Uveal Melanoma Cells Elicit Retinal Pericyte Phenotypical and Biochemical Changes in an in Vitro Model of Coculture
Abstract
1. Introduction
1.1. Pericytes in the Tumor Microenvironment
1.2. Pericyte Markers in Physiological and Pathological Conditions
1.3. PDGF-BB/PDGFRβ Signaling in Pericyte-CAF Transition
1.4. Involvement of Pericytes in Uveal Melanoma Progression
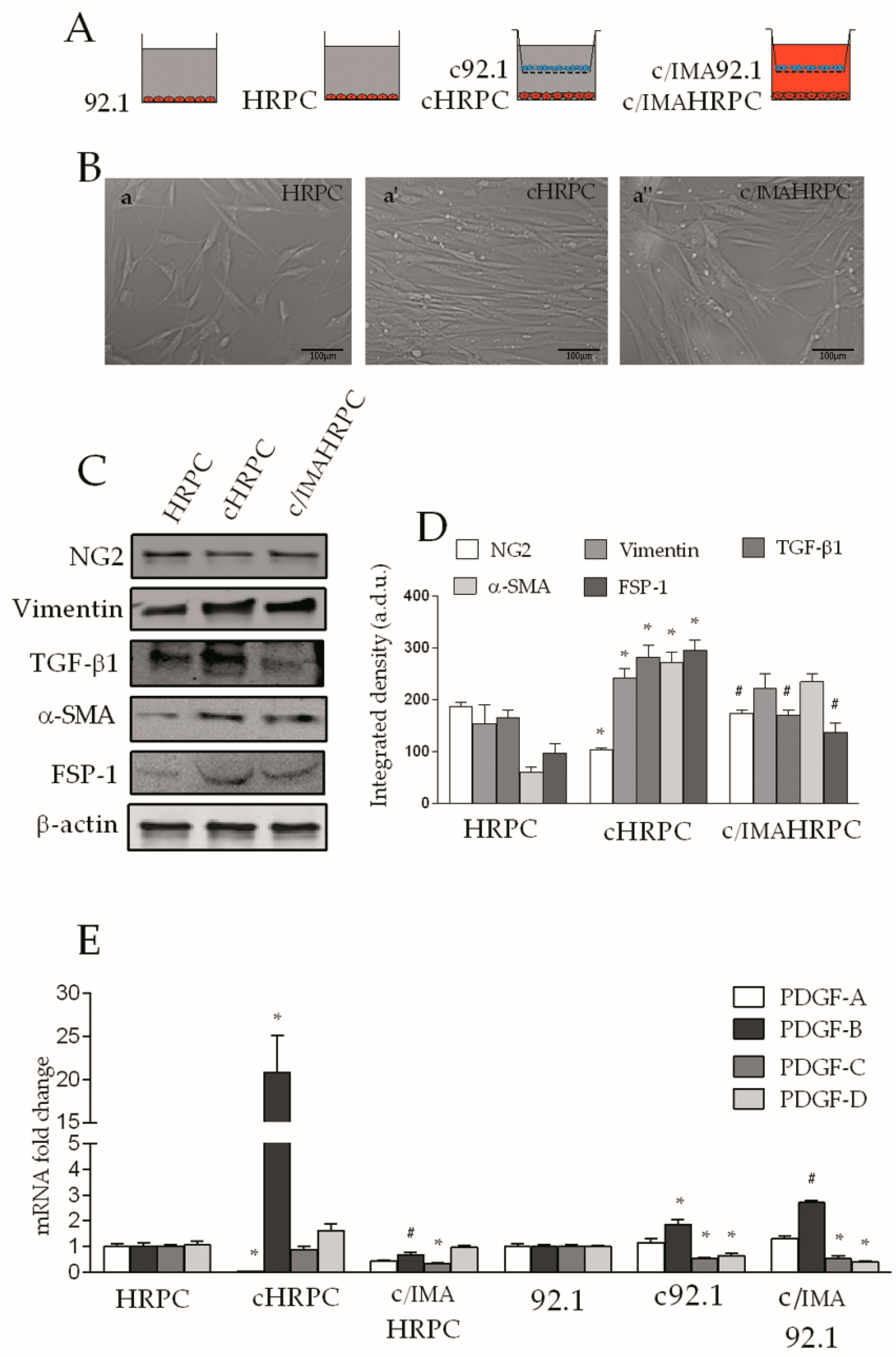
2. Results
2.1. PDGF-BB is Involved in the Modulation of Pericyte Marker Protein Levels in HRPC Conditioned by 92.1UM
2.2. Imatinib Blocked the Enhanced HRPC Migration and Proliferation Induced by 92.1UM
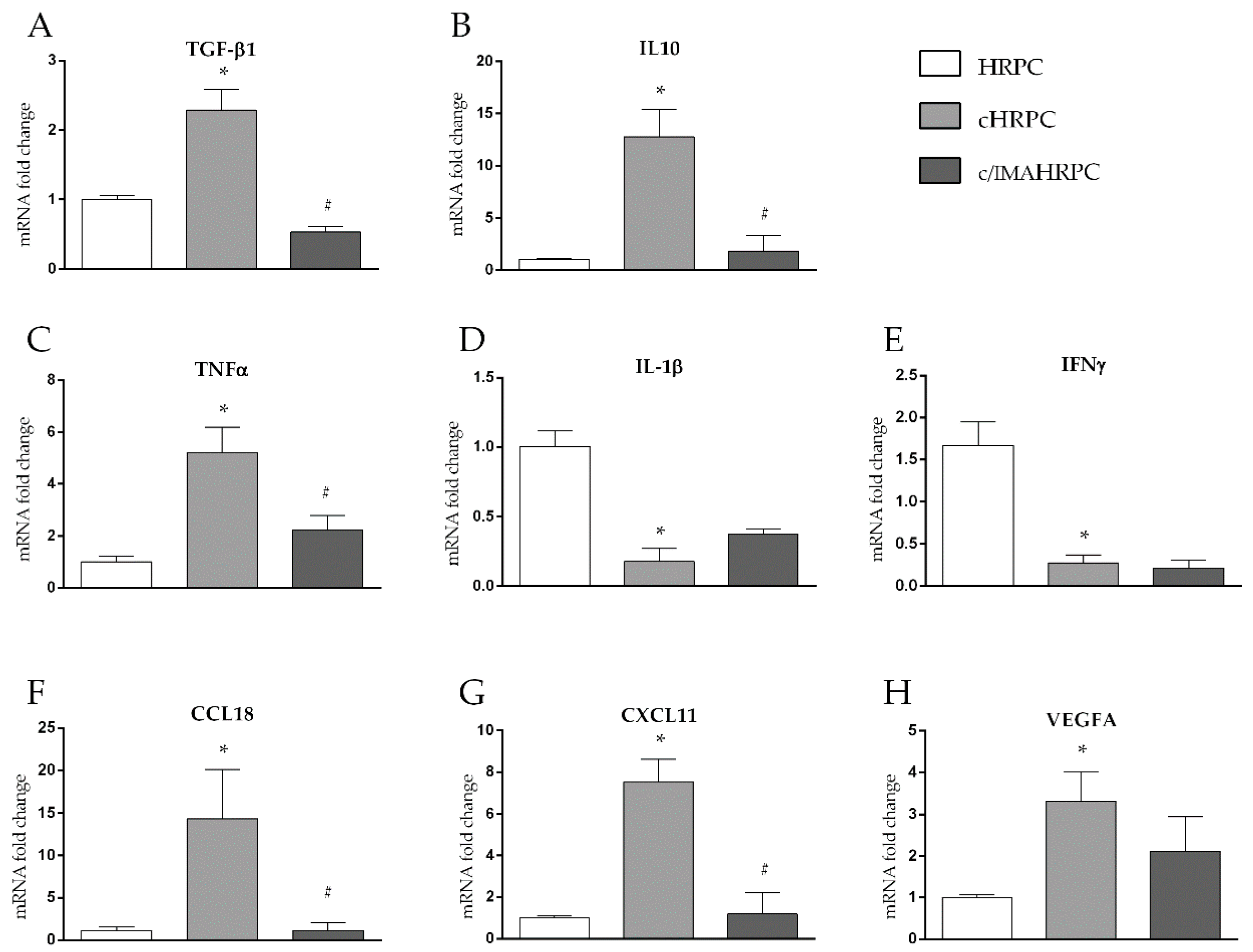
2.3. Long-Lasting Interaction with 92.1UM Induce Pericytes Switch a Pro- to Anti-Inflammatory Phenotype, an Effect Reverted by Imatinib
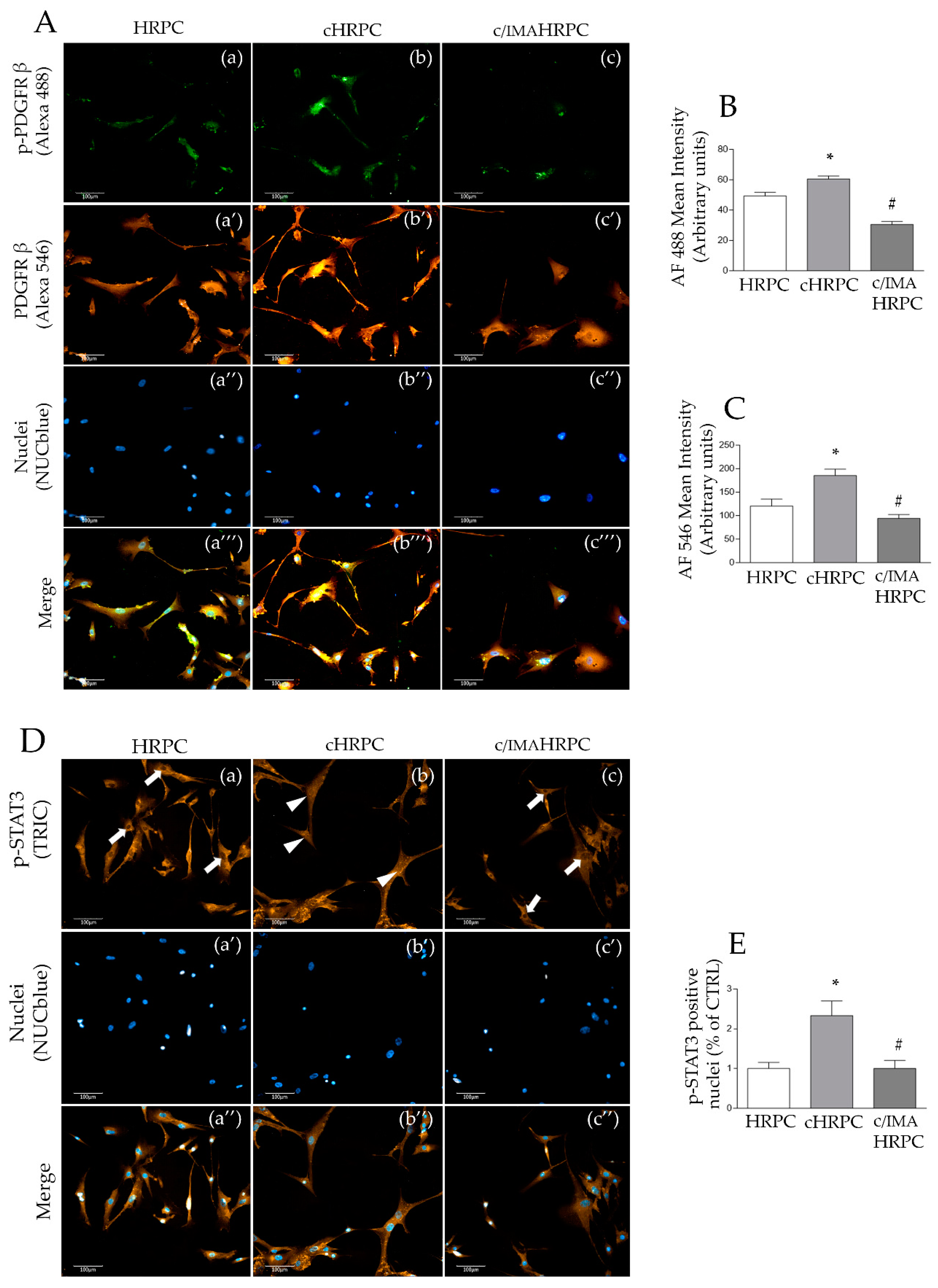
2.4. Imatinib Prevented 92.1UM-Mediated Nuclear Translocation of Phospho-STAT3 and PDGFRβ Activation in HRPC
2.5. Imatinib Prevented 92.1UM-Mediated Nuclear Translocation of Phospho-STAT3 and PDGFRβ Activation in HRPC
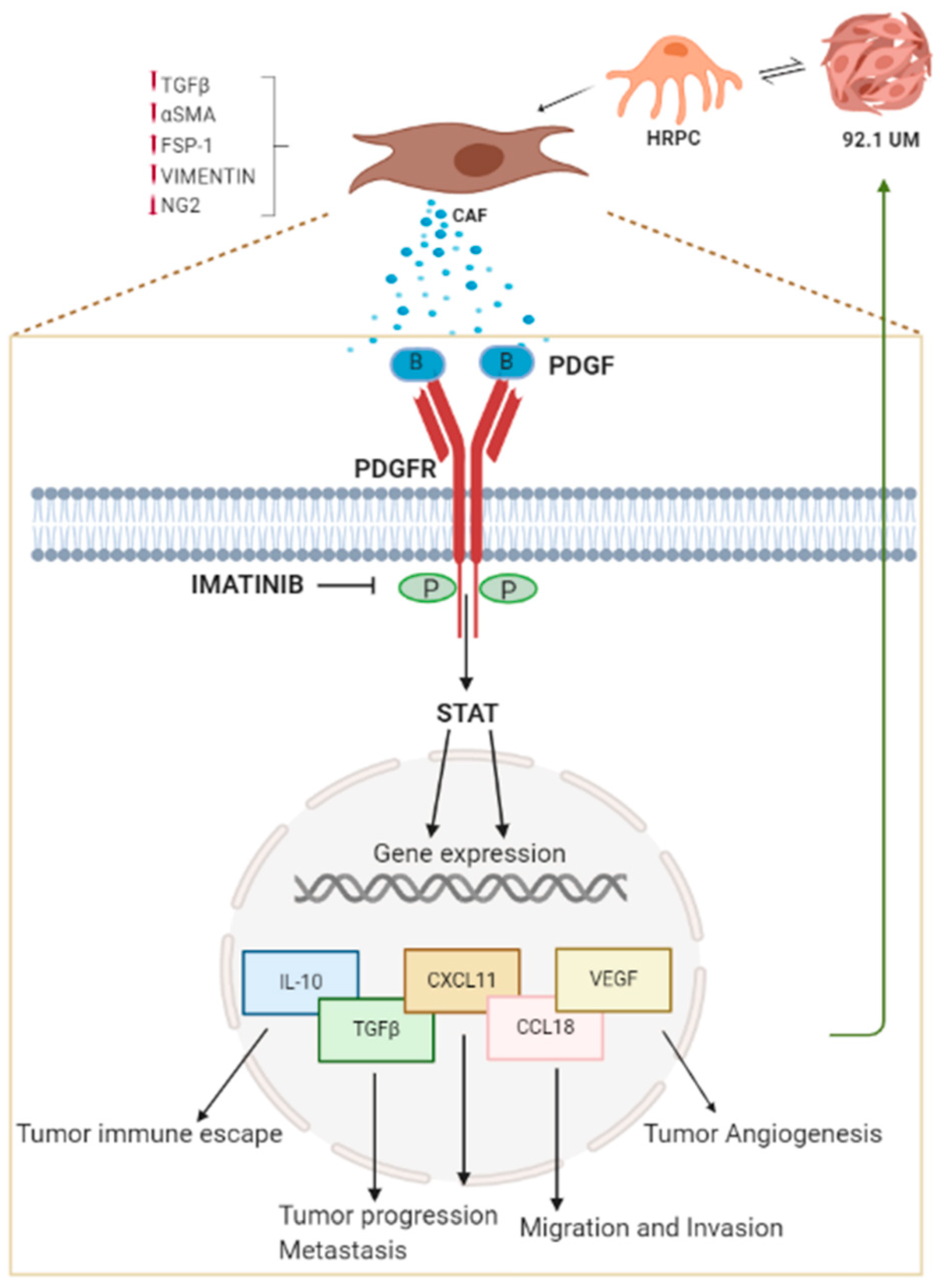
3. Discussion
3.1. 92.1UM Induce Upregulation of Specific CAF Markers in HRPC
3.2. In Vitro Interaction with 92.1UM Modulate mRNA Levels of PDGF Isoforms in Both HRPC and Uveal Melanoma
3.3. HRPC Motility Increase Following In Vitro Interaction with 92.1UM
3.4. High Levels of PDGF-BB Correlate with Tumor Infiltrating Components and Aggressiveness
3.5. 92.1UM-Activated HRPCs are Characterized by a Tumor-Supporting Pattern of Cytokine and Chemokine mRNA Levels
3.6. STAT3 Nuclear Translocation Mediate HRPC-CAF Transition
3.7. Limitations and Weaknesses of the Study
4. Materials and Methods
4.1. Reagents
4.2. Cell Cultures
4.3. HRPC Conditioning by 92.1UM in an “In Vitro” Model of Coculture
4.4. Wound Healing Assay
4.5. Proliferation Assay
4.6. Extraction of Total RNA and Real-Time Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
4.7. High-Content Screening (HCS) and Image Analysis
4.8. Western Blot Analysis
4.9. Invasion Assay of 92.1UM Cells
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Casey, S.C.; Amedei, A.; Aquilano, K.; Azmi, A.S.; Benencia, F.; Bhakta, D.; Bilsland, A.E.; Boosani, C.S.; Chen, S.; Ciriolo, M.R.; et al. Cancer prevention and therapy through the modulation of the tumor microenvironment. Semin. Cancer. Biol. 2015, 35, S199–S223. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Tisty, T.D.; Hein, P.W. Know thy neighbor: Stromal cells can contribute oncogenic signals. Curr. Opin. Genet. Dev. 2001, 11, 54–59. [Google Scholar]
- Bissell, M.J.; Hines, W.C. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 2011, 17, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Han, C.; Wang, S.; Fang, P.; Ma, Z.; Xu, L.; Yin, R. Cancer-associated fibroblasts: An emerging target of anti-cancer immunotherapy. J. Hematol. Oncol. 2019, 12, 86. [Google Scholar] [CrossRef]
- Thakkar, S.; Sharma, D.; Kalia, K.; Tekade, R.K. Tumor microenvironment targeted nanotherapeutics for cancer therapy and diagnosis: A review. Acta. Biomater. 2020, 101, 43–68. [Google Scholar] [CrossRef]
- El-Nikhely, N.; Larzabal, L.; Seeger, W.; Calvo, A.; Savai, R. Tumor-stromal interactions in lung cancer: Novel candidate targets for therapeutic intervention. Expert Opin. Investig. Drugs 2012, 21, 1107–1122. [Google Scholar] [CrossRef]
- Caporarello, N.; D’Angeli, F.; Cambria, M.T.; Candido, S.; Giallongo, C.; Salmeri, M.; Lombardo, C.; Longo, A.; Giurdanella, G.; Anfuso, C.D.; et al. Pericytes in Microvessels: From “Mural” Function to Brain and Retina Regeneration. Int. J. Mol. Sci. 2019, 20, 6351. [Google Scholar] [CrossRef]
- Hellström, M.; Gerhardt, H.; Kalén, M.; Li, X.; Eriksson, U.; Wolburg, H.; Betsholtz, C. Lack of pericytes leads to endothelial hyperplasia and abnormal vascular morphogenesis. J. Cell. Biol. 2001, 153, 543–553. [Google Scholar] [CrossRef]
- Hosaka, K.; Yang, Y.; Seki, T.; Fischer, C.; Dubey, O.; Fredlund, E.; Hartman, J.; Religa, P.; Morikawa, H.; Ishii, Y.; et al. Pericyte-fibroblast transition promotes tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2016, 113, E5618–E5627. [Google Scholar] [CrossRef]
- Armulik, A.; Genové, G.; Betsholtz, C. Pericytes: Developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell. 2011, 21, 193–215. [Google Scholar] [CrossRef]
- Cathery, W.; Faulkner, A.; Maselli, D.; Madeddu, P. Concise Review: The Regenerative Journey of Pericytes Toward Clinical Translation. Stem Cells 2018, 36, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Lin, Y.; Friedrich, C.C.; Neville, C.; Pomerantseva, I.; Sundback, C.A.; Zhang, Z.; Vacanti, J.P.; Hauschka, P.V.; Grottkau, B.E. Bone marrow derived pluripotent cells are pericytes which contribute to vascularization. Stem Cell. Rev. Rep. 2009, 5, 437–445. [Google Scholar] [CrossRef]
- Avolio, E.; Madeddu, P. Discovering cardiac pericyte biology: From physiopathological mechanisms to potential therapeutic applications in ischemic heart disease. Vascul. Pharmacol. 2016, 86, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Giurdanella, G.; Anfuso, C.D.; Olivieri, M.; Lupo, G.; Caporarello, N.; Eandi, C.M.; Drago, F.; Bucolo, C.; Salomone, S. Aflibercept, bevacizumab and ranibizumab prevent glucose-induced damage in human retinal pericytes in vitro, through a PLA2/COX-2/VEGF-A. Biochem. Pharmacol. 2015, 96, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.L.; Okamoto, O.K. Combined effects of pericytes in the tumor microenvironment. Stem Cells Int. 2015, 2015, 868475. [Google Scholar] [CrossRef] [PubMed]
- Salmeri, M.; Motta, C.; Anfuso, C.D.; Amodeo, A.; Scalia, M.; Toscano, M.A.; Alberghina, M.; Lupo, G. VEGF receptor-1 involvement in pericyte loss induced by Escherichia coli in an in vitro model of blood brain barrier. Cell. Microbiol. 2013, 15, 1367–1384. [Google Scholar] [CrossRef]
- Valdor, R.; García-Bernal, D.; Bueno, C.; Ródenas, M.; Moraleda, J.M.; Macian, F.; Martínez, S. Glioblastoma progression is assisted by induction of immunosuppressive function of pericytes through interaction with tumor cells. Oncotarget 2017, 8, 68614–68626. [Google Scholar] [CrossRef]
- D’Arcangelo, E.; Wu, N.C.; Cadavid, J.L.; McGuigan, A.P. The life cycle of cancer-associated fibroblasts within the tumour stroma and its importance in disease outcome. Br. J. Cancer. 2020, 122, 931–942. [Google Scholar] [CrossRef]
- Son, G.M.; Kwon, M.S.; Shin, D.H.; Shin, N.; Ryu, D.; Kang, C.D. Comparisons of cancer-associated fibroblasts in the intratumoral stroma and invasive front in colorectal cancer. Medicine 2019, 98, e15164. [Google Scholar] [CrossRef]
- Aoto, K.; Ito, K.; Aoki, S. Complex formation between platelet-derived growth factor receptor β and transforming growth factor β receptor regulates the differentiation of mesenchymal stem cells into cancer-associated fibroblasts. Oncotarget. 2018, 9, 34090–34102. [Google Scholar] [CrossRef] [PubMed]
- Heldin, C.H.; Eriksson, U.; Ostman, A. New members of the platelet-derived growth factor family of mitogens. Arch. Biochem. Biophys. 2002, 398, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson, L.; Li, H.; Eriksson, U. The PDGF family: Four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 2004, 15, 197–204. [Google Scholar] [CrossRef]
- Ying, H.Z.; Chen, Q.; Zhang, W.Y.; Zhang, H.H.; Ma, Y.; Zhang, S.Z.; Fang, J.; Yu, C.H. PDGF signaling pathway in hepatic fibrosis pathogenesis and therapeutics (Review). Mol. Med. Rep. 2017, 16, 7879–7889. [Google Scholar] [CrossRef] [PubMed]
- Heldin, C.H.; Ostman, A.; Rönnstrand, L. Signal transduction via platelet-derived growth factor receptors. Biochim. Biophys. Acta 1998, 1378, F79–F113. [Google Scholar]
- Heldin, C.H. Targeting the PDGF signaling pathway in tumor treatment. Cell. Commun. Signal. 2013, 11, 97. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, N.; Lennartsson, J. The PDGF/PDGFR pathway as a drug target. J. Mol. Aspects Med. 2018, 62, 75–88. [Google Scholar] [CrossRef]
- Raica, M.; Cimpean, A.M. Platelet-Derived Growth Factor (PDGF)/PDGF Receptors (PDGFR) Axis as Target for Antitumor and Antiangiogenic Therapy. Pharmaceuticals (Basel) 2010, 3, 572–599. [Google Scholar] [CrossRef]
- Dees, C.; Tomcik, M.; Palumbo-Zerr, K.; Distler, A.; Beyer, C.; Lang, V.; Horn, A.; Zerr, P.; Zwerina, J.; Gelse, K.; et al. JAK-2 as a novel mediator of the profibrotic effects of transforminggrowth factorβin systemic sclerosis. Arthritis Rheum. 2012, 64, 3006–3015. [Google Scholar] [CrossRef]
- Albrengues, J.; Bourget, I.; Pons, C.; Butet, V.; Hofman, P.; Tartare-Deckert, S.; Feral, C.C.; Meneguzzi, G.; Gaggioli, C. LIF mediatesproinvasive activation of stromal fibroblasts in cancer. Cell. Rep. 2014, 7, 1664–1678. [Google Scholar] [CrossRef]
- Decker, T.; Kovarik, P. Transcription factor activity of STAT proteins: Structural requirements and regulation by phosphorylation and interacting proteins. Cell. Mol. Life Sci. 1999, 55, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.L.; Darnell, J.E. Mapping of stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic. Acids Res. 1997, 25, 2062–2067. [Google Scholar] [CrossRef] [PubMed]
- Kuzet, S.E.; Gaggioli, C. Fibroblast activation in cancer: When seed fertilizes soil. Cell. Tissue Res. 2016, 365, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Hägglöf, C.; Hammarsten, P.; Josefsson, A.; Stattin, P.; Paulsson, J.; Bergh, A.; Östman, A. Stromal PDGFRbeta expression in prostate tumors and nonmalignant prostate tissue predicts prostate cancer survival. PLoS ONE 2010, 5, e10747. [Google Scholar] [CrossRef]
- Paulsson, J.; Sjöblom, T.; Micke, P.; Pontén, F.; Landberg, G.; Heldin, C.H.; Bergh, J.; Brennan, D.J.; Jirström, K.; Östman, A. Prognostic significance of stromal platelet-derived growth factor β-receptor expression in human breast cancer. Am. J. Pathol. 2009, 175, 334–341. [Google Scholar] [CrossRef]
- Kitadai, Y.; Sasaki, T.; Kuwai, T.; Nakamura, T.; Bucana, C.D.; Hamilton, S.R.; Fidler, I.J. Expression of activated platelet-derived growth factor receptor in stromal cells of human colon carcinomas is associated with metastatic potential. Int. J. Cancer. 2006, 119, 2567–2574. [Google Scholar] [CrossRef]
- Kitadai, Y.; Sasaki, T.; Kuwai, T.; Nakamura, T.; Bucana, C.D.; Fidler, I.J. Targeting the expression of platelet-derived growth factor receptor by reactive stroma inhibits growth and metastasis of human colon carcinoma. Am. J. Pathol. 2006, 169, 2054–2065. [Google Scholar] [CrossRef]
- Yuzawa, S.; Kano, M.R.; Einama, T.; Nishihara, H. PDGFRβ expression in tumor stroma of pancreatic adenocarcinoma as a reliable prognostic marker. Med. Oncol. 2012, 29, 2824–2830. [Google Scholar] [CrossRef]
- Hosaka, K.; Yang, Y.; Seki, T.; Nakamura, M.; Andersson, P.; Rouhi, P.; Yang, X.; Jensen, L.; Lim, S.; Feng, N.; et al. Tumour PDGF-BB expression levels determine dual effects of anti-PDGF drugs on vascular remodelling and metastasis. Nat. Commun. 2013, 4, 2129. [Google Scholar] [CrossRef]
- Virgili, G.; Gatta, G.; Ciccolallo, L.; Capocaccia, R.; Biggeri, A.; Crocetti, E.; Lutz, J.M.; Paci, E. Survival in patients with uveal melanoma in Europe. Arch. Ophthalmol. 2008, 126, 1413–1418. [Google Scholar] [CrossRef]
- Chattopadhyay, C.; Kim, D.W.; Gombos, D.S.; Oba, J.; Qin, Y.; Williams, M.D.; Esmaeli, B.; Grimm, E.A.; Wargo, J.A.; Woodman, S.E.; et al. Uveal Melanoma: From Diagnosis to Treatment and the Science in Between. Cancer 2016, 122, 2299–2312. [Google Scholar] [CrossRef] [PubMed]
- Aronow, M.E.; Topham, A.K.; Singh, A.D. Uveal Melanoma: 5-Year Update on Incidence, Treatment, and Survival (SEER 1973-2013). Ocul. Oncol. Pathol. 2018, 4, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Diener-West, M.; Reynolds, S.M.; Agugliaro, D.J.; Caldwell, R.; Cumming, K.; Earle, J.D.; Hawkins, B.S.; Hayman, J.A.; Jaiyesimi, I.; Jampol, L.M.; et al. Development of metastatic disease after enrollment in the COMS trials for treatment of choroidal melanoma: Collaborative Ocular Melanoma Study Group Report No. 26. Arch. Ophthalmol. 2005, 123, 1639–1643. [Google Scholar] [PubMed]
- Bustamante, P.; Piquet, L.; Landreville, S.; Burnier, J.V. Uveal melanoma pathobiology: Metastasis to the liver. Semin. Cancer. Biol. 2020. [Google Scholar] [CrossRef]
- Bronkhorst, I.H.G.; Jehs, T.M.L.; Dijkgraaf, E.M.; Luyten, G.P.M.; van der Velden, P.A.; van der Burg, S.H.; Jager, M.J. Effect of Hypoxic Stress on Migration and Characteristics of Monocytes in Uveal Melanoma. JAMA Ophthalmol. 2014, 132, 614–621. [Google Scholar] [CrossRef]
- Jager, M.J.; Shields, C.L.; Cebulla, C.M.; Abdel-Rahman, M.H.; Grossniklaus, H.E.; Stern, M.H.; Carvajal, R.D.; Belfort, R.N.; Jia, R.; Shields, J.A.; et al. Uveal melanoma. Nat. Rev. Dis. Primers. 2020, 6, 24. [Google Scholar] [CrossRef]
- Longhitano, L.; Castruccio Castracani, C.; Tibullo, D.; Avola, R.; Viola, M.; Russo, G.; Prezzavento, O.; Marrazzo, A.; Amata, E.; Reibaldi, M.; et al. Sigma-1 and Sigma-2 receptor ligands induce apoptosis and autophagy but have opposite effect on cell proliferation in uveal melanoma. Oncotarget 2017, 8, 91099–91111. [Google Scholar] [CrossRef]
- Ozerdem, U. Targeting pericytes diminishes neovascularization in orthotopic uveal melanoma in nerve/glial antigen 2 proteoglycan knockout mouse. Ophthalmic Res. 2006, 38, 251–254. [Google Scholar] [CrossRef]
- Frank, R.N.; Turczyn, T.J.; Das, A. Pericyte Coverage of Retinal and Cerebral Capillaries Invest. Ophthalmol. Vis. Sci. 1990, 31, 999–1007. [Google Scholar]
- Park, D.Y.; Lee, J.; Kim, J.; Kim, K.; Hong, S.; Han, S.; Kubota, Y.; Augustin, H.G.; Ding, L.; Kim, J.W.; et al. Plastic roles of pericytes in the blood–retinal barrier. Nat. Commun. 2017, 8, 15296. [Google Scholar] [CrossRef]
- Giurdanella, G.; Montalbano, G.; Gennuso, F.; Brancati, S.; Lo Furno, D.; Augello, A.; Bucolo, C.; Drago, F.; Salomone, S. Isolation, cultivation, and characterization of primary bovine cochlear pericytes: A new in vitro model of stria vascularis. J. Cell. Physiol. 2019, 234, 1978–1986. [Google Scholar] [CrossRef] [PubMed]
- Catalano, S.; Panza, S.; Augimeri, G.; Giordano, C.; Malivindi, R.; Gelsomino, L.; Marsico, S.; Giordano, F.; Győrffy, B.; Bonofiglio, D.; et al. Phosphodiesterase 5 (PDE5) Is Highly Expressed in Cancer-Associated Fibroblasts and Enhances Breast Tumor Progression. Cancers (Basel) 2019, 11, 1740. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Xu, Q.; Pan, D.; Xu, D.; Niu, B.; Hong, W.; Zhang, R.; Li, X.; Chen, S. HIC-5 in cancer-associated fibroblasts contributes to esophageal squamous cell carcinoma progression. Cell. Death. Dis. 2019, 10, 873. [Google Scholar] [CrossRef] [PubMed]
- Jehs, T.; Faber, C.; Juel, H.B.; Bronkhorst, I.H.; Jager, M.J.; Nissen, M.H. Inflammation-induced chemokine expression in uveal melanoma cell lines stimulates monocyte chemotaxis. Investig. Ophthalmol. Vis. Sci. 2014, 55, 5169–5175. [Google Scholar] [CrossRef] [PubMed]
- Logan, P.; Burnier, J.; Burnier, M.N., Jr. Vascular endothelial growth factor expression and inhibition in uveal melanoma cell lines. Ecancermedicalscience 2013, 7, 336. [Google Scholar]
- Mueller, M.M.; Fusenig, N.E. Friends or foes—Bipolar effects of the tumour stroma in cancer. Nat. Rev. Cancer 2004, 4, 839–849. [Google Scholar] [CrossRef]
- Singh, A.D.; Turell, M.E.; Topham, A.K. Uveal melanoma: Trends in incidence, treatment, and survival. Ophthalmology 2011, 118, 1881–1885. [Google Scholar] [CrossRef]
- Fretto, L.J.; Snape, A.J.; Tomlinson, J.E.; Seroogy, J.J.; Wolf, D.L.; LaRochelle, W.J.; Giese, N.A. Mechanism of platelet-derived growth factor (PDGF) AA, AB, and BB binding to alpha and beta PDGF receptor. J. Biol. Chem. 1993, 268, 625–631. [Google Scholar]
- Mamer, S.B.; Chen, S.; Weddell, J.C.; Palasz, A.; Wittenkeller, A.; Kumar, M.; Imoukhuede, P.I. Discovery of High-Affinity PDGF-VEGFR Interactions: Redefining RTK Dynamics. Sci. Rep. 2017, 7, 16439. [Google Scholar] [CrossRef]
- Ostman, A.; Heldin, C.H. PDGF receptors as targets in tumor treatment. Adv. Cancer. Res. 2007, 97, 247–274. [Google Scholar]
- Druker, B.J.; Talpaz, M.; Resta, D.J.; Peng, B.; Buchdunger, E.; Ford, J.M.; Lydon, N.B.; Kantarjian, H.; Capdeville, R.; Ohno-Jones, S.; et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 2001, 344, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- Druker, B.J.; Tamura, S.; Buchdunger, E.; Ohno, S.; Segal, G.M.; Fanning, S.; Zimmermann, J.; Lydon, N.B. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat. Med. 1996, 2, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Calipel, A.; Landreville, S.; De La Fouchardière, A.; Mascarelli, F.; Rivoire, M.; Penel, N.; Mouriaux, F. Mechanisms of resistance to imatinib mesylate in KIT-positive metastatic uveal melanoma. Clin. Exp. Metastasis 2014, 31, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Tallquist, M.; Kazlauskas, A. PDGF signaling in cells and mice. Cytokine Growth Factor Rev. 2004, 15, 205–213. [Google Scholar] [CrossRef]
- Salmeri, M.; Motta, C.; Mastrojeni, S.; Amodeo, A.; Anfuso, C.D.; Giurdanella, G.; Morello, A.; Alberghina, M.; Toscano, M.A.; Lupo, G. Involvement of PKCα-MAPK/ERK-phospholipase A2 pathway in the Escherichia coli invasion of brain microvascular endothelial cells. Neurosci. Lett. 2012, 511, 33–37. [Google Scholar] [CrossRef]
- Eriksson, A.; Siegbahn, A.; Westermark, B.; Heldin, C.H.; Claesson-Welsh, L. PDGF alpha- and beta-receptors activate unique and common signal transduction pathways. EMBO J. 1992, 11, 543–550. [Google Scholar] [CrossRef]
- Bronzert, D.A.; Pantazis, P.; Antoniades, H.N.; Kasid, A.; Davidson, N.; Dickson, R.B.; Lippman, M.E. Synthesis and secretion of platelet-derived growth factor by human breast cancer cell lines. Proc. Natl. Acad. Sci. USA 1987, 84, 5763–5767. [Google Scholar] [CrossRef]
- Heldin, C.H.; Westermark, B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 1999, 79, 1283–1316. [Google Scholar] [CrossRef]
- Ning, X.; Zhang, H.; Wang, C.; Song, X. Exosomes Released by Gastric Cancer Cells Induce Transition of Pericytes Into Cancer-Associated Fibroblasts. Med. Sci. Monit. 2018, 24, 2350–2359. [Google Scholar] [CrossRef]
- Gu, J.; Qian, H.; Shen, L.; Zhang, X.; Zhu, W.; Huang, L.; Yan, Y.; Mao, F.; Zhao, C.; Shi, Y.; et al. Gastric cancer exosomes trigger differentiation of umbilical cord derived mesenchymal stem cells to carcinoma-associated fibroblasts through TGF-β/Smad pathway. PLoS ONE 2012, 7, e52465. [Google Scholar] [CrossRef]
- Jin, W. Role of JAK/STAT3 Signaling in the Regulation of Metastasis, the Transition of Cancer Stem Cells, and Chemoresistance of Cancer by Epithelial–Mesenchymal Transition. Cells 2020, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Bordenave, J.; Tu, L.; Berrebeh, N.; Thuillet, R.; Cumont, A.; Le Vely, B.; Fadel, E.; Nadaud, S.; Savale, L.; Humbert, M.; et al. Lineage Tracing Reveals the Dynamic Contribution of Pericytes to the Blood Vessel Remodeling in Pulmonary Hypertension. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 766–782. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Mukouyama, Y.S. Tissue Specific Origin, Development, and Pathological Perspectives of Pericytes. Front. Cardiovasc. Med. 2018, 5, 78. [Google Scholar] [CrossRef] [PubMed]


| Gene | Sequence (5’-3’) | Amplicon (bp) | Accession Number |
|---|---|---|---|
| VEGFA | Fw: ATCTTCAAGCCATCCTGTGTGC | 121 | NM_001025366.3 |
| Rv: GAGGTTTGATCCGCATAATCTG | |||
| CCL18 | Fw: CTCCTTGTCCTCGTCTGCAC | 248 | NM_002988.4 |
| Rv: TCAGGCATTCAGCTTCAGGT | |||
| CXCL11 | Fw: GCCTTGGCTGTGATATTGTG | 235 | NM_001302123.2 |
| Rv: TGATTATAAGCCTTGCTTGCTTCG | |||
| IFN- γ | Fw: AGATGACTTCGAAAAGCTGACT | 95 | NM_000619.3 |
| Rv: ACAGTTCAGCCATCACTTGG | |||
| IL10 | Fw: GACTTTAAGGGTTACCTGGGTTG | 112 | NM_000572.3 |
| Rv: TCACATGCGCCTTGATGTCTG | |||
| IL-1 β | Fw: AGCTACGAATCTCCGACCAC | 186 | NM_000576.3 |
| Rv: CGTTATCCCATGTGTCGAAGAA | |||
| TGF- β1 | Fw: CGTCTGCTGAGGCTCAAGT | 74 | NM_000660.7 |
| Rv: CGCCAGGAATTGTTGCTGTA | |||
| TNF-α | Fw: AGCCCATGTTGTAGCAAA CC | 134 | NM_000594.4 |
| Rv: TGAGGTACAGGCCCTCTGAT | |||
| 18S rRNA | Fw: TAAGTCCCTGCCCTTTGTACACA | 69 | NR_146119 |
| Rv: GATCCGAGGGCCTCACTAAAC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anfuso, C.D.; Longo, A.; Distefano, A.; Amorini, A.M.; Salmeri, M.; Zanghì, G.; Giallongo, C.; Giurdanella, G.; Lupo, G. Uveal Melanoma Cells Elicit Retinal Pericyte Phenotypical and Biochemical Changes in an in Vitro Model of Coculture. Int. J. Mol. Sci. 2020, 21, 5557. https://doi.org/10.3390/ijms21155557
Anfuso CD, Longo A, Distefano A, Amorini AM, Salmeri M, Zanghì G, Giallongo C, Giurdanella G, Lupo G. Uveal Melanoma Cells Elicit Retinal Pericyte Phenotypical and Biochemical Changes in an in Vitro Model of Coculture. International Journal of Molecular Sciences. 2020; 21(15):5557. https://doi.org/10.3390/ijms21155557
Chicago/Turabian StyleAnfuso, Carmelina Daniela, Anna Longo, Alfio Distefano, Angela Maria Amorini, Mario Salmeri, Guido Zanghì, Cesarina Giallongo, Giovanni Giurdanella, and Gabriella Lupo. 2020. "Uveal Melanoma Cells Elicit Retinal Pericyte Phenotypical and Biochemical Changes in an in Vitro Model of Coculture" International Journal of Molecular Sciences 21, no. 15: 5557. https://doi.org/10.3390/ijms21155557
APA StyleAnfuso, C. D., Longo, A., Distefano, A., Amorini, A. M., Salmeri, M., Zanghì, G., Giallongo, C., Giurdanella, G., & Lupo, G. (2020). Uveal Melanoma Cells Elicit Retinal Pericyte Phenotypical and Biochemical Changes in an in Vitro Model of Coculture. International Journal of Molecular Sciences, 21(15), 5557. https://doi.org/10.3390/ijms21155557







