The Glycine- and Proline-Rich Protein AtGPRP3 Negatively Regulates Plant Growth in Arabidopsis
Abstract
1. Introduction
2. Results
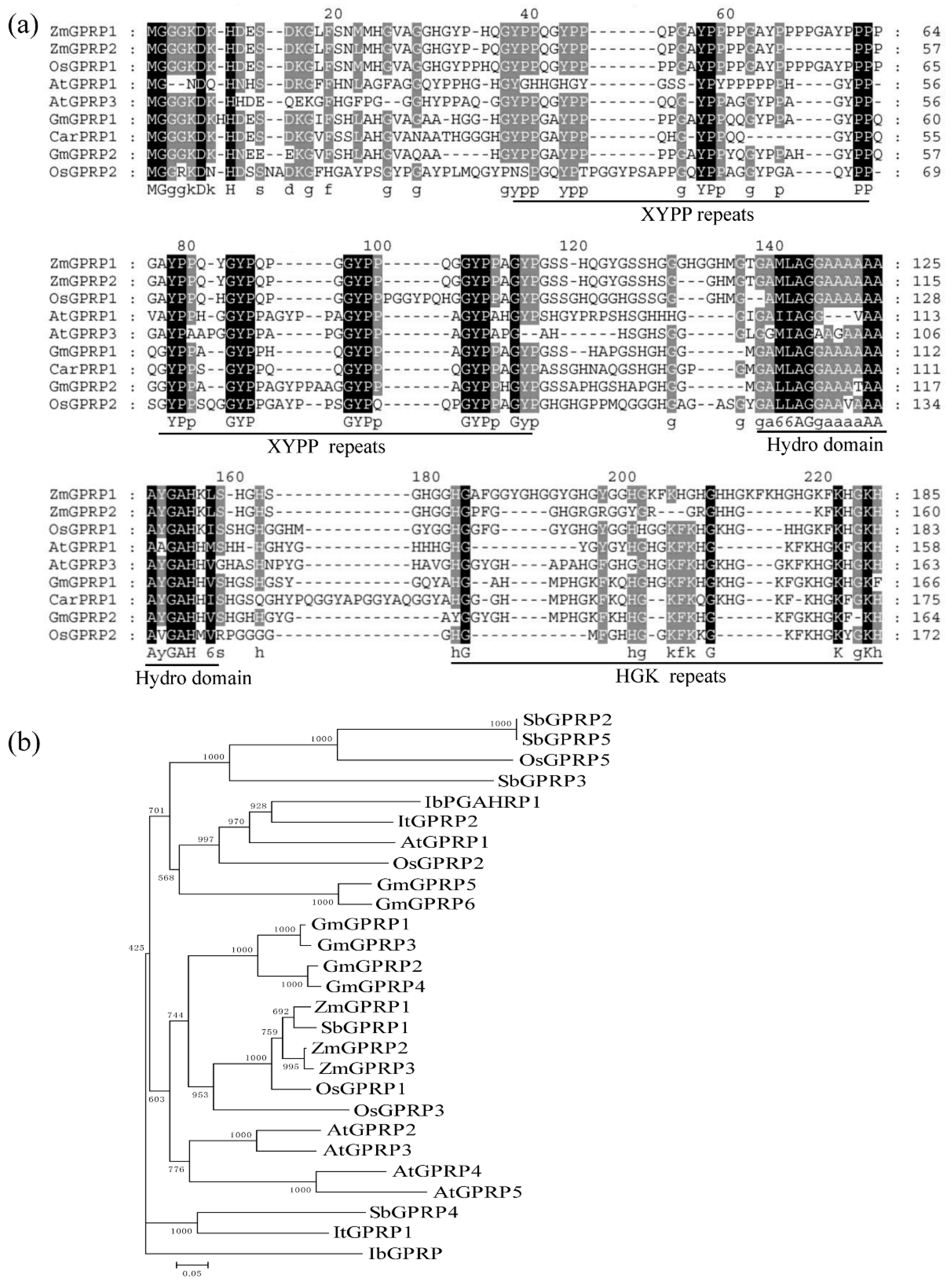
2.1. AtGPRP3 Is a Member of a Small and Conserved Gene Family
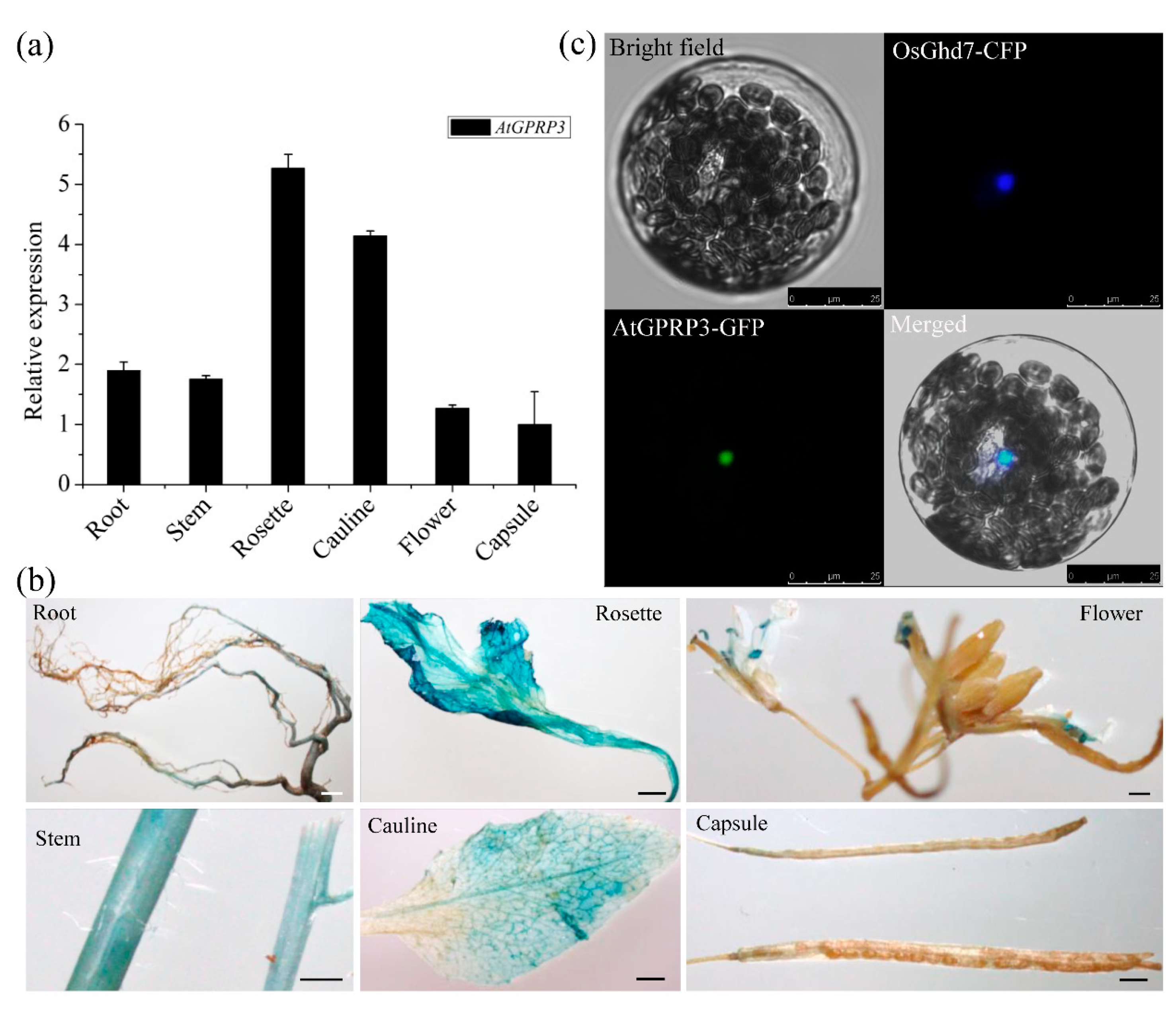
2.2. AtGPRP3 Is Ubiquitously Expressed in Arabidopsis
2.3. Modification of AtGPRP3 Expression Affects Plant Growth
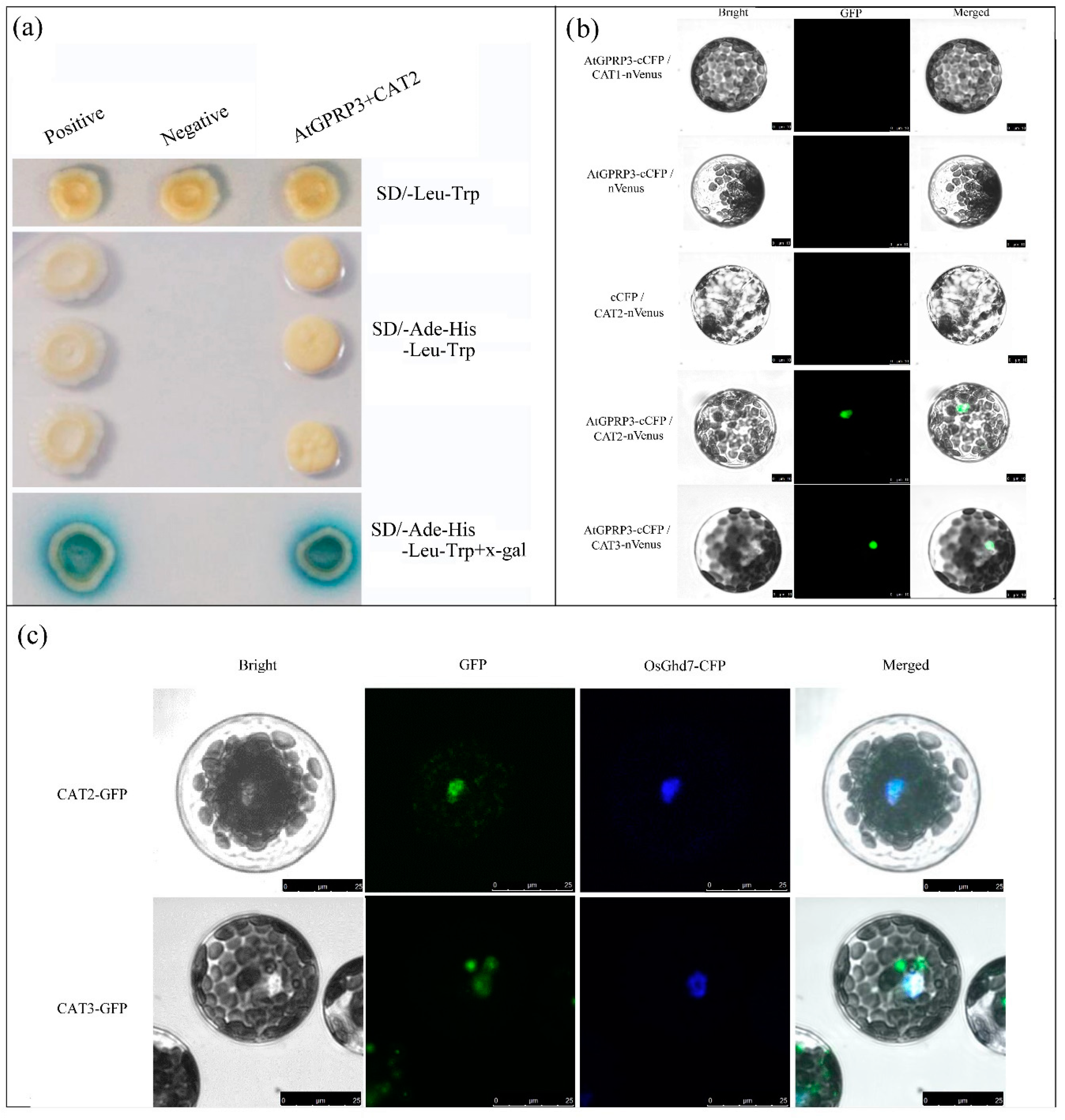
2.4. AtGPRP3 Interacts with CAT2 and 3 But Not CAT1
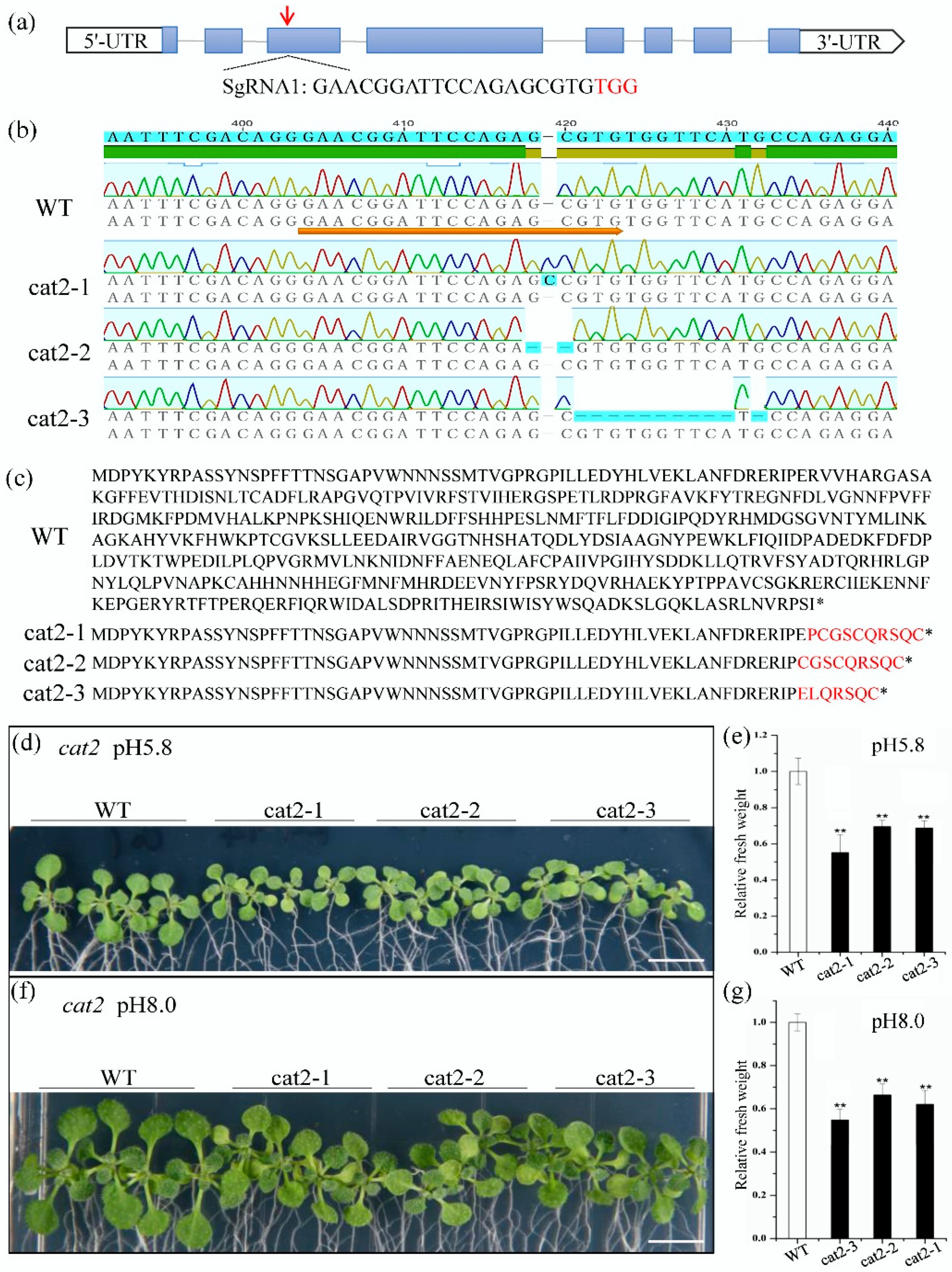
2.5. Knockout of CAT2 Slows the Growth of Arabidopsis Seedlings
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Growth Rate Determination
4.2. Structural and Sequence Analysis of GPRPs
4.3. Plasmid Construction and Plant Transformation
4.4. RNA Isolation and Quantitative RT-PCR Analyses
4.5. ß-Glucuronidase (GUS) Staining
4.6. Subcellular Localization Analysis
4.7. Yeast Two-Hybrid Assay
4.8. BiFC Assay
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Marty, I.; Monfort, A.; Stiefel, V.; Ludevid, D.; Delseny, M.; Puigdomenech, P. Molecular characterization of the gene coding for GPRP, a class of proteins rich in glycine and proline interacting with membranes in Arabidopsis thaliana. Plant Mol. Biol. 1996, 30, 625–636. [Google Scholar] [CrossRef]
- Peng, H.; Feng, Y.; Zhang, H.; Wei, X.; Liang, S. Molecular cloning and characterisation of genes coding for Glycine- and Proline-Rich Proteins (GPRPs) in soybean. Plant Mol. Biol. Rep. 2012, 30, 565–568. [Google Scholar] [CrossRef]
- Maruyama, S.R.; Anatriello, E.; Anderson, J.M.; Ribeiro, J.M.; Brandao, L.G.; Valenzuela, J.G.; Ferreira, B.R.; Garcia, G.R.; Szabo, M.P.; Patel, S.; et al. The expression of genes coding for distinct types of glycine-rich proteins varies according to the biology of three metastriate ticks, Rhipicephalus (Boophilus) microplus, Rhipicephalus sanguineus and Amblyomma cajennense. BMC Genom. 2010, 11, 363. [Google Scholar] [CrossRef]
- Sudhof, T.C.; Lottspeich, F.; Greengard, P.; Mehl, E.; Jahn, R. A synaptic vesicle protein with a novel cytoplasmic domain and four transmembrane regions. Science 1987, 238, 1142–1144. [Google Scholar] [CrossRef]
- Sachetto-Martins, G.; Franco, L.O.; de Oliveira, D.E. Plant glycine-rich proteins: A family or just proteins with a common motif? Biochim. Biophys. Acta Gene Struct. Expr. 2000, 1492, 1–14. [Google Scholar] [CrossRef]
- Blank, M.; Shoenfeld, Y. Histidine-rich glycoprotein modulation of immune/autoimmune, vascular, and coagulation systems. Clin. Rev. Allergy Immunol. 2008, 34, 307–312. [Google Scholar] [CrossRef]
- Al-Salam, A.; Irwin, D.M. Evolution of the vertebrate insulin receptor substrate (Irs) gene family. BMC Evol. Biol. 2017, 17, 148. [Google Scholar] [CrossRef]
- Pommerrenig, B.; Barth, I.; Niedermeier, M.; Kopp, S.; Schmid, J.; Dwyer, R.A.; McNair, R.J.; Klebl, F.; Sauer, N. Common plantain. A collection of expressed sequence tags from vascular tissue and a simple and efficient transformation method. Plant Physiol. 2006, 142, 1427–1441. [Google Scholar] [CrossRef]
- Halder, T.; Upadhyaya, G.; Roy, S.; Biswas, R.; Das, A.; Bagchi, A.; Agarwal, T.; Ray, S. Glycine rich proline rich protein from Sorghum bicolor serves as an antimicrobial protein implicated in plant defense response. Plant Mol. Biol. 2019, 101, 95–112. [Google Scholar] [CrossRef]
- Peng, H. Cloning and Functional Analysis of Seven Chickpea Stress Related Genes (CarNAC1~6 and CarPRP1). Ph.D. Thesis, Nanjing Agricultural University, Nanjing, China, 2010. [Google Scholar]
- Tao, X.; Yong, B.; Shao, H.-H.; Ma, X.-R. Cloning and abiotic stress resistance analyses of a new proline-glycine-alaninehistidine-rich protein gene from Ipomoea batatas (L.) Lam. Turk. J. Biol. 2016, 40, 1148–1157. [Google Scholar] [CrossRef]
- Wit, M.D.; Galvao, V.C.; Fankhauser, C. Light-mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol. 2016, 67, 513–537. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, R.; Xu, D.; Bi, H.; Xia, Z.; Peng, H. Genome-wide identification and analysis of the AP2 transcription factor gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 2019, 10, 1286. [Google Scholar] [CrossRef]
- Weber, K.; Burow, M. Nitrogen—Essential macronutrient and signal controlling flowering time. Physiol. Plant 2018, 162, 251–260. [Google Scholar] [CrossRef]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.; Mylona, P.; Miedema, H.; Torres, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jones, J.D.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.; Wang, G.; Cha, J.Y.; Li, G.; Chen, S.; Li, Z.; Guo, J.; Zhang, C.; Yang, Y.; et al. A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 2015, 27, 908–925. [Google Scholar] [CrossRef]
- Bocca, S.N.; Magioli, C.; Mangeon, A.; Junqueira, R.M.; Cardeal, V.; Margis, R.; Sachetto-Martins, G. Survey of glycine-rich proteins (GRPs) in the Eucalyptus expressed sequence tag database (ForEST). Genet. Mol. Biol. 2005, 28, 608–624. [Google Scholar] [CrossRef]
- Feng, Y.; Peng, H.; Liang, S. Molecular analysis of the PGYRP (proline-, glycine- and tyrosine-rich protein) gene family in soybean. Mol. Biol. Rep. 2011, 38, 2739–2750. [Google Scholar] [CrossRef]
- Smykowski, A.; Zimmermann, P.; Zentgraf, U. G-Box binding factor1 reduces CATALASE2 expression and regulates the onset of leaf senescence in Arabidopsis. Plant Physiol. 2010, 153, 1321–1331. [Google Scholar] [CrossRef]
- Zhang, S.; Li, C.; Ren, H.; Zhao, T.; Li, Q.; Wang, S.; Zhang, Y.; Xiao, F.; Wang, X. BAK1 mediates light intensity to phosphorylate and activate catalases to regulate plant growth and development. Int. J. Mol. Sci. 2020, 21, 1437. [Google Scholar] [CrossRef]
- Li, Y.; Chen, L.; Mu, J.; Zuo, J. LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 2013, 163, 1059–1070. [Google Scholar] [CrossRef]
- Queval, G.; Issakidis-Bourguet, E.; Hoeberichts, F.A.; Vandorpe, M.; Gakiere, B.; Vanacker, H.; Miginiac-Maslow, M.; Van Breusegem, F.; Noctor, G. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 2007, 52, 640–657. [Google Scholar] [PubMed]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. ARF1, a transcription factor that binds to auxin response elements. Science 1997, 276, 1865–1868. [Google Scholar] [CrossRef] [PubMed]
- Waszczak, C.; Carmody, M.; Kangasjarvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed]
- Mhamdi, A.; Hager, J.; Chaouch, S.; Queval, G.; Han, Y.; Taconnat, L.; Saindrenan, P.; Gouia, H.; Issakidis-Bourguet, E.; Renou, J.-P.; et al. Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 2010, 153, 1144–1160. [Google Scholar] [CrossRef] [PubMed]
- Verzili, D.; Zamparelli, C.; Mattei, B.; Noegel, A.A.; Chiancone, E. The sorcin-annexin VII calcium-dependent interaction requires the sorcin N-terminal domain. FEBS Lett. 2000, 471, 197–200. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Yan, L.; Wei, S.; Wu, Y.; Hu, R.; Li, H.; Yang, W.; Xie, Q. High-efficiency genome editing in Arabidopsis Using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 2015, 8, 1820–1823. [Google Scholar] [CrossRef]
- Chen, S.; Liu, Y.; Deng, Y.; Liu, Y.; Dong, M.; Tian, Y.; Sun, H.; Li, Y. Cloning and functional analysis of the VcCXIP4 and VcYSL6 genes as Cd-regulating genes in blueberry. Gene 2019, 686, 104–117. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lung, S.C.; Liao, P.; Yeung, E.C.; Hsiao, A.S.; Xue, Y.; Chye, M.L. Arabidopsis ACYL-COA-BINDING PROTEIN1 interacts with STEROL C4-METHYL OXIDASE1-2 to modulate gene expression of homeodomain-leucine zipper IV transcription factors. New Phytol. 2018, 218, 183–200. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, A.; Yin, H.; Meng, Q.; Yu, X.; Huang, S.; Wang, J.; Ahmad, R.; Liu, B.; Xu, Z.Y. Trithorax-group proteins ARABIDOPSIS TRITHORAX4 (ATX4) and ATX5 function in abscisic acid and dehydration stress responses. New Phytol. 2017, 217, 1582–1597. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Li, X.; Zhong, M.; Yan, J.; Ji, R.; Li, X.; Wang, Q.; Wu, D.; Sun, M.; Tang, D.; et al. A photo-responsive F-box protein FOF2 regulates floral initiation by promoting FLC expression in Arabidopsis. Plant J. 2017, 91, 788–801. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Zhang, D.; Liu, Y.; Ouyang, Y.; Li, J.; Hu, C.; Yao, J. Ectopic expression of EbFIE from apomictic Eulaliopsis binata in rice results in pleiotropic phenotypes likely due to interaction with OsCLF. Plant Sci. 2015, 234, 86–96. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Wang, X.; Yan, X.; Li, S.; Peng, H. The Glycine- and Proline-Rich Protein AtGPRP3 Negatively Regulates Plant Growth in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 6168. https://doi.org/10.3390/ijms21176168
Liu X, Wang X, Yan X, Li S, Peng H. The Glycine- and Proline-Rich Protein AtGPRP3 Negatively Regulates Plant Growth in Arabidopsis. International Journal of Molecular Sciences. 2020; 21(17):6168. https://doi.org/10.3390/ijms21176168
Chicago/Turabian StyleLiu, Xiaojing, Xin Wang, Xin Yan, Shaobo Li, and Hui Peng. 2020. "The Glycine- and Proline-Rich Protein AtGPRP3 Negatively Regulates Plant Growth in Arabidopsis" International Journal of Molecular Sciences 21, no. 17: 6168. https://doi.org/10.3390/ijms21176168
APA StyleLiu, X., Wang, X., Yan, X., Li, S., & Peng, H. (2020). The Glycine- and Proline-Rich Protein AtGPRP3 Negatively Regulates Plant Growth in Arabidopsis. International Journal of Molecular Sciences, 21(17), 6168. https://doi.org/10.3390/ijms21176168




