Characteristics of a Series of Three Bacteriophages Infecting Salmonella enterica Strains
Abstract
1. Introduction
2. Results
2.1. Morphology of Phage Virions and Plaque Morphology
2.2. Lysogenization Ability Assessment
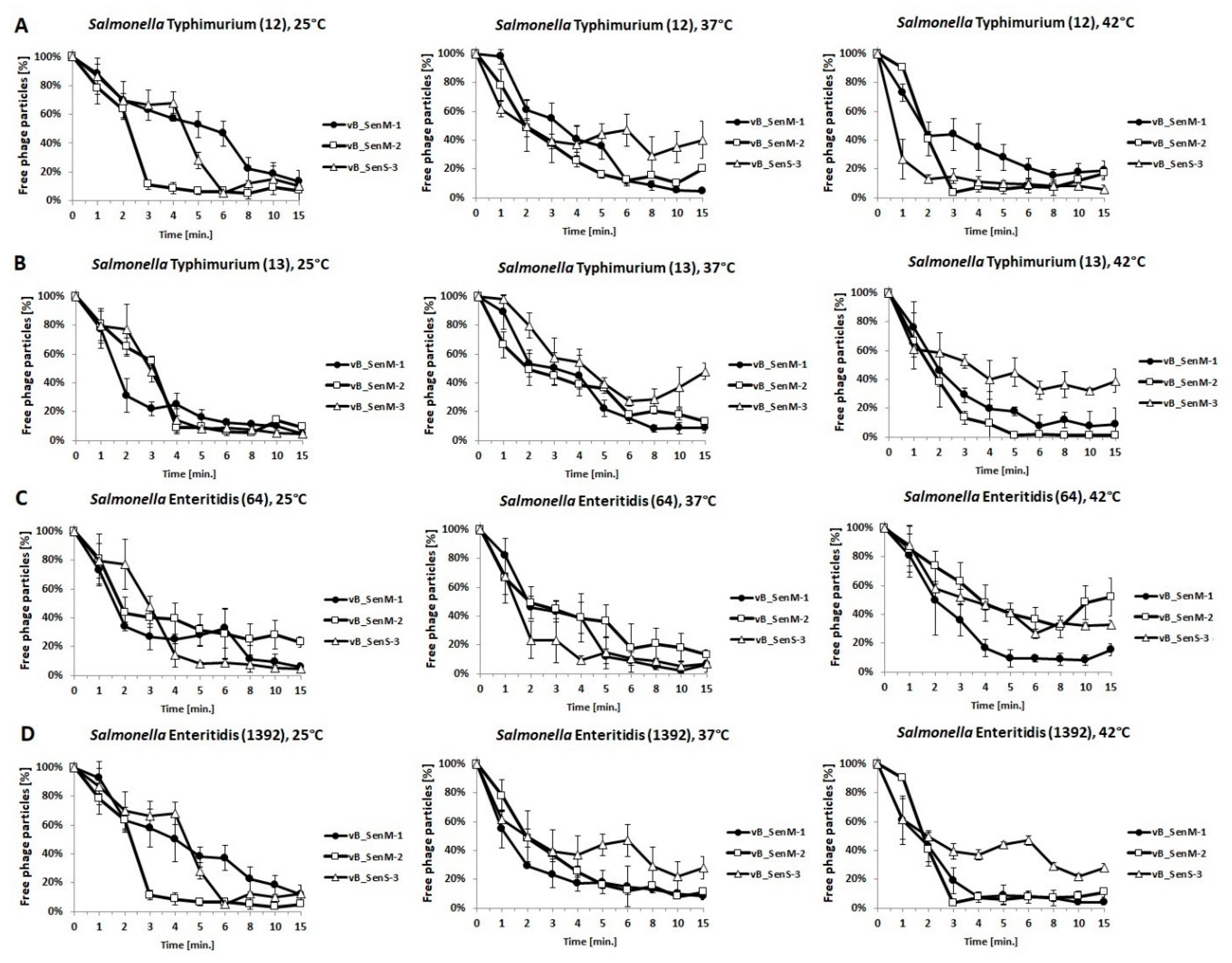
2.3. Host Range and Adsorption on Host Cells
2.4. One-Step Growth Experiments
2.5. Lysis Profile
2.6. pH Sensitivity
2.7. Effects of Phages on Bacterial Biofilm
2.8. Analysis of Phage Genomes
2.9. Phylogenetic Analyses of the Phages
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Bacterial Culture Conditions
4.3. Preparation of Phage Lysates
4.4. Electron Microscopy
4.5. Phage Host Range
4.6. Influence of Low pH on Phage Viability
4.7. Phage Adsorption to Bacterial Host Cells
4.8. One-Step Growth Experiment
4.9. Lysis Profile of Host Bacteria after Phage Infection
4.10. Efficiency of Lysogenization
4.11. Effects of Phages on Bacterial Cells in Biofilm
4.12. Assessment of Biofilm Biomass by Crystal Violet Staining after Phage Infection
4.13. Isolation and Sequencing of Phages vB_SenM-1 and vB_SenS-3 Genomes
4.14. Annotation and Bioinformatic Analysis of Phages vB_SenM-1 and vB_SenS-3
4.15. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Okocha, R.C.; Olatoye, I.O.; Adedeji, O.B. Food safety impacts of antimicrobial use and their residues in aquaculture. Public Health Rev. 2018, 39, 21. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, B.; Greko, C. Antibiotic resistance—Consequences for animal health, welfare, and food production. Upsala J. Med Sci. 2014, 119, 96–102. [Google Scholar] [CrossRef]
- Ventola, C.L. The Antibiotic Resistance Crisis part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Górski, A.; Międzybrodzki, R.; Jończyk-Matysiak, E.; Borysowski, J.; Letkiewicz, S.; Weber-Dąbrowska, B. The fall and rise of phage therapy in modern medicine. Expert Opin. Biol. Ther. 2019, 19, 1115–1117. [Google Scholar] [CrossRef]
- Górski, A.; Międzybrodzki, R.; Węgrzyn, G.; Jończyk-Matysiak, E.; Borysowski, J.; Weber-Dąbrowska, B. Phage therapy: Current status and perspectives. Med. Res. Rev. 2019, 40, 459–463. [Google Scholar] [CrossRef]
- Rohde, C.; Resch, G.; Pirnay, J.-P.; Blasdel, B.G.; Debarbieux, L.; Gelman, D.; Górski, A.; Hazan, R.; Huys, I.; Kakabadze, E.; et al. Expert Opinion on Three Phage Therapy Related Topics: Bacterial phage resistance, phage training and prophages in bacterial production strains. Viruses 2018, 10, 178. [Google Scholar] [CrossRef] [PubMed]
- Torres-Acosta, M.; Clavijo, V.; Vaglio, C.; González-Barrios, A.F.; Vives-Flórez, M.J.; Rito-Palomares, M. Economic evaluation of the development of a phage therapy product for the control of Salmonella in poultry. Biotechnol. Prog. 2019, 35, e2852. [Google Scholar] [CrossRef]
- Clavijo, V.; Flórez, M.J.V. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: A review. Poult. Sci. 2018, 97, 1006–1021. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Boyle, E.C.; Grassl, G.A. Same species, different diseases: How and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 2014, 5, 319. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Liang, L.; Yin, P.; Zhou, Y.; Sharoba, A.M.; Lu, Q.; Dong, X.X.; Liu, K.; Connerton, I.; Li, J. Application of a Novel Phage LPSEYT for Biological Control of Salmonella in Foods. Microorganisms 2020, 8, 400. [Google Scholar] [CrossRef] [PubMed]
- Kakabadze, E.; Makalatia, K.; Grdzelishvili, N.; Bakuradze, N.; Goderdzishvili, M.; Kusradze, I.; Phoba, M.-F.; Lunguya, O.; Lood, C.; Lavigne, R.; et al. Selection of Potential Therapeutic Bacteriophages that Lyse a CTX-M-15 Extended Spectrum β-Lactamase Producing Salmonella enterica Serovar Typhi Strain from the Democratic Republic of the Congo. Viruses 2018, 10, 172. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Virk, S.M.; Shi, J.; Zhou, Y.; Willias, S.P.; Morsy, M.K.; Abdelnabby, H.E.; Liu, J.; Wang, X.; Li, J. Isolation, Characterization, and Application of Bacteriophage LPSE1 Against Salmonella enterica in Ready to Eat (RTE) Foods. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Sritha, K.S.; Bhat, S.G. Genomics of Salmonella phage ΦStp1: Candidate bacteriophage for biocontrol. Virus Genes 2018, 54, 311–318. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, E.; Song, J.; Tong, Y.; Wu, B. Three Salmonella enterica serovar Enteritidis bacteriophages from the Siphoviridae family are promising candidates for phage therapy. Can. J. Microbiol. 2018, 64, 865–875. [Google Scholar] [CrossRef]
- Patil, K.; Zeng, C.; O’Leary, C.; Lessor, L.; Kongari, R.; Gill, J.J.; Liu, M. Complete Genome Sequence of Salmonella enterica Serovar Typhimurium Siphophage Seabear. Microbiol. Resour. Announc. 2019, 8, e01160-19. [Google Scholar] [CrossRef] [PubMed]
- Holguín, A.V.; Cárdenas, P.; Prada-Peñaranda, C.; Leite, L.R.; Buitrago, C.; Clavijo, V.; Oliveira, G.C.; Leekitcharoenphon, P.; Aarestrup, F.M.; Vives, M. Host Resistance, Genomics and Population Dynamics in a Salmonella Enteritidis and Phage System. Viruses 2019, 11, 188. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Shahin, K.; Zhang, Q.; Zhang, H.; Wang, Z.; Zhou, Y.; Zhang, X.; Zhu, S.; Stefan, S.; Wang, R. Morphologic and genomic characterization of a broad host range Salmonella enterica serovar Pullorum lytic phage vB_SPuM_SP116. Microb. Pathog. 2019, 136, 103659. [Google Scholar] [CrossRef]
- Mohamed, A.; Taha, O.; El-Sherif, H.; Connerton, I.; Hooton, S.P.; Bassim, N.; Connerton, I.; El-Shibiny, A. Bacteriophage ZCSE2 is a Potent Antimicrobial against Salmonella enterica Serovars: Ultrastructure, Genomics and Efficacy. Viruses 2020, 12, 424. [Google Scholar] [CrossRef]
- Sevilla-Navarro, S.; Marín, C.; Cortés, V.; García, C.; Vega, S.; Catalá-Gregori, P. Autophage as a control measure for Salmonella in laying hens. Poult. Sci. 2018, 97, 4367–4373. [Google Scholar] [CrossRef] [PubMed]
- Tie, K.; Yuan, Y.; Yan, S.; Yu, X.; Zhang, Q.; Xu, H.; Zhang, Y.; Gu, J.; Sun, C.; Lei, L.; et al. Isolation and identification of Salmonella pullorum bacteriophage YSP2 and its use as a therapy for chicken diarrhea. Virus Genes 2018, 54, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Clavijo, V.; Baquero, D.; Hernandez, S.; Farfan, J.; Arias, J.; Arévalo, A.; Donado-Godoy, P.; Vives, M. Phage cocktail SalmoFREE® reduces Salmonella on a commercial broiler farm. Poult. Sci. 2019, 98, 5054–5063. [Google Scholar] [CrossRef] [PubMed]
- Vaz, C.S.L.; Voss-Rech, D.; Alves, L.; Coldebella, A.; Brentano, L.; Trevisol, I.M. Effect of time of therapy with wild-type lytic bacteriophages on the reduction of Salmonella Enteritidis in broiler chickens. Vet. Microbiol. 2020, 240, 108527. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lin, H.; Jing, Y.; Wang, J. Broad-host-range Salmonella bacteriophage STP4-a and its potential application evaluation in poultry industry. Poult. Sci. 2020, 99, 3643–3654. [Google Scholar] [CrossRef]
- Żbikowska, K.; Michalczuk, M.; Dolka, B. The Use of Bacteriophages in the Poultry Industry. Animals 2020, 10, 872. [Google Scholar] [CrossRef]
- Nikkhahi, F.; Dallal, M.M.S.; Alimohammadi, M.; Foroushani, A.R.; Rajabi, Z.; Fardsanei, F.; Imeni, S.M.; Bonab, P.T. Phage therapy: Assessment of the efficacy of a bacteriophage isolated in the treatment of salmonellosis induced by Salmonella enteritidis in mice. Gastroenterol. Hepatol. Bed Bench 2017, 10, 131–136. [Google Scholar]
- Tang, F.; Zhang, P.; Zhang, Q.; Xue, F.; Ren, J.; Sun, J.; Qu, Z.; Zhuge, X.; Li, D.; Wang, J.; et al. Isolation and characterization of a broad-spectrum phage of multiple drug resistant Salmonella and its therapeutic utility in mice. Microb. Pathog. 2019, 126, 193–198. [Google Scholar] [CrossRef]
- Dallal, M.M.S.; Nikkhahi, F.; Alimohammadi, M.; Douraghi, M.; Rajabi, Z.; Foroushani, A.R.; Azimi, A.; Fardsanei, F. Phage Therapy as an Approach to Control Salmonella enterica serotype Enteritidis Infection in Mice. Rev. Soc. Bras. Med. Trop. 2019, 52, 20190290. [Google Scholar] [CrossRef]
- Bao, H.; Zhou, Y.; Shahin, K.; Zhang, H.; Cao, F.; Pang, M.; Zhu, S.; Olaniran, A.; Schmidt, S.; Wang, R.; et al. The complete genome of lytic Salmonella phage vB_SenM-PA13076 and therapeutic potency in the treatment of lethal Salmonella Enteritidis infections in mice. Microbiol. Res. 2020, 237, 126471. [Google Scholar] [CrossRef]
- Seo, B.-J.; Song, E.-T.; Lee, K.; Kim, J.-W.; Jeong, C.-G.; Moon, S.-H.; Son, J.S.; Kang, S.H.; Cho, H.-S.; Jung, B.Y.; et al. Evaluation of the broad-spectrum lytic capability of bacteriophage cocktails against various Salmonella serovars and their effects on weaned pigs infected with Salmonella Typhimurium. J. Vet. Med Sci. 2018, 80, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Bukhari, S.M.A.U.S.; Andleeb, S.; Ali, M.; Raza, S.; Nawaz, M.A.; Hussain, T.; Rahman, S.U.; Shah, S.S.A. Bacteriophages: An overview of the control strategies against multiple bacterial infections in different fields. J. Basic Microbiol. 2018, 59, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Jurczak-Kurek, A.; Gąsior, T.; Nejman-Faleńczyk, B.; Bloch, S.; Dydecka, A.; Topka, G.; Necel, A.; Jakubowska-Deredas, M.; Narajczyk, M.; Richert, M.; et al. Biodiversity of bacteriophages: Morphological and biological properties of a large group of phages isolated from urban sewage. Sci. Rep. 2016, 6, 34338. [Google Scholar] [CrossRef] [PubMed]
- Fong, K.; Tremblay, D.M.; Delaquis, P.; Goodridge, L.D.; Levesque, R.C.; Moineau, S.; Suttle, C.A.; Wang, S. Wang Diversity and Host Specificity Revealed by Biological Characterization and Whole Genome Sequencing of Bacteriophages Infecting Salmonella enterica. Viruses 2019, 11, 854. [Google Scholar] [CrossRef]
- Liu, A.; Liu, Y.; Peng, L.; Cai, X.; Shen, L.; Duan, M.; Ning, Y.; Liu, S.; Li, C.; Liu, Y.; et al. Characterization of the narrow-spectrum bacteriophage LSE7621 towards Salmonella Enteritidis and its biocontrol potential on lettuce and tofu. LWT 2020, 118, 108791. [Google Scholar] [CrossRef]
- Casjens, S.R.; Jacobs-Sera, D.; Hatfull, G.F.; Hendrix, R.W. Genome Sequence of Salmonella enterica Phage Det7. Genome Announc. 2015, 3, e00279-15. [Google Scholar] [CrossRef]
- Lefkowitz, E.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acid. Res. 2017, 46, D708–D717. [Google Scholar] [CrossRef]
- Pickard, D.; Toribio, A.; Petty, N.K.; Van Tonder, A.J.; Yu, L.; Goulding, D.; Barrell, B.; Rance, R.; Harris, D.; Wetter, M.; et al. A Conserved Acetyl Esterase Domain Targets Diverse Bacteriophages to the Vi Capsular Receptor of Salmonella enterica Serovar Typhi. J. Bacteriol. 2010, 192, 5746–5754. [Google Scholar] [CrossRef]
- Paradiso, R.; Lombardi, S.; Iodice, M.G.; Riccardi, M.G.; Orsini, M.; Censi, S.B.; Galiero, G.; Borriello, G. Complete Genome Sequence of a Lytic Siphoviridae Bacteriophage Infecting Several Serovars of Salmonella enterica. Genome Announc. 2016, 4, e00943-16. [Google Scholar] [CrossRef]
- Paradiso, R.; Lombardi, S.; Iodice, M.G.; Riccardi, M.G.; Orsini, M.; Censi, S.B.; Galiero, G.; Borriello, G. Complete Genome Sequences of Three Siphoviridae Bacteriophages Infecting Salmonella enterica Serovar Enteritidis. Genome Announc. 2016, 4, e00939-16. [Google Scholar] [CrossRef]
- Gencay, Y.E.; Gambino, M.; Prüssing, T.F.; Brøndsted, L. The genera of bacteriophages and their receptors are the major determinants of host range. Environ. Microbiol. 2019, 21, 2095–2111. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Międzybrodzki, R.; Lobocka, M.; Głowacka-Rutkowska, A.; Bednarek, A.; Borysowski, J.; Jończyk-Matysiak, E.; Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Bagińska, N.; et al. Phage Therapy: What Have We Learned. Viruses 2018, 10, 288. [Google Scholar] [CrossRef] [PubMed]
- Romero-Calle, D.; Benevides, R.G.; Góes-Neto, A.; Billington, C. Bacteriophages as Alternatives to Antibiotics in Clinical Care. Antibiotics 2019, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Weber-Dąbrowska, B.; Jończyk-Matysiak, E.; Żaczek, M.; Lobocka, M.; Łusiak-Szelachowska, M.; Górski, A. Bacteriophage Procurement for Therapeutic Purposes. Front. Microbiol. 2016, 7, 1177. [Google Scholar] [CrossRef]
- Imklin, N.; Nasanit, R. Characterization of Salmonella bacteriophages and their potential use in dishwashing materials. J. Appl. Microbiol. 2020, 129, 266–277. [Google Scholar] [CrossRef]
- Wong, C.W.; Delaquis, P.; Goodridge, L.; Lévesque, R.C.; Fong, K.; Wang, S. Inactivation of Salmonella enterica on post-harvest cantaloupe and lettuce by a lytic bacteriophage cocktail. Curr. Res. Food Sci. 2020, 2, 25–32. [Google Scholar] [CrossRef]
- Sambrook, J.F.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory: New York, NY, USA, 2001; Volume 1. [Google Scholar]
- Topka, G.; Bloch, S.; Nejman-Faleńczyk, B.; Gąsior, T.; Jurczak-Kurek, A.; Necel, A.; Dydecka, A.; Richert, M.; Węgrzyn, G.; Węgrzyn, A. Characterization of Bacteriophage vB-EcoS-95, Isolated from Urban Sewage and Revealing Extremely Rapid Lytic Development. Front. Microbiol. 2019, 9, 3326. [Google Scholar] [CrossRef]
- Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Grabski, M.; Topka, G.; Dydecka, A.; Kosznik-Kwaśnicka, K.; Grabowski, Ł.; Jurczak-Kurek, A.; Wolkowicz, T.; et al. Characterization of a bacteriophage, vB_Eco4M-7, that effectively infects many Escherichia coli O157 strains. Sci. Rep. 2020, 10, 3743. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, Z.; Tian, C.; Chen, X.; Hu, L.; Wei, X.; Li, H.; Lin, W.; Jiang, A.; Feng, R.; et al. Characterizing the Biology of Lytic Bacteriophage vB_EaeM_φEap-3 Infecting Multidrug-Resistant Enterobacter aerogenes. Front. Microbiol. 2019, 10, 420. [Google Scholar] [CrossRef]
- Abedon, S.T. Lysis of lysis-inhibited bacteriophage T4-infected cells. J. Bacteriol. 1992, 174, 8073–8080. [Google Scholar] [CrossRef]
- León, M.; Santander, J.; Curtiss, R.; Robeson, J. Natural lysogenization and transduction in Salmonella enterica serovar Choleraesuis by bacteriophage P1. Res. Microbiol. 2013, 164, 1–5. [Google Scholar] [CrossRef]
- Dydecka, A.; Nejman-Faleńczyk, B.; Bloch, S.; Topka, G.; Necel, A.; Donaldson, L.; Węgrzyn, G.; Węgrzyn, A. Roles of orf60a and orf61 in Development of Bacteriophages λ and Φ24B. Viruses 2018, 10, 553. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, G.; Aheto, K.; Shirtliff, M.E.; Tennant, S.M. Poor biofilm-forming ability and long-term survival of invasive Salmonella Typhimurium ST313. Pathog. Dis. 2016, 74, 49. [Google Scholar] [CrossRef] [PubMed]
- Caldeira, J.C.; Peabody, D.S. Stability and assembly in vitro of bacteriophage PP7 virus-like particles. J. Nanobiotechnol. 2007, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acid. Res. 2010, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acid. Res. 2011, 39 (Suppl. 2), W29–W37. [Google Scholar] [CrossRef]
- Stothard, P.; Wishart, D.S. Circular genome visualization and exploration using CGView. Bioinformatics 2004, 21, 537–539. [Google Scholar] [CrossRef]
- Garneau, J.R.; Depardieu, F.; Fortier, L.-C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- McNair, K.; Bailey, B.A.; Edwards, R.A. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 2012, 28, 614–618. [Google Scholar] [CrossRef]
- Naville, M.; Ghuillot-Gaudeffroy, A.; Marchais, A.; Gautheret, D. ARNold: A web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 2011, 8, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Galtier, N.; Gouy, M.; Gautier, C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics 1996, 12, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4; Sinauer Associates: Sunderland, MA, USA, 2003; pp. 160–206. [Google Scholar]










| Bacterial Strain | Sensitivity to Phage c | |||
|---|---|---|---|---|
| Salmonella enterica Serotype a | Source and Identification Number b | vB_SenM-1 | vB_SenM-2 | vB_SenS-3 |
| Heidelberg | DMB UG collection; no. UGSA2 | 100% | 100% | 100% |
| Panama | DMB UG collection; no. UGSA3 | 97.82% ± 3.28% | 93.23% ± 4.21% | 95.12% ± 6.16% |
| Reading | DMB UG collection; no. UGSA4 | <0.01% | <0.01% | 15.21% ± 1.11% |
| London | DMB UG collection; no. UGSA5 | <0.01% | <0.01% | 1.25% ± 0.09% |
| Anatum | NSC, no. 78 | 28.11% ± 3.16% | 95.12% ± 4.11% | <0.01% |
| Typhimurium * | NSC, no. 12 | 67.13% ± 7.28% | 98.20% ± 4.77% | 95.13% ± 2.22% |
| Typhimurium | NSC, no. 13 | 23.28% ± 1.70% | 22.40% ± 3.41% | 98.61% ± 1.17% |
| Stanley * | NSC, no. 15 | 0.80% ± 0.06% | 97.28% ± 2.16% | 0.63% ± 0.11% |
| Heidelberg | NSC, no. 16 | 27.91% ± 8.09% | 98.66% ± 1.06% | 100% ± 1.03% |
| Cholerasuis | NSC, no. 34 | 17.22% ± 0.47% | 97.81% ± 3.22% | 99.82% ± 7.57% |
| Cholerasuis | NSC, no. 39 | 4.80% ± 0.88% | 89.16% ± 7.22% | 93.27% ± 4.21% |
| Cholerasuis var kunznedorf | NSC, no. 37 | 88.46% ± 2.61% | 80.52% ± 7.36% | 93.17% ± 4.16% |
| Cholerasuis | NSC, no. 1439 | <0.01% | <0.01% | 9.11% ± 0.76% |
| Virchow | NSC, no. 41 | <0.01% | <0.01% | <0.01% |
| Newport | NSC, no. 50 | 17.26% ± 2.16% | 10.08% ± 3.25% | <0.01% |
| Newport | NSC, no. 51 | <0.01% | <0.01% | 98.26% ± 3.89% |
| Enteritidis * | NSC, no. 64 | 88.87% ± 6.12% | 95.18% ± 2.16% | 98.26% ± 3.15% |
| Enteritidis * | NSC, no. 1392 | 66.21% ± 4.15% | 81.22% ± 4.33% | 86.36% ± 2.53% |
| Dublin | NSC, no. 65 | 55.21% ± 1.17% | 88.25% ± 3.21% | 97.16% ± 5.21% |
| Gallinarum | NSC, no. 74 | 63.55% ± 2.11% | 91.66% ± 5.11% | 89.21% ± 6.10% |
| Seftenberg | NSC, no. 87 | <0.01% | <0.01% | 94.66% ± 2.22% |
| Infantis * | NSC, no. 155 | 10.71% ± 1.06% | <0.01% | 6.15% ± 0.91% |
| Bovismorbificans * | NSC, no. 300 | <0.01% | <0.01% | 94.27% ± 2.31% |
| Saintpaul | NSC, no. 435 | 63.28% ± 4.95% | 96.21% ± 5.13% | 89.92% ± 2.89% |
| Kentucky * | NSC, no. 1368 | 51.22% ± 7.33% | 49.97% ± 3.47% | 24.25% ± 2.97% |
| Agona * | NSC, no. 1408 | 29.18% ± 3.15% | 98.36% ± 3.67% | 95.22% ± 5.11% |
| Hadar * | NSC, no. 1784 | <0.01% | <0.01% | 7.66% |
| Phage | S. enterica Strain | Yield of Phage Progeny (PFU/Cell) | |||
|---|---|---|---|---|---|
| Serotype | No. | Temperature | |||
| 25 °C | 37 °C | 42 °C | |||
| vB_SenM-1 | Typhimurium | 12 | 275.34 ± 30.27 | 58.31 ± 0.06 | 21.31 ± 5.56 |
| 13 | 375.34 ± 75.85 | 206.20 ± 68.51 | 22.20 ± 3.06 | ||
| Enteritidis | 64 | 13.35 ± 3.05 | 139.40 ± 3.55 | 105.48 ± 20.06 | |
| 1392 | 18.08 ± 6.98 | 499.22 ± 7.16 | 60.78 ± 18.37 | ||
| vB_SenM-2 | Typhimurium | 12 | 109.70 ± 15.98 | 131.24 ± 17.88 | 12.29 ± 2.40 |
| 13 | 12.76 ± 3.01 | 85.07 ± 4.91 | 59.06 ± 7.80 | ||
| Enteritidis | 64 | 23.75 ± 5.34 | 229.79 ± 54.27 | 174.59 ± 10.06 | |
| 1392 | 153.94 ± 34.07 | 283.73 ± 71.35 | 2.41 ± 0.35 | ||
| vB_SenS-3 | Typhimurium | 12 | 50.34 ± 18.98 | 136.21 ± 16.63 | 3.12 ± 0.91 |
| 13 | 1.18 ± 0.21 | 77.15 ± 12.80 | 2.23 ± 0.49 | ||
| Enteritidis | 64 | 28.75 ± 7.35 | 156.45 ± 31.15 | 7.40 ± 1.01 | |
| 1392 | 31.44 ± 19.17 | 117.38 ± 21.38 | 2.91 ± 1.57 | ||
| Phage | Temperature (°C) | Phage Survival (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| pH | ||||||||
| 7.0 | 1.8 | 2.0 | 2.2 | 2.5 | 2.8 | 3.0 | ||
| vB_SenM-1 | 25 °C | 100% ± 0.59% | <0.01% | <0.01% | <0.01% | 3.23% ± 0.03% | 4.83% ± 0.01% | 4.51% ± 0.09% |
| 37 °C | 100% | <0.01% | <0.01% | <0.01% | <0.01% | 2.22% ± 0.01% | 5.71% ± 0.12% | |
| 42 °C | 95.12% ± 2.12% | <0.01% | <0.01% | <0.01% | <0.01% | <0.01% | 3.26% ± 0.01% | |
| vB_SenM-2 | 25 °C | 98.66% ± 1.1% | <0.01% | <0.01% | 3.14% ± 0.02% | 2.23% ± 0.06% | 3.62% ± 0.11% | 12.83% ± 1.13% |
| 37 °C | 100% | <0.01% | <0.01% | 2.13% ± 0.12% | 2.7% ± 0.03% | 2.54% ± 0.06% | 18.27% ± 3.16% | |
| 42 °C | 86.93% ± 6.27% | <0.01% | <0.01% | <0.01% | 1.7% ± 0.01% | 2.13% ± 0.12% | 6.32% ± 0.42% | |
| vB_SenS-3 | 25 °C | 100% ± 2.19% | <0.01% | <0.01% | 1.92% ± 0.24% | 2.43% ± 0.33% | 2.32% ± 0.07% | 7.93% ± 0.04% |
| 37 °C | 100% | <0.01% | <0.01% | <0.01% | 1.71% ± 0.04% | 4.31% ± 0.24% | 6.15% ± 1.13% | |
| 42 °C | 81.26% ± 3.44% | <0.01% | <0.01% | <0.01% | <0.01% | 12.14% ± 3.16% | 13.33% ± 0.72% | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kosznik-Kwaśnicka, K.; Ciemińska, K.; Grabski, M.; Grabowski, Ł.; Górniak, M.; Jurczak-Kurek, A.; Węgrzyn, G.; Węgrzyn, A. Characteristics of a Series of Three Bacteriophages Infecting Salmonella enterica Strains. Int. J. Mol. Sci. 2020, 21, 6152. https://doi.org/10.3390/ijms21176152
Kosznik-Kwaśnicka K, Ciemińska K, Grabski M, Grabowski Ł, Górniak M, Jurczak-Kurek A, Węgrzyn G, Węgrzyn A. Characteristics of a Series of Three Bacteriophages Infecting Salmonella enterica Strains. International Journal of Molecular Sciences. 2020; 21(17):6152. https://doi.org/10.3390/ijms21176152
Chicago/Turabian StyleKosznik-Kwaśnicka, Katarzyna, Karolina Ciemińska, Michał Grabski, Łukasz Grabowski, Marcin Górniak, Agata Jurczak-Kurek, Grzegorz Węgrzyn, and Alicja Węgrzyn. 2020. "Characteristics of a Series of Three Bacteriophages Infecting Salmonella enterica Strains" International Journal of Molecular Sciences 21, no. 17: 6152. https://doi.org/10.3390/ijms21176152
APA StyleKosznik-Kwaśnicka, K., Ciemińska, K., Grabski, M., Grabowski, Ł., Górniak, M., Jurczak-Kurek, A., Węgrzyn, G., & Węgrzyn, A. (2020). Characteristics of a Series of Three Bacteriophages Infecting Salmonella enterica Strains. International Journal of Molecular Sciences, 21(17), 6152. https://doi.org/10.3390/ijms21176152









