Formation of Persister Cells in Stationary-Phase Cultures of Escherichia Coli Is Associated with the Aggregation of Endogenous Proteins. PLoS ONE 2013, 8, e54737. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Wood, T.K. ppGpp ribosome dimerization model for bacterial persister formation and resuscitation. Biochem. Biophys. Res. Commun. 2020, 523, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Barth, V.C.; Rodrigues, B.A.; Bonatto, G.D.; Gallo, S.W.; Pagnussatti, V.E.; Ferreira, C.A.S.; De Oliveira, S.D. Heterogeneous persister cells formation in Acinetobacter baumannii. PLoS ONE 2013, 8, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.H.S.; Cai, Y.; Leck, H.; Lim, T.P.; Teo, J.Q.M.; Lee, W.; Koh, T.H.; Tan, T.T.; Tan, K.W.; Kwa, A.L.H. Determining the development of persisters in extensively drug-resistant Acinetobacter baumannii upon exposure to polymyxin B-based antibiotic combinations using flow cytometry. Antimicrob. Agents Chemother. 2020, 64, e01712–e01719. [Google Scholar] [CrossRef] [PubMed]
- Neou, E.; Michail, G.; Tsakris, A.; Pournaras, S. Virulence of Acinetobacter baumannii exhibiting phenotypic heterogeneous growth against meropenem in a murine thigh infection model. Antibiotics 2013, 2, 73–82. [Google Scholar] [CrossRef]
- Alkasir, R.; Ma, Y.; Liu, F.; Li, J.; Lv, N.; Xue, Y.; Hu, Y.; Zhu, B. Characterization and transcriptome analysis of Acinetobacter baumannii persister cells. Microb. Drug Resist. 2018, 24, 1466–1474. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Garcia, L. Relationship between tolerance and persistence mechanisms in Acinetobacter baumannii strains with AbkAB Toxin-Antitoxin Systen. Antimicrob. Agents Chemother. 2018, 62, e00250-18. [Google Scholar] [CrossRef]
- Zou, J.; Kou, S.; Xie, R.; Vannieuwenhze, M.S.; Qu, J. Non-walled spherical Acinetobacter baumannii is an important type of persisters upon β -lactam antibiotics treatment. Emerg. Microbes Infect. 2020, 9, 1149–1159. [Google Scholar] [CrossRef]
- Santos-Lopez, A.; Marshall, C.W.; Scribner, M.R.; Snyder, D.J.; Cooper, V.S. Evolutionary pathways to antibiotic resistance are dependent upon environmental structure and bacterial lifestyle. eLife 2019, 8, e47612. [Google Scholar] [CrossRef]
- Penesyan, A.; Nagy, S.S.; Kjelleberg, S.; Gillings, M.R.; Paulsen, I.T. Rapid microevolution of biofilm cells in response to antibiotics. NPJ Biofilms Microb. 2019, 5, 34. [Google Scholar] [CrossRef]
- Tipton, K.A.; Dimitrova, D.; Rather, P.N. Phase-variable control of multiple phenotypes in Acinetobacter baumannii strain AB5075. J. Bacteriol. 2015, 197, 2593–2599. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Karah, N.; Nadeem, A.; Wai, S.N.; Uhlin, B.E. Analysis of colony phase variation switch in Acinetobacter baumannii clinical isolates. PLoS ONE 2019, 14, e0210082. [Google Scholar] [CrossRef] [PubMed]
- Clark, N.M.; Zhanel, G.G.; Lynch, J.P. Emergence of antimicrobial resistance among Acinetobacter species: A global threat. Curr. Opin. Crit. Care 2016, 22, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Abdi, S.N.; Ghotaslou, R.; Ganbarov, K.; Mobed, A.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Kafil, H.S. Acinetobacter baumannii efflux pumps and antibiotic resistance. Infect. Drug Resist. 2020, 13, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, present, and future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef]
- Bonnin, R.A.; Poirel, L.; Naas, T.; Pirs, M.; Seme, K.; Schrenzel, J.; Nordmann, P. Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 2012, 18, E362–E365. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Bonomo, R.A.; Tolmasky, M.E. Carbapenemases: Transforming Acinetobacter baumannii into a yet more dangerous menace. Biomolecules 2020, 10, 720. [Google Scholar] [CrossRef]
- Moulana, Z.; Babazadeh, A.; Eslamdost, Z.; Shokri, M.; Ebrahimpour, S. Phenotypic and genotypic detection of metallo-beta-lactamases in carbapenem resistant Acinetobacter baumannii. Casp. J. Intern. Med. 2020, 11, 171–176. [Google Scholar]
- Sun, X.; Liu, B.; Chen, Y.; Huang, H.; Wang, G.; Li, F.; Ni, Z. Molecular characterization of Ambler class A to D β-lactamases, ISAba1, and integrons reveals multidrug-resistant Acinetobacter spp. isolates in northeastern China. J. Chemother. 2016, 28, 469–475. [Google Scholar] [CrossRef]
- Périchon, B.; Goussard, S.; Walewski, V.; Krizova, L.; Cerqueira, G.; Murphy, C.; Feldgarden, M.; Wortman, J.; Clermont, D.; Nemec, A.; et al. Identification of 50 class D β-lactamases and 65 Acinetobacter-derived cephalosporinases in Acinetobacter spp. Antimicrob. Agents Chemother. 2014, 58, 936–949. [Google Scholar] [CrossRef] [PubMed]
- Bansal, G.; Allen-McFarlane, R.; Eribo, B. Antibiotic Susceptibility, Clonality, and Molecular Characterization of Carbapenem-Resistant Clinical Isolates of Acinetobacter baumannii from Washington DC. Int. J. Microbiol. 2020, 2020, 2120159. [Google Scholar] [CrossRef] [PubMed]
- Yin Wong, M.H.; Wai Chan, B.K.; Chi Chan, E.W.; Chen, S. Over-Expression of ISAba1-Linked Intrinsic and Exogenously Acquired OXA Type Carbapenem-Hydrolyzing-Class D-ß-Lactamase-Encoding Genes Is Key Mechanism Underlying Carbapenem Resistance in Acinetobacter baumannii. Front. Microbiol. 2019, 10, 2809. [Google Scholar] [CrossRef] [PubMed]
- Hamidian, M.; Holt, K.E.; Pickard, D.; Dougan, G.; Hall, R.M. A GC1 Acinetobacter baumannii isolate carrying AbaR3 and the aminoglycoside resistance transposon TnaphA6 in a conjugative plasmid. J. Antimicrob. Chemother. 2014, 69, 955–958. [Google Scholar] [CrossRef]
- Graña-Miraglia, L.; Evans, B.A.; López-Jácome, L.E.; Hernández-Durán, M.; Colín-Castro, C.A.; Volkow-Fernández, P.; Cevallos, M.A.; Franco-Cendejas, R.; Castillo-Ramírez, S. Origin of OXA-23 Variant OXA-239 from a Recently Emerged Lineage of Acinetobacter baumannii International Clone V. mSphere 2020, 5, 1–9. [Google Scholar] [CrossRef]
- Karah, N.; Jolley, K.A.; Hall, R.M.; Uhlin, B.E. Database for the ampC alleles in Acinetobacter baumannii. PLoS ONE 2017, 12, e0176695. [Google Scholar] [CrossRef]
- Fernández, L.; Hancock, R.E.W. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef]
- Singh, H.; Thangaraj, P.; Chakrabarti, A. Acinetobacter baumannii: A brief account of mechanisms of multidrug resistance and current and future therapeutic management. J. Clin. Diagn. Res. 2013, 7, 2602–2605. [Google Scholar] [CrossRef]
- Beabout, K.; Hammerstrom, T.G.; Perez, A.M.; Magalhães, B.F.; Prater, A.G.; Clements, T.P.; Arias, C.A.; Saxer, G.; Shamoo, Y. The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob. Agents Chemother. 2015, 59, 5561–5566. [Google Scholar] [CrossRef]
- De Silva, P.M.; Kumar, A. Signal transduction proteins in Acinetobacter baumannii: Role in antibiotic resistance, virulence, and potential as drug targets. Front. Microbiol. 2019, 10, 49. [Google Scholar] [CrossRef]
- De Silva, P.M.; Kumar, A. Effect of Sodium Chloride on Surface-Associated Motility of Acinetobacter baumannii and the Role of AdeRS Two-Component System. J. Membr. Biol. 2018, 251, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.R.; Perng, C.L.; Chan, M.C.; Morita, Y.; Lin, J.C.; Su, C.M.; Wang, W.Y.; Chang, T.Y.; Chiueh, T.S. A Truncated AdeS Kinase Protein Generated by ISAba1 Insertion Correlates with Tigecycline Resistance in Acinetobacter baumannii. PLoS ONE 2012, 7, e49534. [Google Scholar] [CrossRef] [PubMed]
- Coyne, S.; Courvalin, P.; Périchon, B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 2011, 55, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Fournier, P.E.; Richet, H.; Weinstein, R.A. The Epidemiology and Control of Acinetobacter baumannii in Health Care Facilities. Clin. Infect. Dis. 2006, 42, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, G.; Domingues, S. Insights on the Horizontal Gene Transfer of Carbapenemase Determinants in the Opportunistic Pathogen Acinetobacter baumannii. Microorganisms 2016, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Roca, I.; Espinal, P.; Vila-Fanés, X.; Vila, J. The Acinetobacter baumannii oxymoron: Commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 2012, 3, 148. [Google Scholar] [CrossRef]
- Salgado-Camargo, A.D.; Castro-Jaimes, S.; Gutierrez-Rios, R.-M.; Lozano, L.F.; Altamirano-Pacheco, L.; Silva-Sanchez, J.; Pérez-Oseguera, Á.; Volkow, P.; Castillo-Ramírez, S.; Cevallos, M.A. Structure and Evolution of Acinetobacter baumannii Plasmids. Front. Microbiol. 2020, 11, 1283. [Google Scholar] [CrossRef]
- Bertini, A.; Poirel, L.; Mugnier, P.D.; Villa, L.; Nordmann, P.; Carattoli, A. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 4168–4177. [Google Scholar] [CrossRef]
- Lean, S.S.; Yeo, C.C. Small, enigmatic plasmids of the nosocomial pathogen, Acinetobacter baumannii: Good, bad, who knows? Front. Microbiol. 2017, 8, 1547. [Google Scholar] [CrossRef]
- Cameranesi, M.M.; Morán-Barrio, J.; Limansky, A.S.; Repizo, G.D.; Viale, A.M. Site-specific recombination at XerC/D sites mediates the formation and resolution of plasmid co-integrates carrying a blaOXA-58- and TnaphA6-resistance module in Acinetobacter baumannii. Front. Microbiol. 2018, 9, 66. [Google Scholar] [CrossRef]
- Towner, K.J.; Evans, B.; Villa, L.; Levi, K.; Hamouda, A.; Amyes, S.G.B.; Carattoli, A. Distribution of intrinsic plasmid replicase genes and their association with carbapenem-hydrolyzing class D β-lactamase genes in European clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 2154–2159. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Turton, J.F.; Ward, M.E.; Woodford, N.; Kaufmann, M.E.; Pike, R.; Livermore, D.M.; Pitt, T.L. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 2006, 258, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Nigro, S.J.; Hall, R.M. Structure and Context of acinetobacter transposons carrying the oxa23 carbapenemase gene. J. Antimicrob. Chemother. 2016, 71, 1135–1147. [Google Scholar] [CrossRef]
- Mcgann, P.; Courvalin, P.; Snesrud, E.; Clifford, R.J.; Yoon, E.; Onmus-leone, F.; Ong, A.C. Amplification of Aminoglycoside Resistance Gene aphA1 in Acinetobacter baumannii Results in Tobramycin Therapy Failure. mBio 2014, 5, 1–8. [Google Scholar] [CrossRef]
- Liu, L.L.; Ji, S.J.; Ruan, Z.; Fu, Y.; Fu, Y.Q.; Wang, Y.F.; Yu, Y.S. Dissemination of blaOXA-23 in Acinetobacter spp. in China: Main roles of conjugative plasmid pAZJ221 and transposon Tn2009. Antimicrob. Agents Chemother. 2015, 59, 1998–2005. [Google Scholar] [CrossRef]
- Cerezales, M.; Xanthopoulou, K.; Wille, J.; Krut, O.; Seifert, H.; Gallego, L.; Higgins, P.G. Mobile Genetic Elements Harboring Antibiotic Resistance Determinants in Acinetobacter baumannii Isolates From Bolivia. Front. Microbiol. 2020, 11, 919. [Google Scholar] [CrossRef]
- Simo Tchuinte, P.L.; Rabenandrasana, M.A.N.; Kowalewicz, C.; Andrianoelina, V.H.; Rakotondrasoa, A.; Andrianirina, Z.Z.; Enouf, V.; Ratsima, E.H.; Randrianirina, F.; Collard, J.M. Phenotypic and molecular characterisations of carbapenem-resistant Acinetobacter baumannii strains isolated in Madagascar. Antimicrob. Resist. Infect. Control 2019, 8, 31. [Google Scholar] [CrossRef]
- Hamidian, M.; Hall, R.M. The AbaR antibiotic resistance islands found in Acinetobacter baumannii global clone 1—Structure, origin and evolution. Drug Resist. Update 2018, 41, 26–39. [Google Scholar] [CrossRef]
- Krizova, L.; Dijkshoorn, L.; Nemec, A. Diversity and evolution of AbaR genomic resistance islands in Acinetobacter baumannii strains of European clone I. Antimicrob. Agents Chemother. 2011, 55, 3201–3206. [Google Scholar] [CrossRef]
- Ploy, M.C.; Gassama, A.; Chainier, D.; Denis, F. Integrons: An antibiotic resistance gene capture system. Immuno-Anal. Biol. Spec. 2005, 20, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Long, Q.; Qian, K.; Fu, T.; Zhang, Z.; Liao, P.; Xie, J. Resistance and integron characterization of Acinetobacter baumannii in a teaching hospital in Chongqing, China. New Microbes New Infect. 2015, 8, 103–108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pagano, M.; Martins, A.F.; Barth, A.L. Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz. J. Microbiol. 2016, 47, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Zarrilli, R.; Pournaras, S.; Giannouli, M.; Tsakris, A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 2013, 41, 11–19. [Google Scholar] [CrossRef]
- Fleitas, O.; Franco, O.L. Induced bacterial cross-resistance toward host antimicrobial peptides: A worrying phenomenon. Front. Microbiol. 2016, 7, 381. [Google Scholar] [CrossRef]
- Bazzi, W.; Abou Fayad, A.G.; Nasser, A.; Haraoui, L.P.; Dewachi, O.; Abou-Sitta, G.; Nguyen, V.K.; Abara, A.; Karah, N.; Landecker, H.; et al. Heavy Metal Toxicity in Armed Conflicts Potentiates AMR in A. baumannii by Selecting for Antibiotic and Heavy Metal Co-resistance Mechanisms. Front. Microbiol. 2020, 11, 68. [Google Scholar] [CrossRef]
- Hood, M.I.; Jacobs, A.C.; Sayood, K.; Dunman, P.M.; Skaar, E.P. Acinetobacter baumannii increases tolerance to antibiotics in response to monovalent cations. Antimicrob. Agents Chemother. 2010, 54, 1029–1041. [Google Scholar] [CrossRef]
- Norton, M.D.; Spilkia, A.J.; Godoy, V.G. Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii. J. Bacteriol. 2013, 195, 1335–1345. [Google Scholar] [CrossRef]
- Aranda, J.; Bardina, C.; Beceiro, A.; Rumbo, S.; Cabral, M.P.; Barbé, J.; Bou, G. Acinetobacter baumannii reca protein in repair of DNA damage, antimicrobial resistance, general stress response, and virulence. J. Bacteriol. 2011, 193, 3740–3747. [Google Scholar] [CrossRef]
- Poole, K. Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria. Trends Microbiol. 2012, 20, 227–234. [Google Scholar] [CrossRef]
- Levin-Reisman, I.; Ronin, I.; Gefen, O.; Braniss, I.; Shoresh, N.; Balaban, N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science 2017, 355, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Van Den Bergh, B.; Michiels, J.E.; Wenseleers, T.; Windels, E.M.; Vanden Boer, P.; Kestemont, D.; De Meester, L.; Verstrepen, K.J.; Verstraeten, N.; Fauvart, M.; et al. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat. Microbiol. 2016, 1, 16020. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N.R.; Lobritz, M.A.; Collins, J.J. Microbial persistence and the road to drug resistance. Cell Host Microbe 2013, 13, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Gefen, O.; Ronin, I.; Bar-Meir, M.; Balaban, N.Q. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 2020, 367, 200–204. [Google Scholar] [PubMed]
- Windels, E.M.; Michiels, J.E.; Fauvart, M.; Wenseleers, T.; Van den Bergh, B.; Michiels, J. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 2019, 13, 1239–1251. [Google Scholar] [CrossRef]
- El-Halfawy, O.M.; Valvano, M.A. Antimicrobial heteroresistance: An emerging field in need of clarity. Clin. Microbiol. Rev. 2015, 28, 191–207. [Google Scholar] [CrossRef]
- Andersson, D.I.; Nicoloff, H.; Hjort, K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat. Rev. Microbiol. 2019, 17, 479–496. [Google Scholar] [CrossRef]
- Li, J.; Rayner, C.R.; Nation, R.L.; Owen, R.J.; Spelman, D.; Tan, K.E.; Liolios, L. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2006, 50, 2946–2950. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Adler, B.; Nation, R.L.; Li, J.; Boyce, J.D. Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2011, 55, 3022–3024. [Google Scholar] [CrossRef]
- Pournaras, S.; Ikonomidis, A.; Markogiannakis, A.; Maniatis, A.N.; Tsakris, A. Heteroresistance to carbapenems in Acinetobacter baumannii. J. Antimicrob. Chemother. 2005, 55, 1055–1056. [Google Scholar] [CrossRef]
- Anderson, S.E. Aminoglycoside Heteroresistance in Acinetobacter baumannii AB5075. mSphere 2018, 3, e00271-18. [Google Scholar] [CrossRef] [PubMed]
- Nicoloff, H.; Hjort, K.; Levin, B.R.; Andersson, D.I. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat. Microbiol. 2019, 4, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Rittershaus, E.S.C.; Baek, S.H.; Sassetti, C.M. The normalcy of dormancy: Common themes in microbial quiescence. Cell Host Microbe 2013, 13, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Sambanthamoorthy, K.; Luo, C.; Pattabiraman, N.; Feng, X.; Koestler, B.; Waters, C.M.; Palys, T.J. Identification of small molecules inhibiting diguanylate cyclases to control bacterial biofilm development. Biofouling 2014, 30, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Nait Chabane, Y.; Mlouka, M.B.; Alexandre, S.; Nicol, M.; Marti, S.; Pestel-Caron, M.; Vila, J.; Jouenne, T.; Dé, E. Virstatin inhibits biofilm formation and motility of Acinetobacter baumannii. BMC Microbiol. 2014, 14, 62. [Google Scholar] [CrossRef] [PubMed]
- Stacy, D.M.; Welsh, M.A.; Rather, P.N.; Blackwell, H.E. Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native N -acyl homoserine lactones. ACS Chem. Biol. 2012, 7, 1719–1728. [Google Scholar] [CrossRef]
- Chow, J.Y.; Yang, Y.; Tay, S.B.; Chua, K.L.; Yew, W.S. Disruption of biofilm formation by the human pathogen Acinetobacter baumannii using engineered quorum-quenching lactonases. Antimicrob. Agents Chemother. 2014, 58, 1802–1805. [Google Scholar] [CrossRef]
- Nicol, M.; Alexandre, S.; Luizet, J.B.; Skogman, M.; Jouenne, T.; Salcedo, S.P.; Dé, E. Unsaturated fatty acids affect quorum sensing communication system and inhibit motility and biofilm formation of Acinetobacter baumannii. Int. J. Mol. Sci. 2018, 19, 214. [Google Scholar] [CrossRef]
- Gallo, S.W.; Ferreira, C.A.S.; de Oliveira, S.D. Combination of polymyxin B and meropenem eradicates persister cells from Acinetobacter baumannii strains in exponential growth. J. Med. Microbiol. 2017, 66, 1257–1260. [Google Scholar] [CrossRef]
- De Breij, A.; Riool, M.; Cordfunke, R.A.; Malanovic, N.; De Boer, L.; Koning, R.I.; Ravensbergen, E.; Franken, M.; Van Der Heijde, T.; Boekema, B.K.; et al. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 2018, 10, eaan4044. [Google Scholar] [CrossRef]
- Moon, K.H.; Weber, B.S.; Feldman, F. Subinhibitory Concentrations of Trimethoprim and Sulfamethoxazole Acinetobacter baumannii through Inhibition of Csu Pilus Expression. Antimicrob. Agents Chemother. 2017, 61, e00778-17. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, J.; Yin, Y.; Wang, G.; Yang, M.; Li, Y.; Zhang, Z.; Lai, R. The antimicrobial peptide ZY4 combats multidrugresistant Pseudomonas aeruginosa and Acinetobacter baumannii infection. Proc. Natl. Acad. Sci. USA 2019, 116, 26516–26522. [Google Scholar] [CrossRef] [PubMed]
- Nicol, M.; Mlouka, M.A.B.; Berthe, T.; Di Martino, P.; Jouenne, T.; Brunel, J.M.; Dé, E. Anti-persister activity of squalamine against Acinetobacter baumannii. Int. J. Antimicrob. Agents 2019, 53, 337–342. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).