N-Terminus Does Not Govern Protein Turnover of Schizosaccharomyces pombe CENP-A
Abstract
:1. Introduction
2. Results
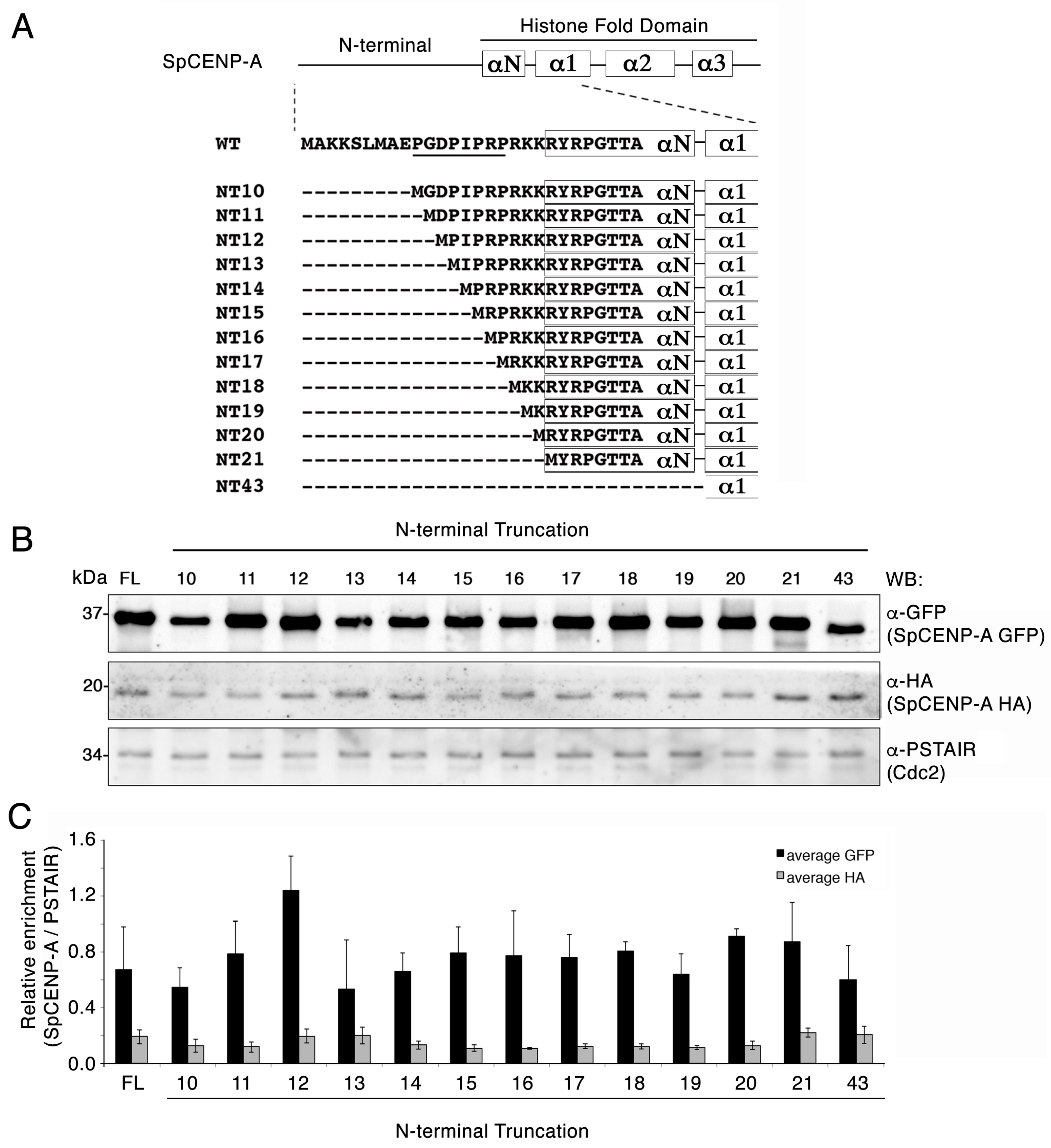
2.1. Creation of SpCENP-A N-Terminal Truncation (NT) Mutants
2.2. Truncation of the NTD Does Not Destabilize the SpCENP-A Protein
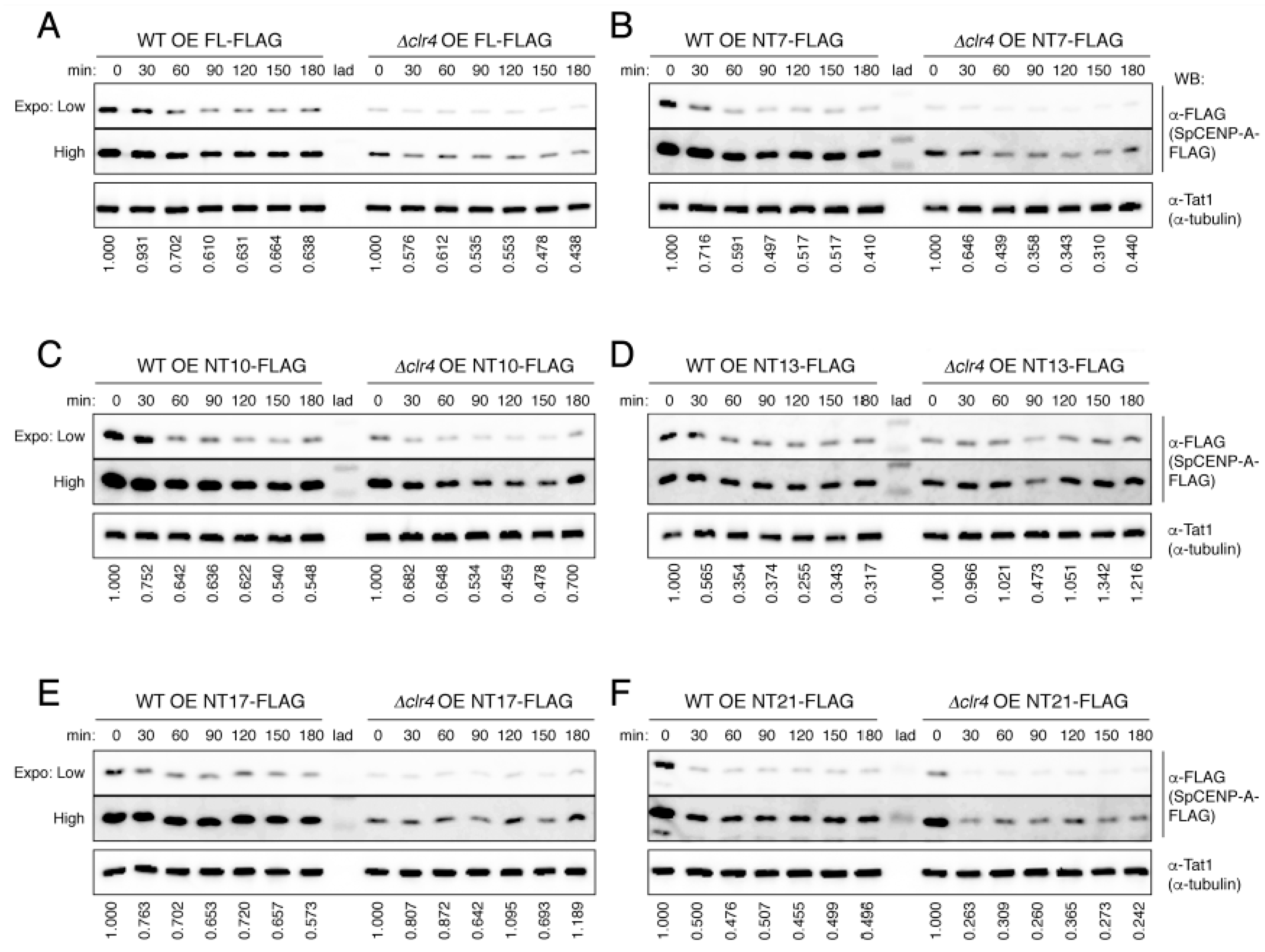
2.3. Heterochromatin Synergistically Acts with NTD to Safeguard the Integrity of SpCENP-A Protein
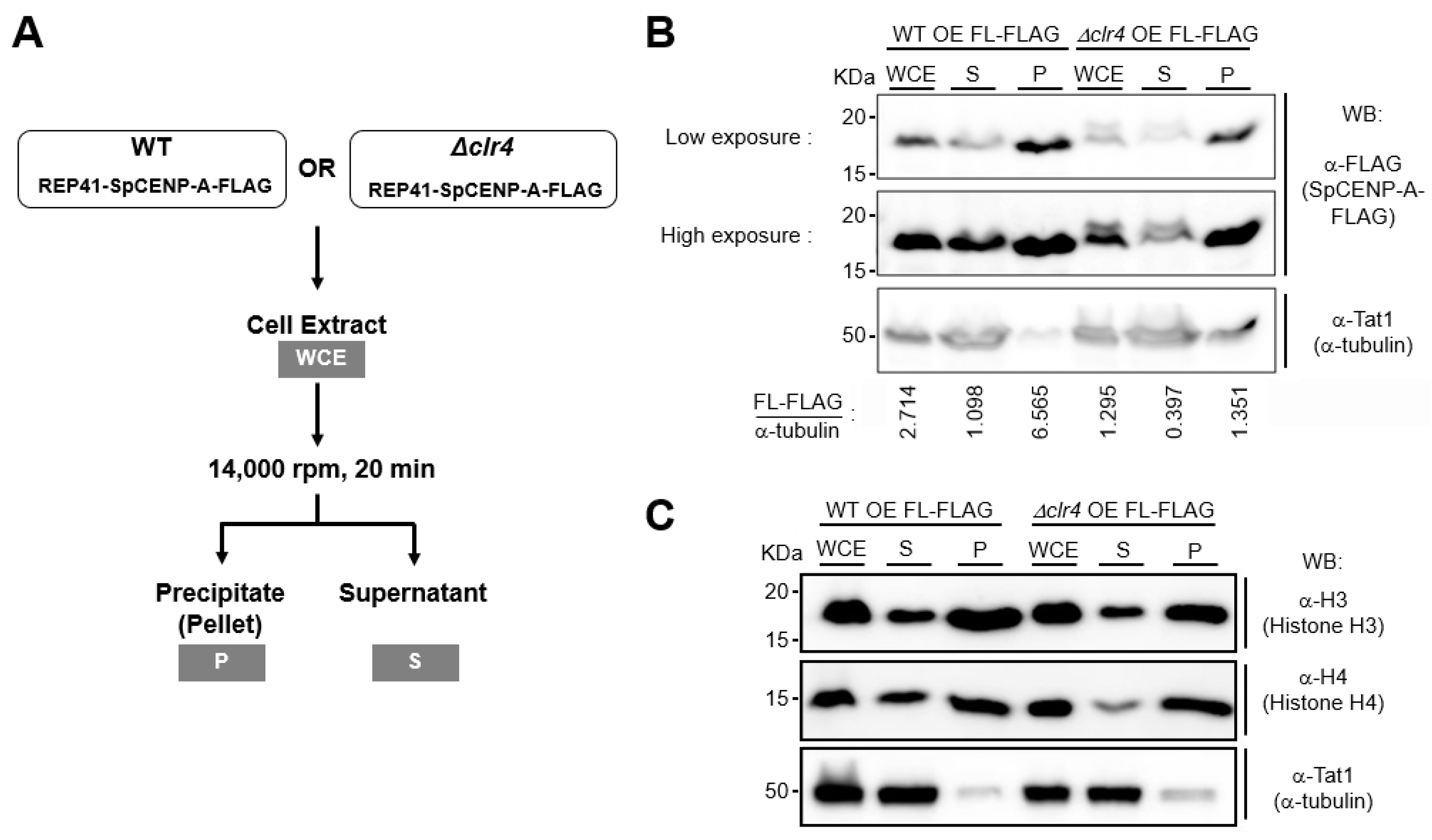
2.4. Effect of Heterochromatin on SpCENP-A in Sub-Cellular Fractions
3. Discussion
4. Materials and Methods
4.1. Schizosaccharomyces pombe Growth Media and Strains
4.2. Contruction of SpCENP-A N-Terminal Truncation Plasmids and Escherichia coli Cell Transformation
4.3. Construction of Exogenously Expressed SpCENP-A NT Mutant Strains
4.4. Cycloheximide Chase Assay
4.5. Sub-Cellular Fractionation
4.6. Statistical Tests for Significance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CCAN | Constitutive centromere-associated network (of proteins) |
| CENP-A | Centromere protein A |
| CHX | Cycloheximide |
| clr4 | Cryptic Loci Regulator 4 (histone lysine H3 methyltransferase Clr4) |
| Cnp1 | CENP-A in Schizosaccharomyces pombe |
| Cse4 | CENP-A in Saccharomyces cerevisiae |
| DMSO | Dimethyl sulfoxide |
| E3 | Ubiquitin ligase E3 |
| EMM (-leu) | Edinburgh minimal media (without leucine) |
| FL | Full-length |
| FLAG | FLAG (tag) |
| GFP | Green fluorescence protein (tag) |
| GRANT | Genomic stability-Regulating site within CENP-A N-Terminus motif |
| H3 | Histone H3 |
| HA | Hemagglutinin (tag) |
| kan | Kanamycin |
| KMN | KNL-1/Mis12 complex/Ndc80 complex |
| LB | Lysogeny broth, Luria–Bertani medium |
| leu | Leucine |
| max | Maximum |
| min | Minimum |
| N-terminal | NH2 (amino) terminal |
| NH2 | Amino |
| NT | NH2-terminal truncation |
| NTD | NH2 (amino)-terminal domain |
| OE | Overexpression |
| PCR | Polymerase chain reaction |
| PEG-LiOAC-TE | Polyethylene glycol 4000-Lithium acetate-Tris-ethylenediaminetetraacetic acid |
| S. pombe | Schizosaccharomyces pombe (fission yeast) |
| S. cerevisiae | Saccharomyces cerevisiae |
| Sim3 | Silencing in the Middle of the centromere 3 (NASP family CENP-A chaperone) |
| SpCENP-A | CENP-A in Schizosaccharomyces pombe |
| Tat1 | α-Tubulin |
| TCA | Trichloroacetic acid |
| TE | Tris-EDTA buffer |
| WB | Western blot |
| WT | Wild-type |
| YEA | Yeast extract adenine |
References
- Ganmore, I.; Smooha, G.; Izraeli, S. Constitutional aneuploidy and cancer predisposition. Hum. Mol. Genet. 2009, 18, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.A.; Nguyen, M.L.; Barrett, A.N.; Tan, Y.Y.; Choolani, M.A.; Chen, E.S. Synthetic combinations of missense polymorphic genetic changes underlying Down syndrome susceptibility. Cell Mol. Life Sci. 2016, 73, 4001–4017. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Desai, A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008, 9, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Santaguida, S.; Musacchio, A. The life and miracles of kinetochores. EMBO J. 2009, 28, 2511–2531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherwin, D.; Wang, Y. The opposing functions of protein kinases and phosphatases in chromosome bipolar attachment. Int. J. Mol. Sci. 2019, 20, 6182. [Google Scholar] [CrossRef] [Green Version]
- Hara, M.; Fukagawa, T. Dynamics of kinetochore structure and its regulations during mitotic progression. Cell Mol. Life Sci. 2020. Epub ahead of print. [Google Scholar] [CrossRef]
- Wimbish, R.T.; DeLuca, J.G. Hec1/Ndc80 tail domain function at the kinetochore-microtubule interface. Front. Cell Div. Biol. 2020, 8, 43. [Google Scholar] [CrossRef] [Green Version]
- Musacchio, A.; Desai, A. A molecular view of kinetochore assembly and function. Biology 2017, 6, 5. [Google Scholar] [CrossRef] [Green Version]
- McGovern, S.L.; Qi, Y.; Pusztai, L.; Symmans, W.F.; Buchholz, T.A. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 2012, 14, R72. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Qian, Y.; Zhao, X.; Wang, S.; Feng, X.; Chen, X.; Zhang, S. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer 2012, 77, 407–414. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Z.; Zhang, S.; Yu, D.; Yu, H.; Liu, L.; Cao, X.; Wang, L.; Gao, H.; Zhu, M. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS ONE 2011, 3, e17794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, X.M.; Fu, J.; Feng, X.J.; Huang, X.; Wang, S.M.; Chen, X.F.; Zhu, M.H.; Zhang, S.H. Expression and prognostic relevance of centromere protein A in primary osteosarcoma. Pathol. Res. Pract. 2014, 210, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Guo, J.; Lv, T.; Jin, H.; Ding, J.; Feng, W.; Zhang, Y.; Hua, K. Prognostic value of centromere protein-A expression in patients with epithelial ovarian cancer. Tumor Biol. 2013, 34, 2971–2975. [Google Scholar] [CrossRef]
- Tomonaga, T.; Matsushita, K.; Yamaguchi, S.; Oohashi, T.; Shimada, H.; Ochiai, T.; Yoda, K.; Nomura, F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003, 63, 3511–3516. [Google Scholar]
- Amato, A.; Schillaci, T.; Lentini, L.; Di Leonardo, A. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol. Cancer 2009, 8, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arimura, Y.; Shirayama, K.; Horikoshi, N.; Fujita, R.; Taguchi, H.; Kagawa, W.; Fukagawa, T.; Almouzni, G.; Kurumizaka, H. Cystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. Sci. Rep. 2014, 4, 7115. [Google Scholar] [CrossRef] [Green Version]
- Tomonaga, T.; Matsushita, K.; Ishibashi, M.; Nezu, M.; Shimada, H.; Ochiai, T.; Yoda, K.; Nomura, F. Centromere protein H is up-regulated in primary human colorectal cancer and its overexpression induces aneuploidy. Cancer Res. 2005, 65, 4685–4689. [Google Scholar] [CrossRef] [Green Version]
- Heun, P.; Erhardt, S.; Blower, M.D.; Weiss, S.; Skora, A.D.; Karpen, G.H. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell 2006, 10, 303–315. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, M.; He, H.; Dong, Q.; Sun, S.; Li, F. Ectopic centromere nucleation by CENP-A in fission yeast. Genetics 2014, 198, 1433–1446. [Google Scholar] [CrossRef]
- Hilderbrand, E.M.; Biggins, S. Regulation of budding yeast CENP-A levels prevents misincorportion at promoter nucleosomes and transcriptional defects. PLoS Genet. 2016, 12, e1005930. [Google Scholar]
- Zhang, W.; Mao, J.H.; Zhu, W.; Jain, A.K.; Liu, K.; Brown, J.B.; Karpen, G.H. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat. Commun. 2016, 7, 12619. [Google Scholar] [CrossRef] [PubMed]
- Ranjitkar, P.; Press, M.O.; Yi, X.; Baker, R.; MacCoss, M.J.; Biggins, S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell 2010, 40, 455–464. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Moreno, O.; Torras-Llort, M.; Azorin, F. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila to centromere. Nucleic Acids Res. 2006, 34, 6247–6255. [Google Scholar] [CrossRef]
- Mouysset, J.; Gilberto, S.; Meier, M.G.; Lampert, F.; Belwal, M.; Meraldi, P.; Peter, M. CRL4RBBP7 is required for efficient CENP-A deposition at centromeres. J. Cell Sci. 2015, 128, 1732–1745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Au, W.C.; Dawson, A.R.; Rawson, D.W.; Taylor, S.B.; Baker, R.E.; Basrai, M. A novel role of the N terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics 2013, 194, 513–518. [Google Scholar] [CrossRef] [Green Version]
- Ohkuni, K.; Levy-Myers, R.; Warren, J.; Au, W.C.; Takahashi, Y.; Baker, R.E.; Basrai, M.A. N-terminal sumoylation of centromeric histone H3 variant Cse4 regulates its proteolysis to prevent mislocalization to non-centromeric chromatin. G3(Bethesda) 2018, 8, 1215–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohkuni, K.; Abdulle, R.; Kitagawa, K. Degradation of centromeric histone H3 variant Cse4 requires the Fpr3 peptidyl-prolyl cis-trans isomerase. Genetics 2014, 196, 1041–1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, H.L.; Lim, K.K.; Yang, Q.; Fan, J.S.; Sayed, A.M.M.; Low, L.S.; Ren, B.; Lim, T.K.; Lin, Q.; Mok, Y.K.; et al. Prolyl isomerization of the CENP-A N-terminus regulates centromeric integrity in fission yeast. Nucleic Acids Res. 2018, 46, 1167–1179. [Google Scholar] [CrossRef]
- Yang, J.; Sun, S.; Zhang, S.; Gonzalez, M.; Dong, Q.; Chi, Z.; Chen, Y.H.; Li, F. Heterochromatin and RNAi regulate centromeres by protecting CENP-A from ubiquitin-mediated degradation. PLoS Genet. 2018, 14, e1007572. [Google Scholar] [CrossRef]
- Folco, H.D.; Pidoux, A.L.; Urano, T.; Allshire, R.C. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 2008, 319, 94–97. [Google Scholar] [CrossRef] [Green Version]
- Kagansky, A.; Folco, H.D.; Almeida, R.; Pidoux, A.L.; Boukaba, A.; Simmer, F.; Urano, T.; Hamilton, G.L.; Allshire, R.C. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 2009, 324, 1716–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, E.S.; Zhang, K.; Nicolas, E.; Cam, H.P.; Zofall, M.; Grewal, S.I. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 2008, 451, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Kobayashi, R.; Grewal, S.I. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 2005, 7, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- Collins, K.A.; Furuyama, S.; Biggins, S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 2004, 14, 1968–1972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takayama, Y.; Toda, T. Coupling histone homeostasis to centromere integrity via the ubiquitin-proteasome system. Cell Div. 2010, 5, 18. [Google Scholar] [CrossRef] [Green Version]
- Bassett, E.A.; DeNizio, J.; Barnhart-Dailey, M.C.; Panchenko, T.; Sekulic, N.; Rogers, D.J.; Foltz, D.R.; Black, B.E. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev. Cell 2012, 22, 749–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Feng, H.; Zhou, B.R.; Ghirlando, R.; Hu, K.; Zwolak, A.; Miller Jenkins, L.M.; Xiao, H.; Tjandra, N.; Wu, C.; et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 2011, 472, 234–237. [Google Scholar] [CrossRef] [Green Version]
- Le Goff, S.; Keceli, B.N.; Jarabkova, H.; Heckmann, S.; Rutten, T.; Cotterell, S.; Schubert, V.; Roitinger, E.; Mechtler, K.; Franklin, F.C.H.; et al. The H3 histone chaperone NASPSim3 escorts CenH3 in Arabidopsis. Plant J. 2020, 101, 71–86. [Google Scholar] [CrossRef] [Green Version]
- Folco, H.D.; Campbell, C.S.; May, K.M.; Espinoza, C.A.; Oegema, K.; Hardwick, K.G.; Grewal, S.I.S.; Desai, A. The CENP-A N-tail confers epigenetic stability to centromeres via the CENP-T branch of the CCAN in fission yeast. Curr. Biol. 2015, 25, 348–356. [Google Scholar] [CrossRef] [Green Version]
- Fachinetti, D.; Folco, H.D.; Nechemia-Arbely, Y.; Valente, L.P.; Nguyen, K.; Wong, A.J.; Zhu, Q.; Holland, A.J.; Desai, A.; Jensen, L.E.; et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 2013, 15, 1056–1066. [Google Scholar] [CrossRef] [Green Version]
- Smith, K.M.; Phatale, P.A.; Sullivan, C.M.; Pomraning, K.R.; Freitag, M. Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol. Cell Biol. 2011, 31, 2528–2542. [Google Scholar] [CrossRef] [Green Version]
- Castillo, A.G.; Pidoux, A.L.; Catania, S.; Durand-Dubief, M.; Choi, E.S.; Hamilton, G.; Ekwall, K.; Allshire, R.C. Telomeric repeats facilitate CENP-ACnp1 incorporation via telomere binding protein. PLoS ONE 2013, 8, e69673. [Google Scholar] [CrossRef]
- Ogiyama, Y.; Ohno, Y.; Kubota, Y.; Ishii, K. Epigenetically induced paucity of histone H2A stabilizes fission yeast ectopic centromeres. Nat. Struct. Mol. Biol. 2013, 20, 1397–1406. [Google Scholar] [CrossRef]
- Boyarchuk, E.; Filipescu, D.; Vassias, I.; Cantoloube, S.; Almouzni, G. The histone variant composition of centromeres is controlled by the pericentric heterochromatin state during cell cycle. J. Cell Sci. 2014, 127, 3347–3359. [Google Scholar] [CrossRef] [Green Version]
- Swartz, S.Z.; McKay, L.S.; Su, K.C.; Bury, L.; Padeganeh, A.; Maddox, P.S.; Knouse, K.A.; Cheeseman, I.M. Quiescent cells actively replenish CENP-A nucleosomes to maintain centromere identity and proliferative potential. Dev. Cell 2019, 51, 25–48. [Google Scholar] [CrossRef]
- Venkatesh, S.; Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 178–189. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Lim, J.S.; Tang, R.M.; Zhang, L.; Chen, E.S. Fitness profiling links topoisomerase II regulation of centromeric integrity to doxorubicin resistance in fission yeast. Sci. Rep. 2015, 5, 8400. [Google Scholar] [CrossRef] [Green Version]
- Forsburg, S.L.; Rhind, N. Basic methods for fission yeast. Yeast 2006, 23, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Funakoshi, M.; Hochstrasser, M. Small epitope-linker modules for PCR-based C-terminal tagging in Saccharomyces cerevisiae. Yeast 2009, 26, 185–192. [Google Scholar] [CrossRef] [Green Version]
- Bahler, J.; Wu, J.Q.; Longtine, M.S.; Shah, N.G.; McKenzie, A., 3rd; Steever, A.B.; Wach, A.; Philippsen, P.; Pringle, J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 1998, 14, 943–951. [Google Scholar] [CrossRef]
- Lim, K.K.; Ong, T.Y.; Tan, Y.R.; Yang, E.G.; Ren, B.; Seah, K.S.; Yang, Z.; Tan, T.S.; Dymock, B.W.; Chen, E.S. Mutation of histone H3 serine 86 disrupts GATA factor Ams2 expression and precise chromosome segregation in fission yeast. Sci. Rep. 2015, 5, 14064. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, H.L.; Zeng, Y.B.; Chen, E.S. N-Terminus Does Not Govern Protein Turnover of Schizosaccharomyces pombe CENP-A. Int. J. Mol. Sci. 2020, 21, 6175. https://doi.org/10.3390/ijms21176175
Tan HL, Zeng YB, Chen ES. N-Terminus Does Not Govern Protein Turnover of Schizosaccharomyces pombe CENP-A. International Journal of Molecular Sciences. 2020; 21(17):6175. https://doi.org/10.3390/ijms21176175
Chicago/Turabian StyleTan, Hwei Ling, Yi Bing Zeng, and Ee Sin Chen. 2020. "N-Terminus Does Not Govern Protein Turnover of Schizosaccharomyces pombe CENP-A" International Journal of Molecular Sciences 21, no. 17: 6175. https://doi.org/10.3390/ijms21176175




