Combinational Analyses with Multiple Methods Reveal the Existence of Several Forms of Polysialylated Neural Cell Adhesion Molecule in Mouse Developing Brains
Abstract
:1. Introduction
2. Results
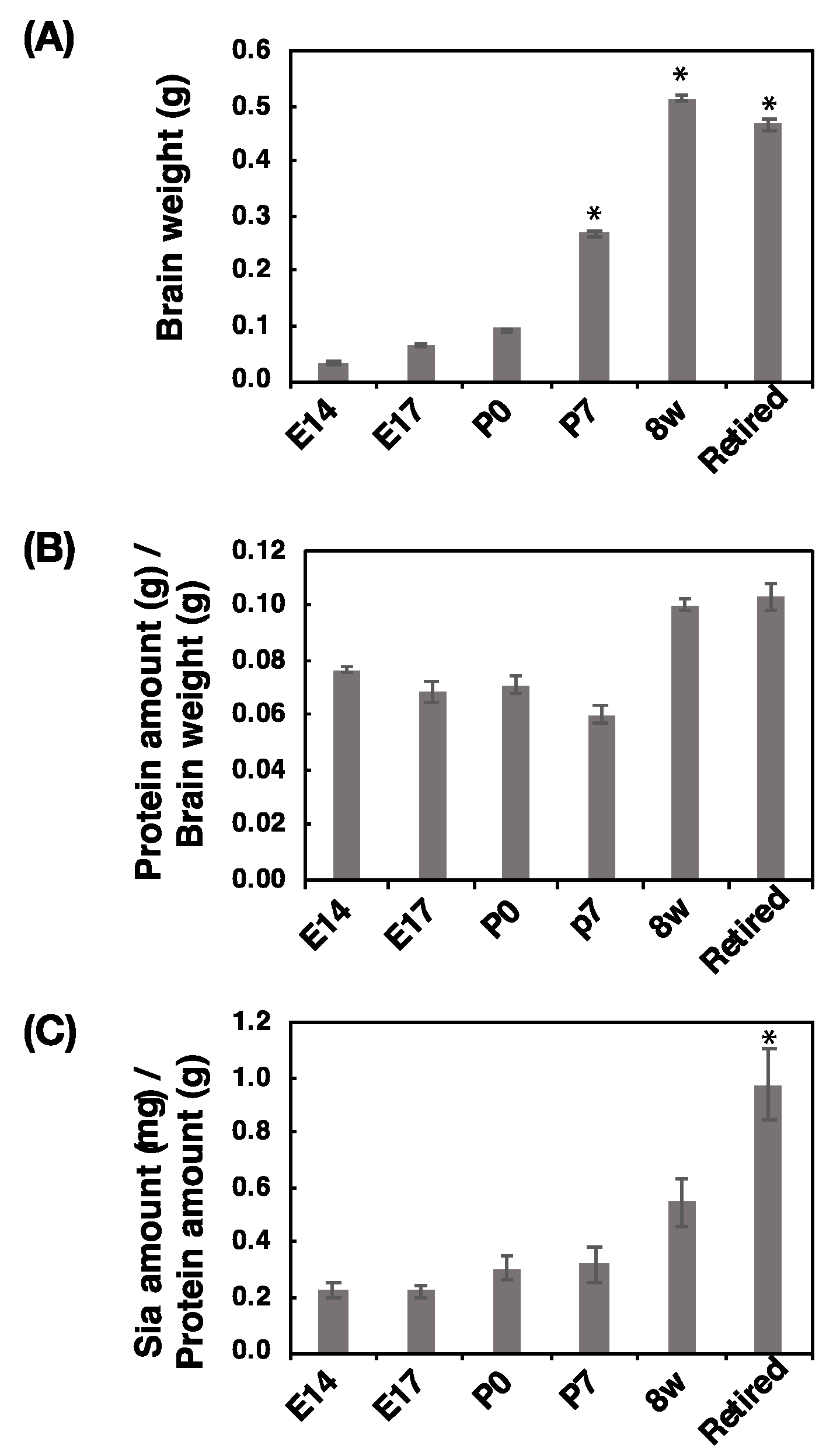
2.1. Brain Size, Protein Amount, and Sialic Acid Amount
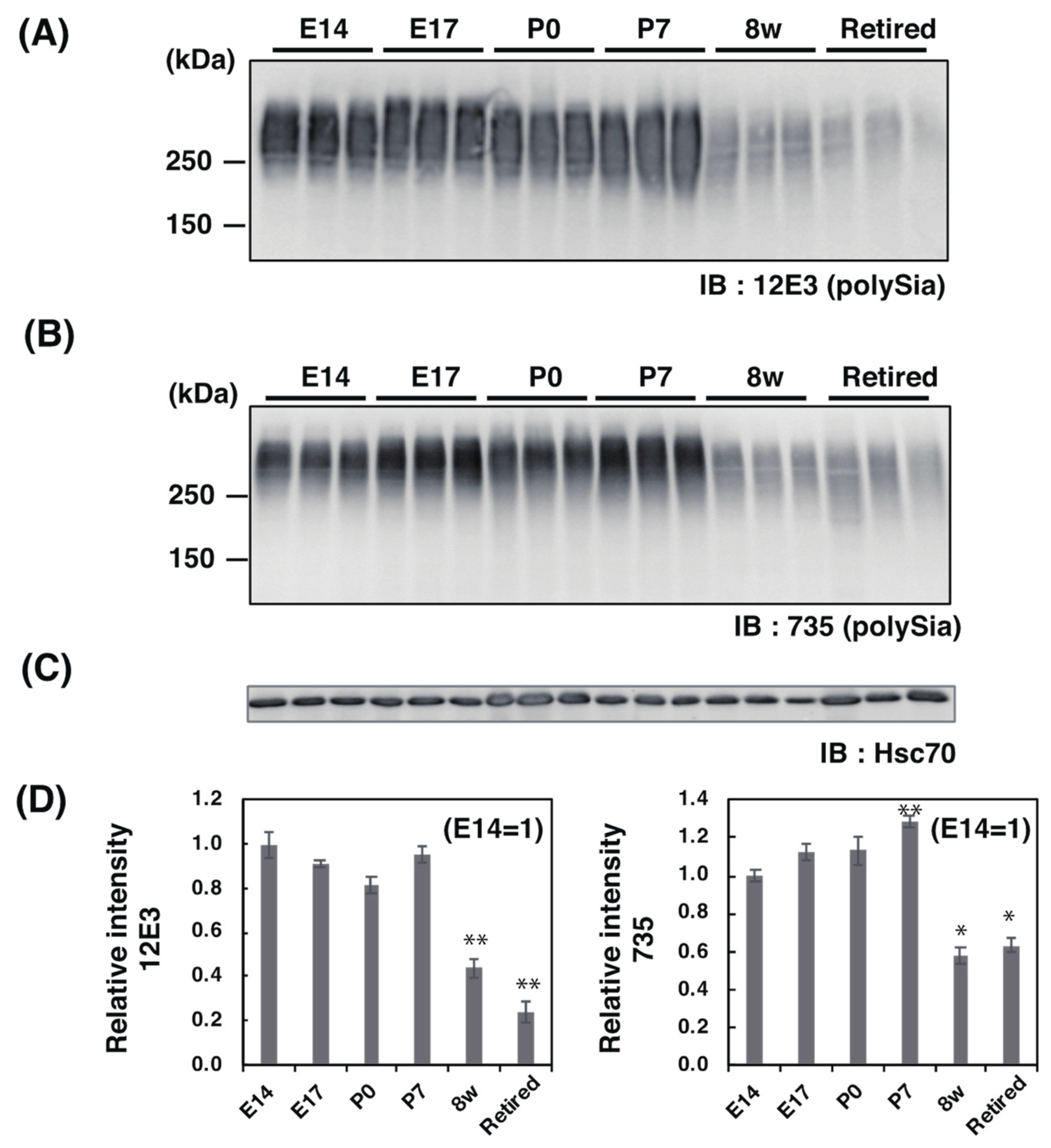
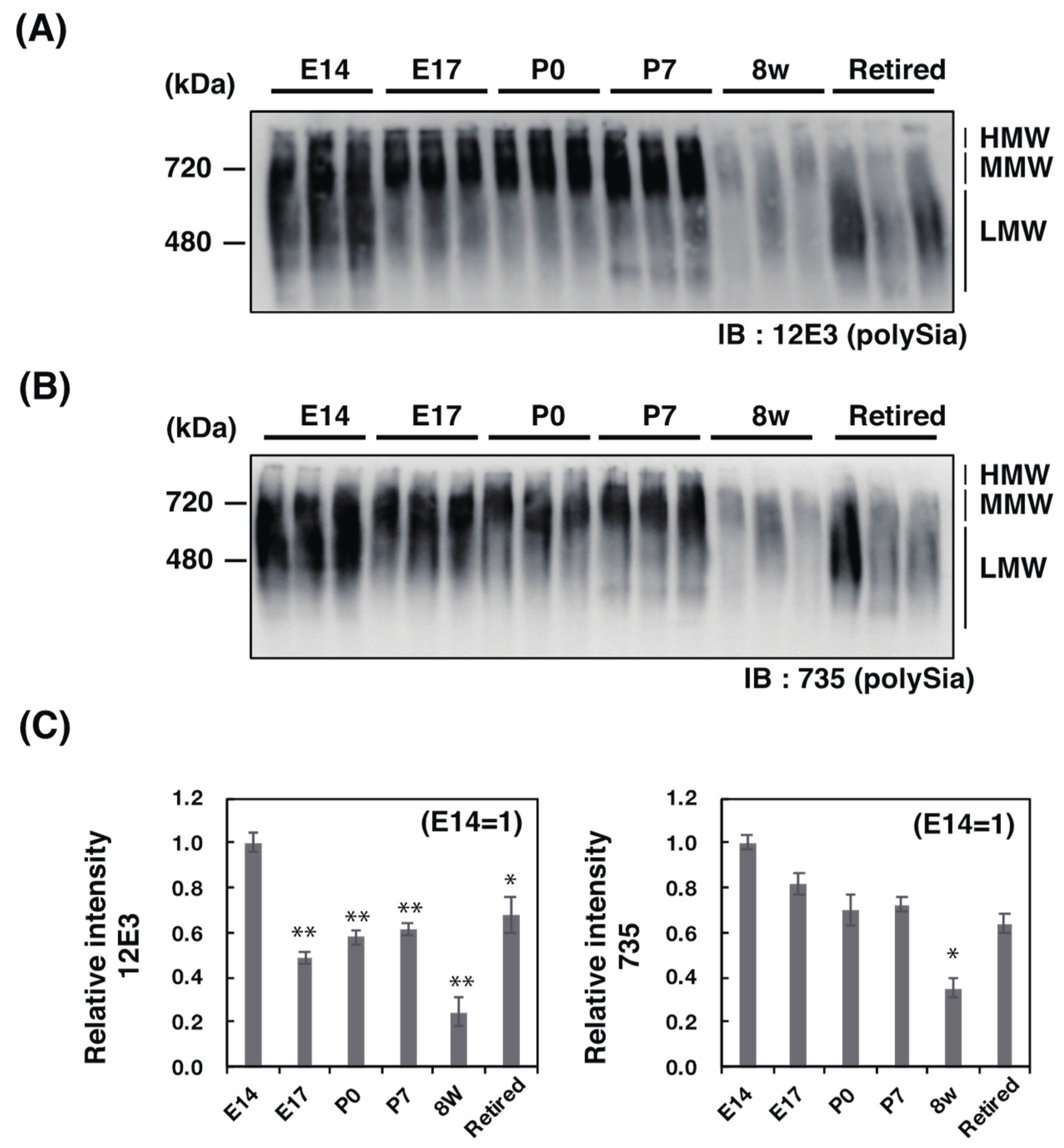
2.2. Western Blotting Analysis of PolySia–NCAM
2.2.1. SDS–PAGE/Western Blotting Analysis
2.2.2. Native PAGE/Western Blotting Analysis
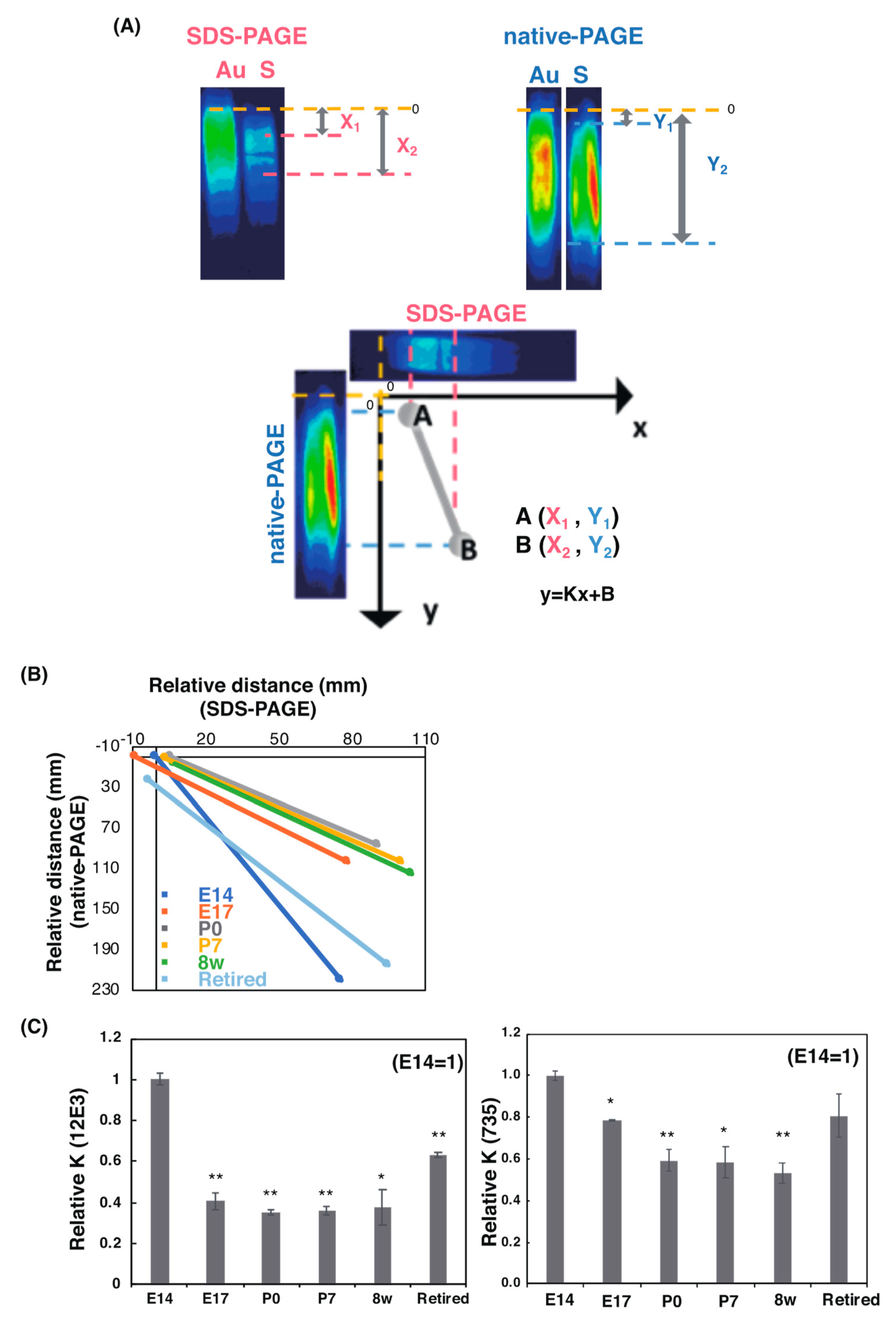
2.2.3. SDS–PAGE/Native PAGE (S/N) MAP Analysis
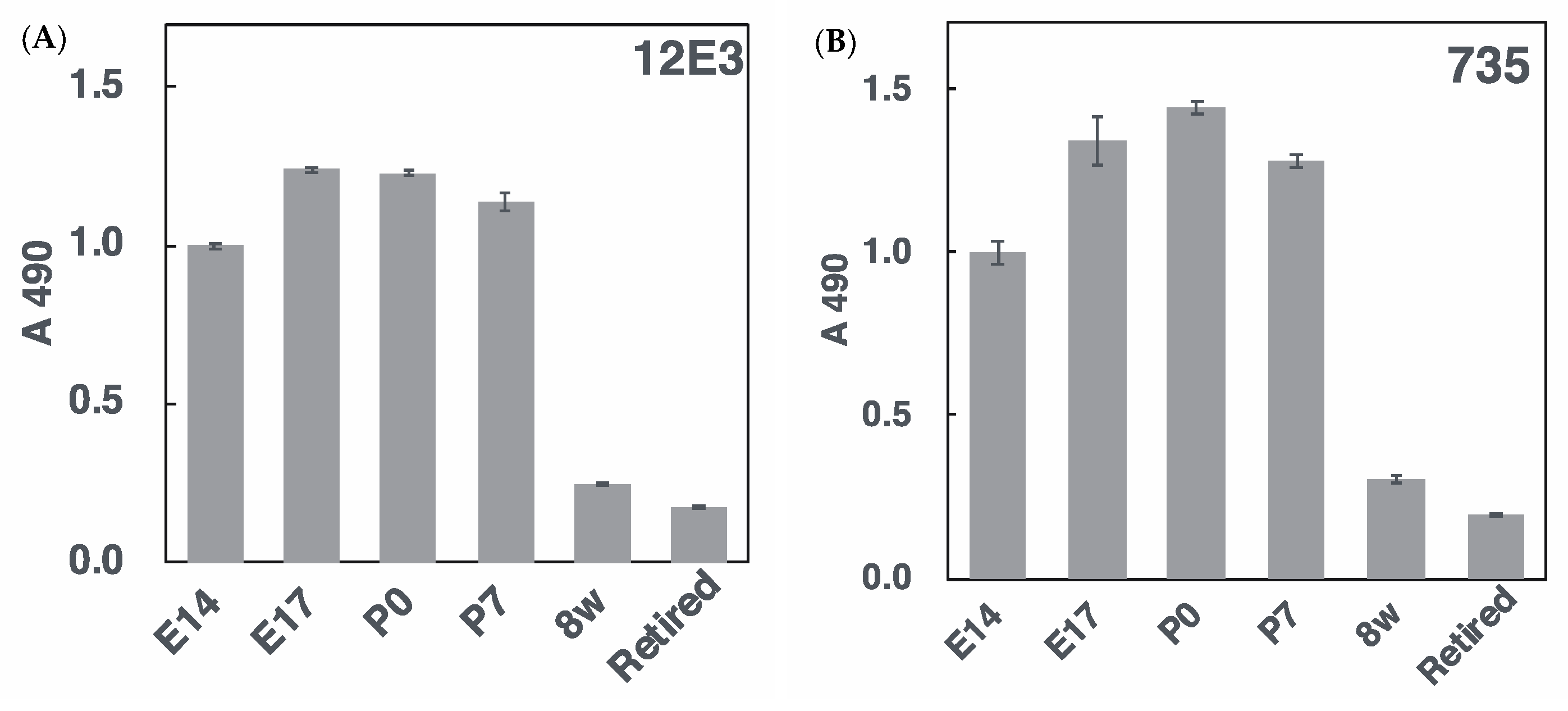
2.2.4. ELISA Analysis
2.3. ChemicalAanalysis of PolySia–NCAM
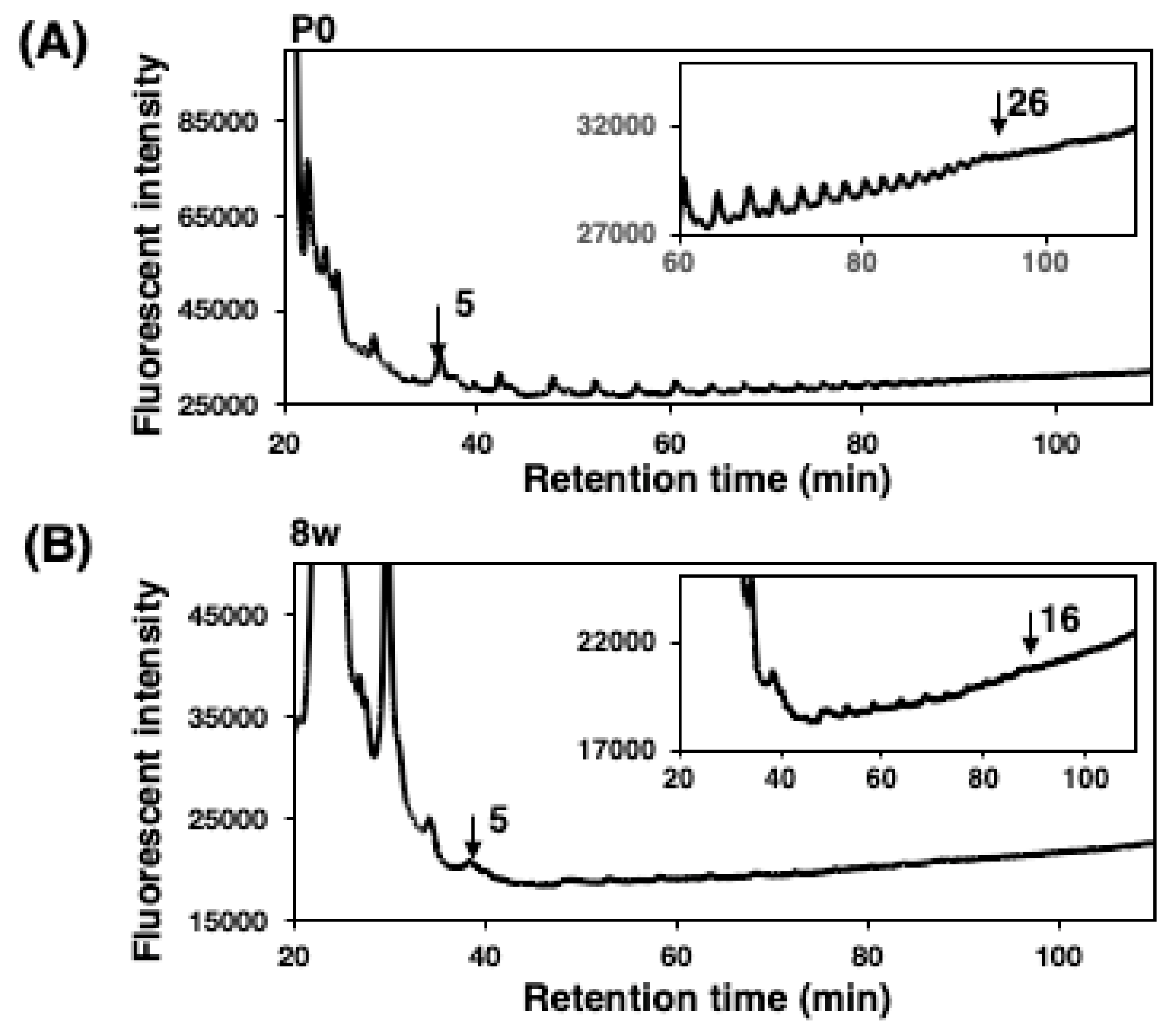
2.3.1. Mild Acid Hydrolysis–Fluorometric Anion-Exchange Chromatography Method (MH–FAEC, Oligo-Analysis)
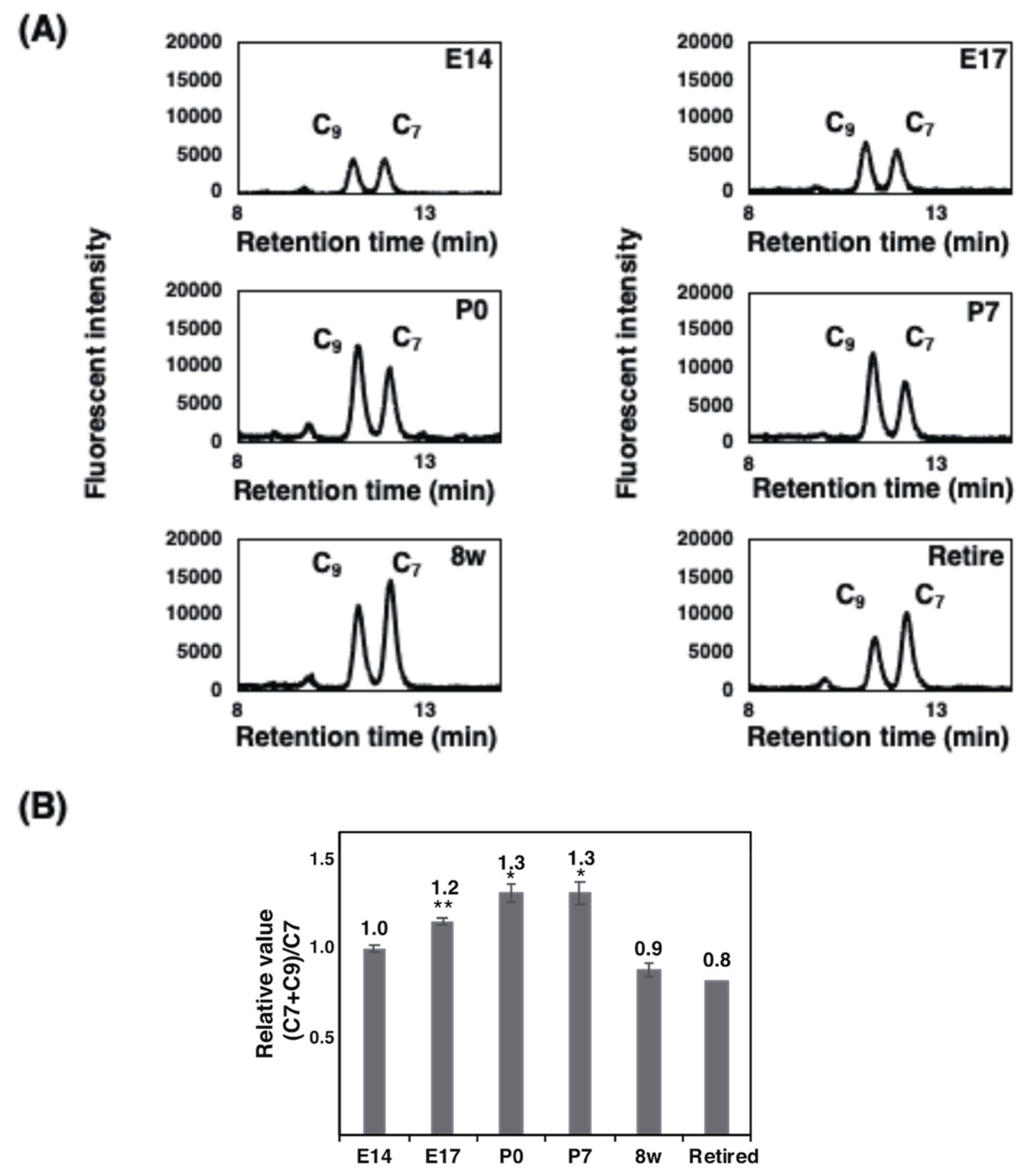
2.3.2. Fluorometric C7/C9 Analysis
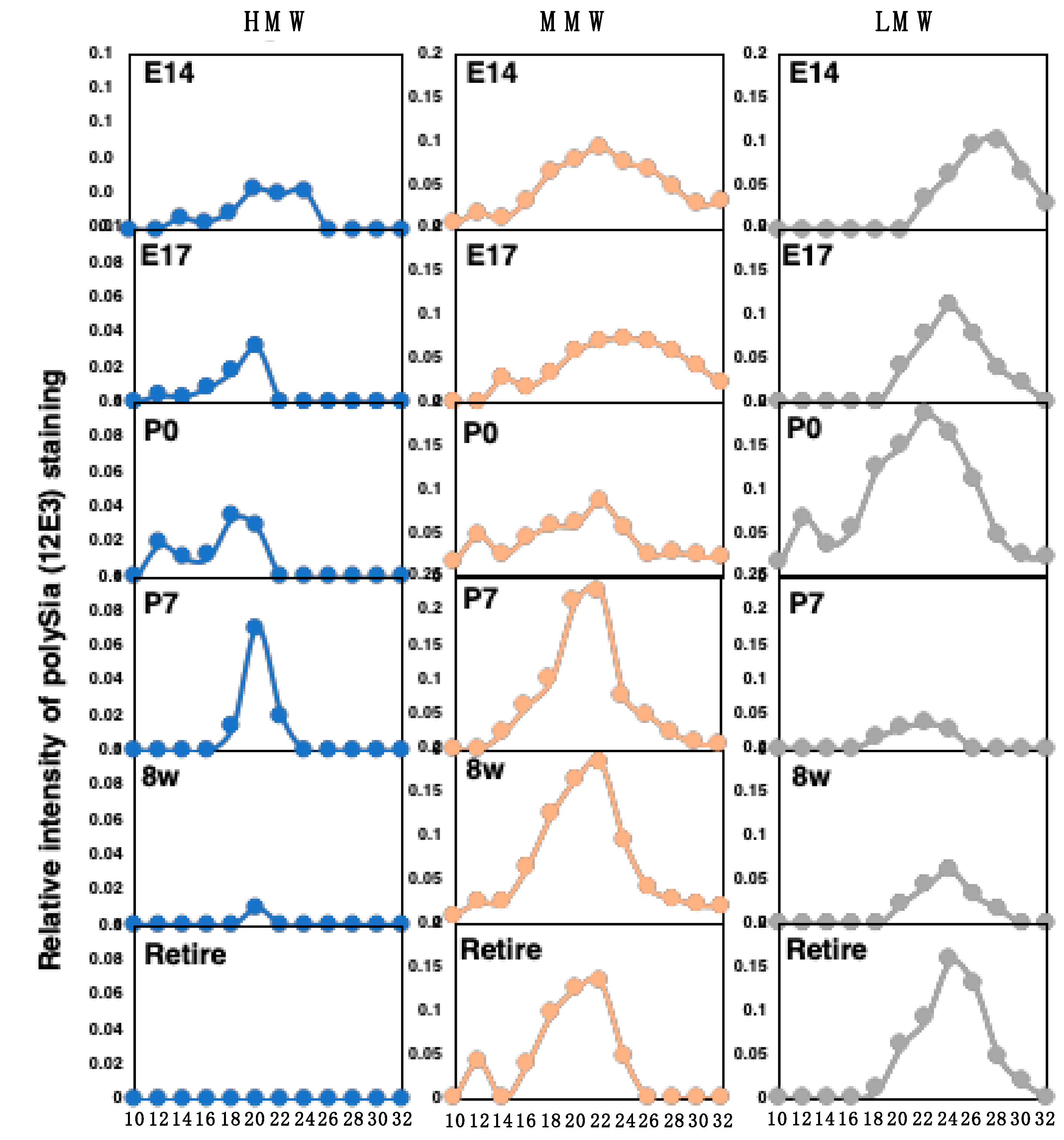
2.4. Chromatographical Analysis of PolySia–NCAM
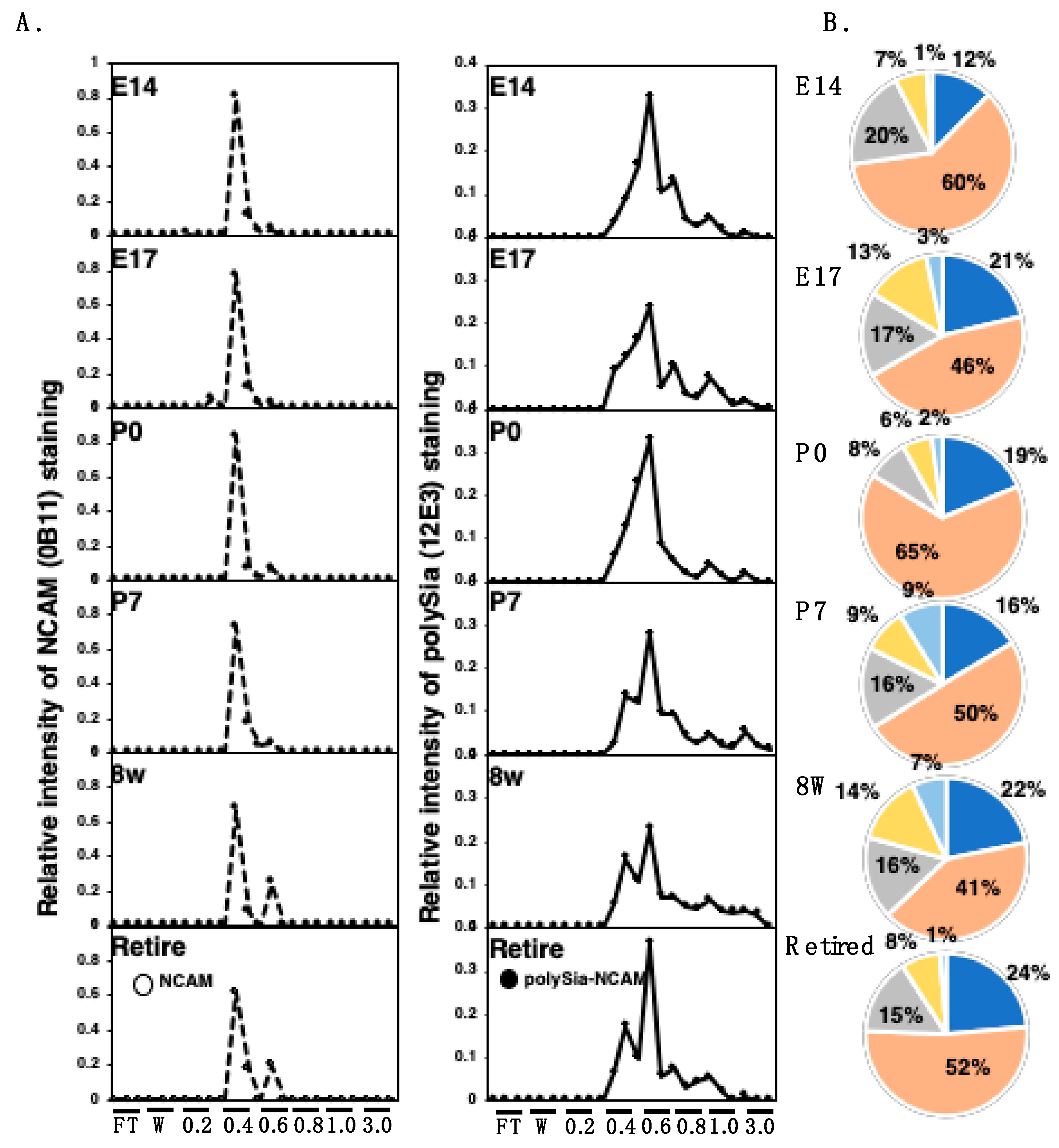
2.4.1. Anion-Exchange Chromatography of PolySia–NCAM and NCAM
2.4.2. Gel Filtration
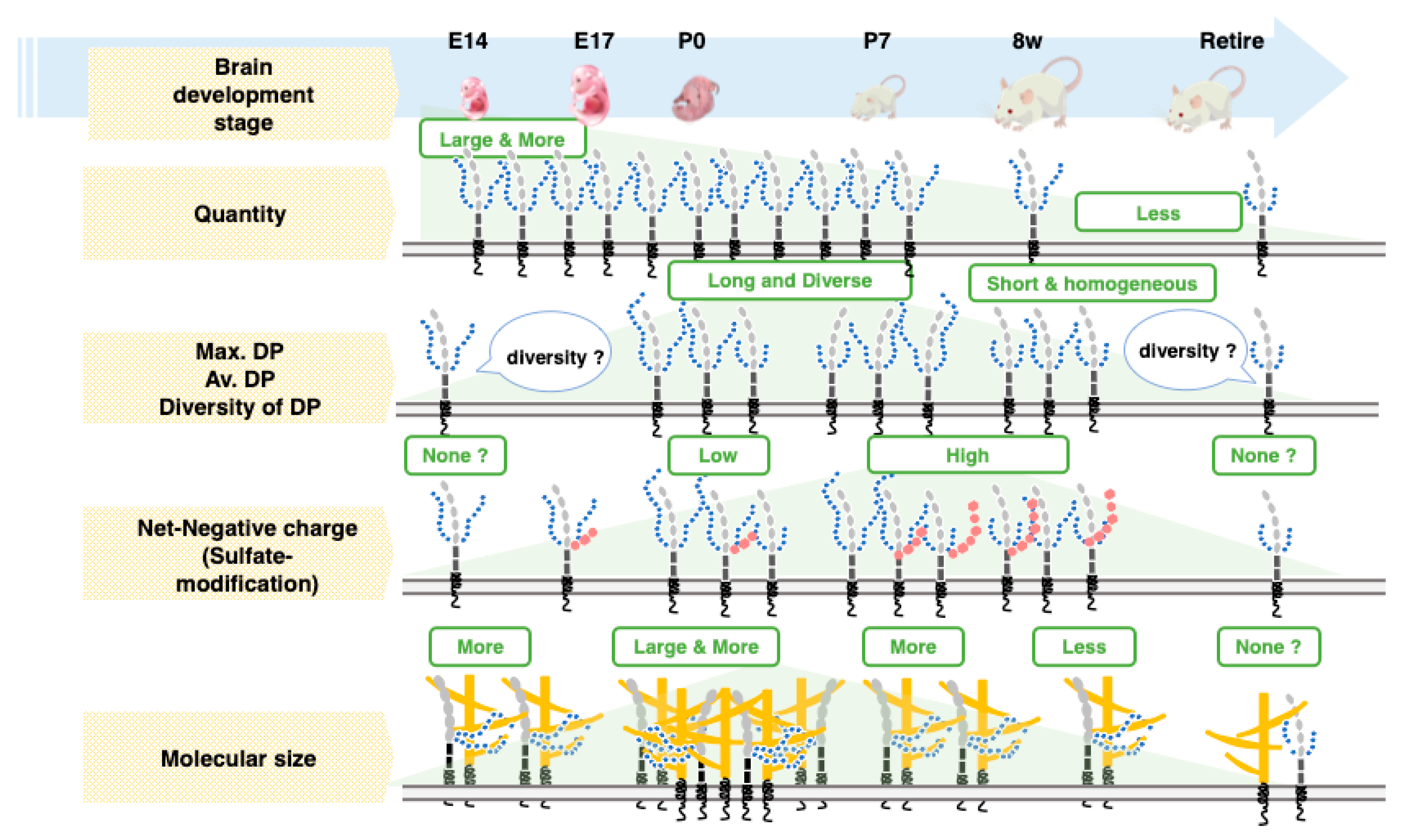
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Animals and Ethics Statement
4.3. Sample Preparation
4.4. Content of Sialic Acid
4.5. Western Blotting
4.6. SDS–PAGE/Native PAGE Map
4.7. ELISA Analysis
4.8. Mild Acid Hydrolysis–Fluorometric Anion-Exchange Chromatography Analysis
4.9. Chemical Analysis of α2,8-Linked Oligo/PolySia Chains on Glycoproteins Blotted onto PVDF Membranes
4.10. Analysis of PolySia–NCAM from Brain Homogenates Using Anion-Exchange Chromatography
4.11. Analysis of PolySia–NCAM from Brain Homogenates Using Gel-Filtration Chromatography
4.12. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DP | Degree of polymerization |
| FGF | Fibroblast growth factor |
| FGFR | Fibroblast growth factor receptor |
| GAG | Glycosaminoglycan |
| HPLC | High performance liquid chromatography |
| Ig | Immunoglobulin |
| IgG | Immunoglobulin G |
| IgM | Immunoglobulin M |
| KO | Knockout |
| NCAM | Neural cell adhesion molecule |
| PAGE | Polyacrylamide gel electrophoresis |
| polySia | Polysialic acid |
| polySia–NCAM | Polysialylated neural cell adhesion molecule |
| Sia | Sialic acid |
| SNPs | Single nucleotide polymorphisms |
References
- Sato, C.; Kitajima, K. Disialic, oligosialic and polysialic acids: Distribution, functions and related disease. J. Biochem. 2013, 154, 115–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barry, G.T.; Goebel, W.F. Colominic acid, a substance of bacterial origin related to sialic acid. Nature 1957, 179, 206. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, A.K.; Jennings, H.J.; Kenny, C.P.; Martin, A.; Smith, I.C. Structural determination of the sialic acid polysaccharide antigens of Neisseria meningitidis serogroups B and C with carbon 13 nuclear magnetic resonance. J. Biol. Chem. 1975, 250, 1926–1932. [Google Scholar]
- Inoue, S.; Iwasaki, M. Isolation of a novel glycoprotein from the eggs of rainbow trout: Occurrence of disialosyl groups on all carbohydrate chains. Biochem. Biophys. Res. Commun. 1978, 83, 1018–1023. [Google Scholar] [CrossRef]
- Finne, J. Occurrence of unique polysialosyl carbohydrate units in glycoproteins of developing brain. J. Biol. Chem. 1982, 257, 11966–11970. [Google Scholar]
- Cremer, H.; Lange, R.; Christoph, A.; Plomann, M.; Vopper, G.; Roes, J.; Brown, R.; Baldwin, S.; Kraemer, P.; Scheff, S. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature 1994, 367, 455–459. [Google Scholar] [CrossRef]
- Galuska, S.P.; Rollenhagen, M.; Kaup, M.; Eggers, K.; Oltmann-Norden, I.; Schiff, M.; Hartmann, M.; Weinhold, B.; Hildebrandt, H.; Geyer, R.; et al. Synaptic cell adhesion molecule SynCAM 1 is a target for polysialylation in postnatal mouse brain. Proc. Natl. Acad. Sci. USA 2010, 107, 10250–10255. [Google Scholar] [CrossRef] [Green Version]
- Zuber, C.; Lackie, P.; Catterall, W.; Roth, J. Polysialic acid is associated with sodium channels and the neural cell adhesion molecule N-CAM in adult rat brain. J. Biol. Chem. 1992, 267, 9965–9971. [Google Scholar] [PubMed]
- Yabe, U.; Sato, C.; Matsuda, T.; Kitajima, K. Polysialic acid in human milk. CD36 is a new member of mammalian polysialic acid-containing glycoprotein. J. Biol. Chem. 2003, 278, 13875–13880. [Google Scholar] [CrossRef] [Green Version]
- Curreli, S.; Arany, Z.; Gerardy-Schahn, R.; Mann, D.; Stamatos, N. Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J. Biol. Chem. 2007, 282, 30346–30356. [Google Scholar] [CrossRef] [Green Version]
- Kiermaier, E.; Moussion, C.; Veldkamp, C.T.; Gerardy-Schahn, R.; de Vries, I.; Williams, L.G.; Chaffee, G.R.; Phillips, A.J.; Freiberger, F.; Imre, R.; et al. Polysialylation controls dendritic cell trafficking by regulating chemokine recognition. Science 2016, 351, 186–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumida, M.; Hane, M.; Yabe, U.; Shimoda, Y.; Pearce, O.M.; Kiso, M.; Miyagi, T.; Sawada, M.; Varki, A.; Kitajima, K.; et al. Rapid Trimming of Cell Surface Polysialic Acid (PolySia) by Exovesicular Sialidase Triggers Release of Preexisting Surface Neurotrophin. J. Biol. Chem. 2015, 290, 13202–13214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werneburg, S.; Buettner, F.F.; Erben, L.; Mathews, M.; Neumann, H.; Mühlenhoff, M.; Hildebrandt, H. Polysialylation and lipopolysaccharide-induced shedding of E-selectin ligand-1 and neuropilin-2 by microglia and THP-1 macrophages. Glia 2016, 64, 1314–1330. [Google Scholar] [CrossRef] [PubMed]
- Mühlenhoff, M.; Eckhardt, M.; Bethe, A.; Frosch, M.; Gerardy-Schahn, R. Autocatalytic polysialylation of polysialyltransferase-1. EMBO J. 1996, 15, 6943–6950. [Google Scholar] [CrossRef]
- Rutishauser, U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci. 2008, 9, 26–35. [Google Scholar] [CrossRef]
- Seki, T.; Arai, Y. Distribution and possible roles of the highly polysialylated neural cell adhesion molecule (NCAM-H) in the developing and adult central nervous system. Neurosci. Res. 1993, 17, 265–290. [Google Scholar] [CrossRef]
- Seki, T.; Arai, Y. Highly polysialylated neural cell adhesion molecule (NCAM-H) is expressed by newly generated granule cells in the dentate gyrus of the adult rat. J. Neurosci. 1993, 13, 2351–2358. [Google Scholar] [CrossRef] [Green Version]
- Bonfanti, L.; Olive, S.; Poulain, D.A.; Theodosis, D.T. Mapping of the distribution of polysialylated neural cell adhesion molecule throughout the central nervous system of the adult rat: An immunohistochemical study. Neuroscience 1992, 49, 419–436. [Google Scholar] [CrossRef]
- Sato, C.; Kitajima, K. Sialic Acids in Neurology. Adv. Carbohydr. Chem. Biochem. 2019, 76, 1–64. [Google Scholar]
- Angata, K.; Fukuda, M. Polysialyltransferases: Major players in polysialic acid synthesis on the neural cell adhesion molecule. Biochimie 2003, 85, 195–206. [Google Scholar] [CrossRef]
- Angata, K.; Long, J.M.; Bukalo, O.; Lee, W.; Dityatev, A.; Wynshaw-Boris, A.; Schachner, M.; Fukuda, M.; Marth, J.D. Sialyltransferase ST8Sia-II assembles a subset of polysialic acid that directs hippocampal axonal targeting and promotes fear behavior. J. Biol. Chem. 2004, 279, 32603–32613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckhardt, M.; Bukalo, O.; Chazal, G.; Wang, L.; Goridis, C.; Schachner, M.; Gerardy-Schahn, R.; Cremer, H.; Dityatev, A. Mice deficient in the polysialyltransferase ST8SiaIV/PST-1 allow discrimination of the roles of neural cell adhesion molecule protein and polysialic acid in neural development and synaptic plasticity. J. Neurosci. 2000, 20, 5234–5244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnaar, R.L.; Gerardy-Schahn, R.; Hildebrandt, H. Sialic acids in the brain: Gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 2014, 94, 461–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanato, Y.; Kitajima, K.; Sato, C. Direct binding of polysialic acid to a brain-derived neurotrophic factor depends on the degree of polymerization. Glycobiology 2008, 18, 1044–1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, S.; Hane, M.; Kitajima, K.; Sato, C. Novel regulation of fibroblast growth factor 2 (FGF2)-mediated cell growth by polysialic acid. J. Biol. Chem. 2012, 287, 3710–3722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isomura, R.; Kitajima, K.; Sato, C. Structural and functional impairments of polysialic acid by a mutated polysialyltransferase found in schizophrenia. J. Biol. Chem. 2011, 286, 21535–21545. [Google Scholar] [CrossRef] [Green Version]
- Mori, A.; Hane, M.; Niimi, Y.; Kitajima, K.; Sato, C. Different properties of polysialic acids synthesized by the polysialyltransferases ST8SIA2 and ST8SIA4. Glycobiology 2017, 27, 834–846. [Google Scholar] [CrossRef] [Green Version]
- Barbeau, D.; Liang, J.; Robitalille, Y.; Quirion, R.; Srivastava, L. Decreased expression of the embryonic form of the neural cell adhesion molecule in schizophrenic brains. Proc. Natl. Acad. Sci. USA 1995, 92, 2785–2789. [Google Scholar] [CrossRef] [Green Version]
- Varea, E.; Guirado, R.; Gilabert-Juan, J.; Martí, U.; Castillo-Gomez, E.; Blasco-Ibáñez, J.M.; Crespo, C.; Nacher, J. Expression of PSA-NCAM and synaptic proteins in the amygdala of psychiatric disorder patients. J. Psychiatr. Res. 2012, 46, 189–197. [Google Scholar] [CrossRef]
- Sato, C.; Inoue, S.; Matsuda, T.; Kitajima, K. Development of a highly sensitive chemical method for detecting alpha2-->8-linked oligo/polysialic acid residues in glycoproteins blotted on the membrane. Anal. Biochem. 1998, 261, 191–197. [Google Scholar] [CrossRef]
- Sato, C.; Inoue, S.; Matsuda, T.; Kitajima, K. Fluorescent-assisted detection of oligosialyl units in glycoconjugates. Anal. Biochem. 1999, 266, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Sato, C.; Fukuoka, H.; Ohta, K.; Matsuda, T.; Koshino, R.; Kobayashi, K.; Troy, F.A.; Kitajima, K. Frequent occurrence of pre-existing alpha 2-->8-linked disialic and oligosialic acids with chain lengths up to 7 Sia residues in mammalian brain glycoproteins. Prevalence revealed by highly sensitive chemical methods and anti-di-, oligo-, and poly-Sia antibodies specific for defined chain lengths. J. Biol. Chem. 2000, 275, 15422–15431. [Google Scholar] [PubMed] [Green Version]
- Hane, M.; Matsuoka, S.; Ono, S.; Miyata, S.; Kitajima, K.; Sato, C. Protective effects of polysialic acid on proteolytic cleavage of FGF2 and proBDNF/BDNF. Glycobiology 2015, 25, 1112–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hane, M.; Sumida, M.; Kitajima, K.; Sato, C. Structural and functional impairments of polysialic acid (polySia)-neural cell adhesion molecule (NCAM) synthesized by a mutated polysialyltransferase of a schizophrenic patient. Pure Appl. Chem. 2012, 84, 1895–1906. [Google Scholar] [CrossRef]
- Arai, M.; Yamada, K.; Toyota, T.; Obata, N.; Haga, S.; Yoshida, Y.; Nakamura, K.; Minabe, Y.; Ujike, H.; Sora, I.; et al. Association between polymorphisms in the promoter region of the sialyltransferase 8B (SIAT8B) gene and schizophrenia. Biol. Psychiatry 2006, 59, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Abe, C.; Yi, Y.; Hane, M.; Kitajima, K.; Sato, C. Acute stress-induced change in polysialic acid levels mediated by sialidase in mouse brain. Sci. Rep. 2019, 9, 9950. [Google Scholar] [CrossRef]
- Abe, C.; Nishimura, S.; Mori, A.; Niimi, Y.; Yang, Y.; Hane, M.; Kitajima, K.; Sato, C. Chlorpromazine Increases the Expression of Polysialic Acid (PolySia) in Human Neuroblastoma Cells and Mouse Prefrontal Cortex. Int. J. Mol. Sci. 2017, 18, 1123. [Google Scholar] [CrossRef] [Green Version]
- Sato, C.; Kitajima, K. Impact of structural aberrancy of polysialic acid and its synthetic enzyme ST8SIA2 in schizophrenia. Front. Cell. Neurosci. 2013, 7, 61. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, S.; Hane, M.; Niimi, Y.; Shinji, M.; Ken, K.; Chihiro, S. Comparison of analytical methods to detect polysialic acid. J. Glycomics Lipidomics 2014, 4, 113. [Google Scholar]
- Sato, C.; Kitajima, K.; Inoue, S.; Seki, T.; Troy, F.A.; Inoue, Y. Characterization of the antigenic specificity of four different anti-(alpha 2-->8-linked polysialic acid) antibodies using lipid-conjugated oligo/polysialic acids. J. Biol. Chem. 1995, 270, 18923–18928. [Google Scholar] [CrossRef] [Green Version]
- Nagae, M.; Ikeda, A.; Hane, M.; Hanashima, S.; Kitajima, K.; Sato, C.; Yamaguchi, Y. Crystal Structure of Anti-polysialic Acid Antibody Single Chain Fv Fragment Complexed with Octasialic Acid INSIGHT INTO THE BINDING PREFERENCE FOR POLYSIALIC ACID. J. Biol. Chem. 2013, 288, 33784–33796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudo, M.; Kitajima, K.; Inoue, S.; Shiokawa, K.; Morris, H.; Dell, A.; Inoue, Y. Characterization of the major core structures of the alpha2-->8-linked polysialic acid-containing glycan chains present in neural cell adhesion molecule in embryonic chick brains. J. Biol. Chem. 1996, 271, 32667–32677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, B.; von der Ohe, M.; Schulze, C.; Bian, S.; Makhina, T.; Loers, G.; Kleene, R.; Schachner, M. Functional role of the interaction between polysialic acid and extracellular histone H1. J. Neurosci. 2010, 30, 12400–12413. [Google Scholar] [CrossRef] [PubMed]
- Theis, T.; Mishra, B.; von der Ohe, M.; Loers, G.; Prondzynski, M.; Pless, O.; Blackshear, P.J.; Schachner, M.; Kleene, R. Functional role of the interaction between polysialic acid and myristoylated alanine-rich C kinase substrate at the plasma membrane. J. Biol. Chem. 2013, 288, 6726–6742. [Google Scholar] [CrossRef] [Green Version]
- McAuley, E.Z.; Scimone, A.; Tiwari, Y.; Agahi, G.; Mowry, B.J.; Holliday, E.G.; Donald, J.A.; Weickert, C.S.; Mitchell, P.B.; Schofield, P.R.; et al. Identification of sialyltransferase 8B as a generalized susceptibility gene for psychotic and mood disorders on chromosome 15q25-26. PLoS ONE 2012, 7, e38172. [Google Scholar] [CrossRef]
- Hane, M.; Kitajima, K.; Sato, C. Effects of intronic single nucleotide polymorphisms (iSNPs) of a polysialyltransferase, ST8SIA2 gene found in psychiatric disorders on its gene products. Biochem. Biophys. Res. Commun. 2016, 478, 1123–1129. [Google Scholar] [CrossRef]
- Hallenbeck, P.; Vimr, E.; Yu, F.; Bassler, B.; Troy, F. Purification and properties of a bacteriophage-induced endo-N-acetylneuraminidase specific for poly-alpha-2,8-sialosyl carbohydrate units. J. Biol. Chem. 1987, 262, 3553–3561. [Google Scholar]
- Gattenlöhner, S.; Stühmer, T.; Leich, E.; Reinhard, M.; Etschmann, B.; Völker, H.U.; Rosenwald, A.; Serfling, E.; Bargou, R.C.; Ertl, G.; et al. Specific detection of CD56 (NCAM) isoforms for the identification of aggressive malignant neoplasms with progressive development. Am. J. Pathol. 2009, 174, 1160–1171. [Google Scholar] [CrossRef] [Green Version]
| Name of Methods | Abbreviation Name | Quality | Quantity | Figure | References |
|---|---|---|---|---|---|
| SDS–PAGE/Western blotting | d-Western | + * (MW of denaturated polySia–NCAM) | + | Figure 2 | [1,19], Many, This paper |
| Native PAGE/Western-blotting | n-Western | +++ (Intact MW charge, conformation) | + | Figure 3 | This paper |
| SDS-PAGE/native PAGE map | S/N map | +++ | − | Figure 4 | This paper |
| ELISA | ELISA | + * | + | Figure 5 | [1,19,39], This paper |
| MH–FAEC | Oligo-analysis | ++ (minimum DP of the sample) | + | Figure 6 | [1,27,31,34,39], This paper |
| Fluorometric C7/C9 analysis | C7/C9 analysis | ++ (Estimated DP, average chain length) | ++ (amounts of α2,8-linked Sia) | Figure 7 | [1,26,30,32,36], This paper |
| Anion-exhange chromatography | NNC-analysis | +++ (Net negative charge of intact polySia–NCAM) | + | Figure 8 | [26,27], This paper |
| Gel filtration | Size-analysis | +++ (observed MW of intact polySia–NCAM) | + | Figure 9 | This paper |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mori, A.; Yang, Y.; Takahashi, Y.; Hane, M.; Kitajima, K.; Sato, C. Combinational Analyses with Multiple Methods Reveal the Existence of Several Forms of Polysialylated Neural Cell Adhesion Molecule in Mouse Developing Brains. Int. J. Mol. Sci. 2020, 21, 5892. https://doi.org/10.3390/ijms21165892
Mori A, Yang Y, Takahashi Y, Hane M, Kitajima K, Sato C. Combinational Analyses with Multiple Methods Reveal the Existence of Several Forms of Polysialylated Neural Cell Adhesion Molecule in Mouse Developing Brains. International Journal of Molecular Sciences. 2020; 21(16):5892. https://doi.org/10.3390/ijms21165892
Chicago/Turabian StyleMori, Airi, Yi Yang, Yuka Takahashi, Masaya Hane, Ken Kitajima, and Chihiro Sato. 2020. "Combinational Analyses with Multiple Methods Reveal the Existence of Several Forms of Polysialylated Neural Cell Adhesion Molecule in Mouse Developing Brains" International Journal of Molecular Sciences 21, no. 16: 5892. https://doi.org/10.3390/ijms21165892






