Abstract
Ethylene plays an important role in stress adaptation and fruit ripening. Acireductone dioxygenase (ARD) is pivotal for ethylene biosynthesis. However, the response of ARD to fruit ripening or cold stress is still unclear. In this study, we identified three members of Malus ARD family, and expression profile analysis revealed that the transcript level of MdARD4 was induced during apple fruit ripening and after apple plants were being treated with cold stress. To investigate its function in cold tolerance and fruit ripening, MdARD4 was ectopically expressed in Solanum lycopersicum cultivar ‘Micro-Tom’, which has been considered as an excellent model plant for the study of fruit ripening. At the cellular level, the MdARD protein expressed throughout Nicotiana benthamiana epidermal cells. Overexpression of MdARD4 in tomato demonstrated that MdARD4 regulates the ethylene and carotenoid signaling pathway, increases ethylene and carotenoid concentrations, and accelerates fruit ripening. Furthermore, MdARD4 increased the antioxidative ability and cold hardiness in tomato. To conclude, MdARD4 may potentially be used in apple breeding to accelerate fruit ripening and increase cold hardiness.
1. Introduction
Ripening of fleshy fruits is a complex process that involves biochemical and metabolic changes. Ethylene, the natural plant hormone, regulates various physiological and developmental events in plants and is important for the ripening of climacteric fruits [1,2]. Ethylene and carotenoid concentrations are relevant to this stud related to fruit ripening. Tomato (Lycopersicon esculentum Mill.) has been considered as an excellent model plant for the study of fruit ripening [3]. Several studies have investigated the molecular basis of fruit ripening. Naeem et al. identified that SmCOP1 (CONSTITUTIVE PHOTOMORPHOGENIC) regulates ethylene biosynthesis and affects fruit ripening in tomato [4]. Studies have also shown that ethylene triggers fruit ripening via the ethylene signaling pathway [5,6,7,8].
Plants are challenged by both biotic and abiotic stresses, including pathogen infection, salt stress, drought, and cold stress that limit their growth and development. These stresses pose significant threats to crop production worldwide [9,10,11]. As plants are sessile, they cannot escape from stress. Therefore, they utilize complex physiological and genetic regulatory networks to react to the continuously changing environmental conditions [9,10,12,13,14,15,16]. Apple (Malus domestica Borkh.) is one of the most important climacteric fruit crops worldwide. Extreme temperature, especially cold stress is one of the adverse environmental conditions that limit apple cultivation, growth, and development [17,18]. Low temperature influences the geographical distribution of apple and negatively impacts the productivity and quality of the fruit.
Ethylene, the fruit ripening hormone, also plays a key role in plant growth and development and regulates the plant response to stress. Different abiotic stress factors such as flooding, drought, high salinity, low temperature, and mechanical damage induce ethylene change in plants [19]. Researchers have investigated the association between enhanced ethylene levels and cold and freezing tolerance [20,21,22,23,24,25,26]. Ethylene is necessary in the normal response to cold stress in plants [27]. Overexpression of tomato ERF (ethylene-responsive factor) gene TERF2/LeERF2 significantly activated stress-related genes and increased ethylene levels under the cold condition and thus enhanced cold tolerance [28]. Ethylene response factors such as VaERF080 and VaERF087 have been validated as cold-responsive factors. Transgenic Arabidopsis overexpression of VaERF080 and VaERF087 showed enhanced cold tolerance via lower malonyldialdehyde (MDA) content and higher superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) activities [29].
Similar findings have been reported in bermudagrass; CdERF1 improved cold tolerance by regulating stress-associated gene expression levels, relieving reactive oxygen species (ROS) damage, and improving the antioxidant system [30]. These earlier findings suggest that cold tolerance of a plant is particularly dependent on ethylene.
Different factors regulate ethylene syntheses. In plants, the Met cycle is required to sustain high rates of ethylene synthesis [31]. Methionine (Met) cycle exists in prokaryotes and eukaryotes. Acireductone dioxygenase (ARD) catalyzes the production of 2-keto-4-methylthiobuty-rate (KMTB) from acireductone and dioxygen and that affects ethylene synthesis [32]. ARD plays an important role in plant growth and development and stress response by participating in the Met cycle. It has been reported that OsARD4 improves root architecture in rice by promoting development of secondary roots [33]. TaARD may be involved in ethylene synthesis and ethylene signaling in response to biotic and abiotic stresses [34]. However, unlike the role of ethylene in fruit ripening and stress resistance, the function of ARDs, locates upstream of the ethylene biosynthetic pathway, in plants is not clear. Therefore, this study investigates the expression of MdARD in apple under the cold condition and during fruit ripening. To better understand the role of ARD4 in regulating ethylene synthesis, fruit ripening, and cold hardiness, we ectopically expressed MdARD4 in Solanum lycopersicum cultivar ‘Micro-Tom’, and then compared the differences of wild-type (WT) and transgenic plants in growth development and low temperature tolerance. These findings will provide valuable insights into ARD4 regulation of cold hardiness and fruit ripening.
2. Results
2.1. Identification of MdARD Genes and Phylogenetic Analysis and Location Determination of MdARD Proteins
To identify the members of the ARD family in apple, previously reported ARD proteins of Arabidopsis were used as TBLASTN query to search the apple genome (version 1.0). Three proteins were identified using SMART and Pfam tools. To evaluate the evolutionary relationship between ARD proteins in apple, Arabidopsis, rice, pear, strawberry, potato, and grape, a phylogenetic tree was constructed based on the full length ARD protein sequences from these species. ARD of apple showed a close evolutionary relationship with Arabidopsis (Figure 1A). The ARD family proteins of apple were named based on their homologues in Arabidopsis. We identified and cloned three Malus ARD genes, namely MdARD1, MdARD2, and MdARD4 with GenBank accession numbers MH481533, MH481534, and MH481535, respectively.
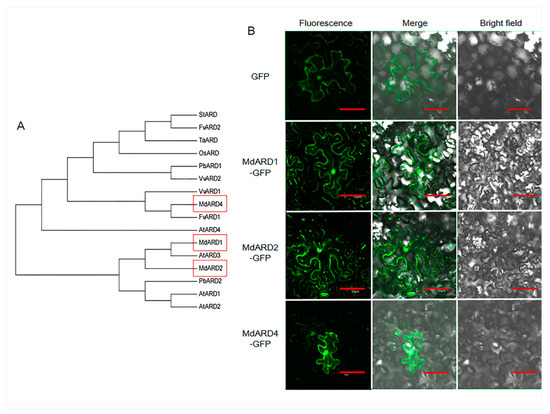
Figure 1.
(A) Phylogenetic analysis among plant acireductone dioxygenase (ARD) proteins and (B) subcellular localization of GFP (green fluorescent protein) and MdARD-GFP. GFP or MdARD-GFP fusion protein were transiently expressed in Nicotiana benthamiana epidermal cells and visualized by confocal microscopy. The fluorescent green signal in the dark field or merged field shows the localization of GFP or MdARD-GFP fusion protein. Bars, 50 μm. Md, Malus domestica; MdARD1 (MH481533), MdARD2 (MH481534), MdARD4 (MH481535). Pb, Pyrus bretschneideri; PbARD1 (XP_009343958), PbARD2 (XP_009375983). Fv, Fragaria vesca; FvARD1 (XP_004296576), FvARD2 (XP_004296575). Vv, Vitis vinifera; VvARD1 (XP_010649963), VvARD2 (XP_010649962). Os, Oryza sativa; OsARD (AAX55895). Ta, Triticum aestivum; TaARD (ACQ99820). St, Solanum tuberosum; StARD (NP_001274845). At, Arabidopsis thaliana; AtARD1 (NP_567443), AtARD2 (NP_001190730), AtARD3 (NP_180208), AtARD4 (NP_568630).
The MdARD proteins were expressed driven by a 35S promoter. The subcellular localization of GFP and MdARD-GFP proteins was examined by confocal microscopy. As shown in Figure 1B, the 35S-driven GFP protein showed expression throughout cell, and MdARD protein after fusion with GFP also showed expression throughout the cell.
2.2. Expression Pattern of MdARD Genes under Cold Condition and during Fruit Ripening in the Apple
To evaluate the role of MdARD genes under cold stress, their expression levels in the leaves of a ‘Golden delicious’ apple under cold stress were determined. MdARD1 and MdARD4 were significantly induced by cold while MdARD2 did not change significantly under cold condition (Figure 2A). To evaluate the role of MdARD genes in fruit ripening, their expression levels in the fruit of the ‘Honey crisp’ apple during fruit ripening were determined. At 90 and 120 days after bloom, MdARD genes were significantly induced; MdARD1 and MdARD4 were increased during the fruit development process (Figure 2B). MdARD4 showed higher expression under cold stress and during fruit ripening. Therefore, we chose MdARD4 for further study.
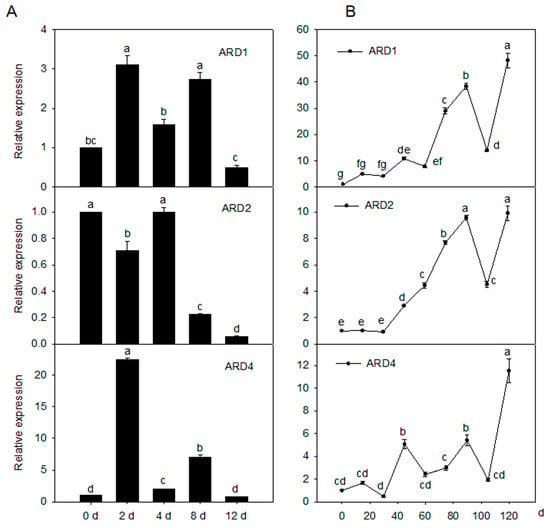
Figure 2.
The expression pattern of MdARDs. (A) Change in the relative transcript level of MdARDs in response to the cold treatment, the expression level of MdARD1, MdARD2, or MdARD4 in response to cold on Day 0 was set as 1 and (B) trends in relative expression of MdARDs at different developmental stages of the apple fruit. Samples from 0 days after bloom (DAB) were set to 1. Expression levels were calculated relative to that of ELONGATION FACTOR1α (EF-1α). Bars represent the mean value ± SE (n ≥ 3). Different letters indicate significant differences of MdARDs expression level on the different day of treatments, according to one-way ANOVA Tukey’s multiple range tests (p < 0.05).
2.3. Overexpression of MdARD4 Promotes Fruit Ripening in Tomato
Ethylene plays a key role in plant growth and development. ARD catalyzes the step of acireductone reacts with dioxygen to produce 2-keto-4-methylthiobuty-rate (KMTB) in the Met cycle and that affects the synthesis of ethylene. What about the function of MdARD4 in the fruit ripening process? We ectopically expressed MdARD4 in Solanum lycopersicum cultivar ‘Micro-Tom’, and found that fruits of MdARD4 transgenic tomato plants ripened faster than those of WT plants grown for the same duration under the same conditions (Figure 3A). Transgenic fruits differed from WT fruits in color. This change in color occurred at breaker stage; transgenic fruits turned yellow while WT remained green (Figure 3B). These results indicated that the ripening period was longer for WT plants than that for the transgenic plants (Figure 3C).
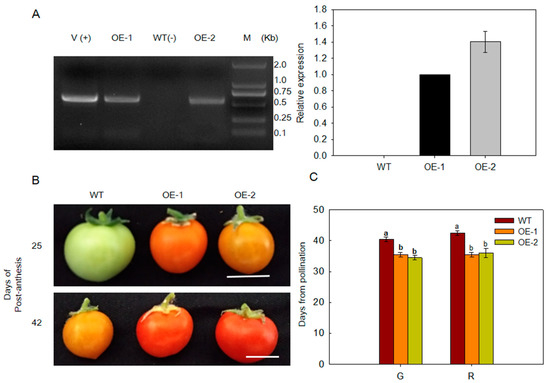
Figure 3.
(A) PCR confirmation for transgenic tomato plants. Left panel: PCR with DNA; lanes: M, molecular marker DL2000; V(+), positive vector containing pCambia2300-MdARD4 plasmid; WT(−), non-transformed wild type; OE-1 and OE-2, MdARD4 transgenic tomato lines; Right panel: Quantitative RT-PCR analysis of MdARD4 expression in leaves of WT and MdARD4 transgenic lines OE-1 and OE-2, the expression level of MdARD4 in line OE-1 was set as 1; (B) phenotype comparisons between WT and MdARD4 transgenic fruits. Bar, 2 cm; and (C) days from pollination to green (G) and red (R) stages. Data are means of three replicates with SD. Different letters indicate significant differences between WT and MdARD4 transgenic plants on the same day of different treatments, according to one-way ANOVA Tukey’s multiple range tests (p < 0.05).
2.4. Overexpression of MdARD4 Improves Ethylene Concentration in Tomato
At the maturation stages corresponding to green and red fruit stages of tomato, ARD4 relative expression level and ethylene release were higher in transgenic tomato fruits than those in WT fruits (Figure 4A,B). The expression levels of ethylene biosynthetic genes (ACS2, ACO3, ACO1, and RIN) and ethylene-responsive genes (E4, E8, and ERF1) were more also higher in MdARD4 transgenic tomato fruits than those in WT fruits (Figure 4C,D).
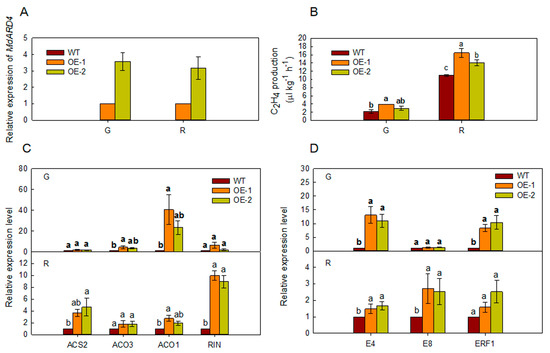
Figure 4.
(A) Relative expression level of ARD4 from WT and MdARD4 transgenic tomato fruits, the expression level of MdARD4 in line OE-1 was set as 1 and (B) ethylene release level of ARD4 from WT and MdARD4 transgenic tomato fruits. Expression of ethylene biosynthesis genes (C) and ethylene-responsive genes (D) in fruits from WT and MdARD4 transgenic tomato plants. Expression levels were calculated relative to that of CAC, the expression level of each gene in line WT was set as 1. Bars represent the mean value ± SE (n ≥ 3). Data are means of three replicates with SD. Different letters indicate significant differences between WT and transgenic plants on the same day of different treatments, according to one-way ANOVA Tukey’s multiple range tests (p < 0.05).
2.5. Overexpression of MdARD4 Improves Carotenoid Concentration in Tomato
The color of transgenic tomato fruits differed from that of WT fruits at the same stage of maturation. Carotenoid concentration was higher in transgenic tomato fruits compared to that in WT fruits (Figure 5A). The expression of carotenoid biosynthetic genes (PSY1, PDS, and ZDS) was higher in the fruits of transgenic tomato (Figure 5B).
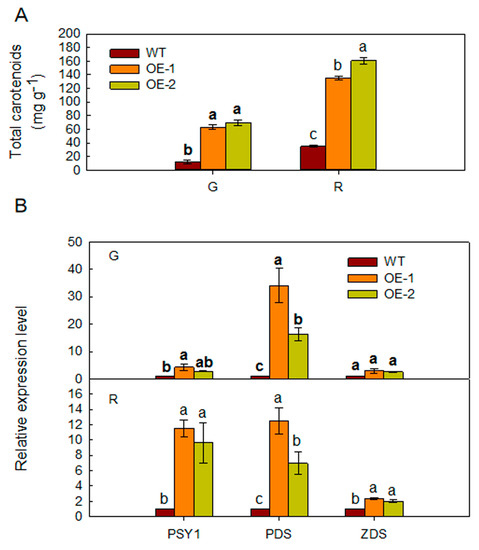
Figure 5.
(A) The carotenoid concentrations in fruits from WT and MdARD4 transgenic tomato plants and (B) expression of carotenoid biosynthesis genes in fruits from WT and MdARD4 transgenic tomato plants, the expression level of each gene in line WT was set as 1. Data are means of three replicates with SD. Different letters indicate significant differences between WT and transgenic plants on the same day of different treatments, according to one-way ANOVA Tukey’s multiple range tests (p < 0.05).
2.6. MdARD4-Overexpressing Tomato Lines Behave Better under the Cold Condition
To determine the role of MdARD4 in plant cold resistance, we further compared the tomato plants grown under normal (25 °C) temperature and low (4 °C) temperature conditions. After 3 days of treatment, transgenic plants performed better than the WT plants (Figure 6A). Electrolyte leakage and malonyldialdehyde (MDA) concentration of leaves were measured to assess membrane damage under stress conditions. Under normal temperature, the relative electrolyte leakage and MDA concentration did not differ between the transgenic lines and the WT. However, they were significantly lower in the transgenic plants compared with the WT under cold condition (Figure 6B). The survival rates of the transgenic plant lines under low temperature conditions were considerably higher than that of the WT plants (Figure 6C). These results suggested that tomato plants overexpressing MdARD4 sustained less damage and were cold tolerant.
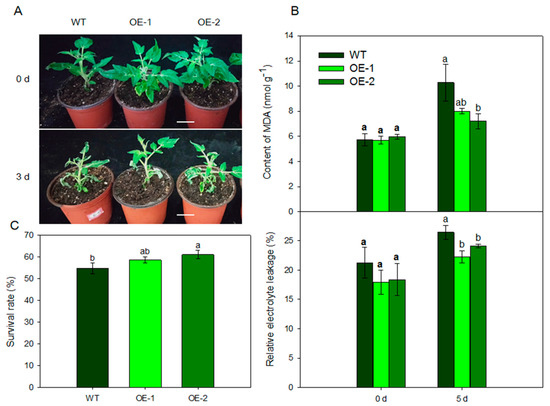
Figure 6.
Effects of MdARD4 overexpression in tomato on the degree of plant damage under cold conditions. (A) Phenotype comparisons between WT and MdARD4 transgenic plants. Bar, 5 cm; (B) relative electrolyte leakage and MDA concentrations of WT and MdARD4-overexpression plants; and (C) survival rates of WT and MdARD4-overexpression plants. Data are means of three replicates with SD. Different letters indicate significant differences between WT and transgenic plants on the same day of different treatments, according to one-way ANOVA Tukey’s multiple range tests (p < 0.05).
2.7. Overexpression of MdARD4 Activates the Antioxidant System in Tomato under Cold Conditions
Stress-induced accumulation of highly toxic reactive oxygen species (ROS) can lead to oxidative stress, which damages various cell components. We monitored H2O2 levels of transgenic and WT plants leaves under both normal temperature and low temperature conditions. As shown in Figure 7, there were no visible differences in any parameter for any genotype grown under normal temperature conditions. However, after 5 days of 4 °C low temperature, transgenic plants showed significantly lesser H2O2 concentration compared with the WT plants (Figure 7A). There were no obvious differences in the antioxidant enzyme (SOD, CAT, and POD) involved in scavenging H2O2 activities in any genotype grown under normal temperature conditions, while the activities in transgenic plants were obviously more than that in the WT (Figure 7B–D). These results suggested that overexpressing MdARD4 activates the antioxidant system under cold conditions.
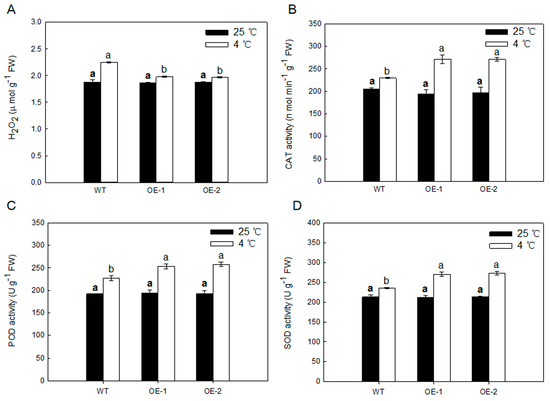
Figure 7.
(A) Results of H2O2 concentrations in leaves from WT and MdARD4 transgenic plants under both normal temperature and low temperature conditions and (B–D) antioxidative enzyme activities in leaves from WT and MdARD4 transgenic plants under both normal temperature and low temperature conditions: CAT (B), POD (C), and SOD (D). Data are means of three replicates with SD. Different letters indicate significant differences between WT and transgenic plants on the same day of different treatments, according to one-way ANOVA Tukey’s multiple range tests (p < 0.05).
3. Discussion
Ethylene plays a key role in plant growth, development, and stress resistance. The methionine (Met) cycle is required to sustain high rates of ethylene synthesis [31]. ARD catalyzes the step where acireductone reacts with dioxygen to produce 2-keto-4-methylthiobuty-rate (KMTB) and that affects the synthesis of ethylene [32]. The functions of ethylene in fruit ripening and stress and the underlying mechanisms have been reported in model plants. However, ARD’s roles in fruit ripening and abiotic stresses, such as cold stress, have been unexplored. In this study, we identified MdARD of Malus domestica. The ectopic expression of MdARD4 in transgenic tomato suggested that MdARD4 accelerates fruit ripening and increases cold hardiness.
Ethylene, which regulates various physiological and developmental events, is important for the overall fruit ripening process [1,2]. Ethylene biosynthesis is catalyzed by two key biosynthetic enzymes ACO and ACS [35,36,37,38]. In tomato, SlACO1 and SlACO3 were significantly induced during fruit ripening [6]. RNA interference of SlACS2 in tomato fruits repressed ethylene production and fruit ripening [39]. Additionally, RIPENING INHIBITOR (RIN) and ethylene, via ethylene response factors (ERFs), are required for the complete expression of ripening genes such as E4 and E8, the classic ethylene-induced genes [8]. ERFs regulate ethylene-dependent transcription and ethylene-inducible gene expression that play an important role in ripening [5,7,38,40,41,42,43,44]. Suppression of E4 produced low-ethylene-producing phenotypes and increase in E8 level induced ethylene production [45]. These results indicated that ethylene triggers fruit ripening via the ethylene signaling pathway. ARD locates upstream of the ethylene biosynthetic pathway. However, the function of MdARD4 in fruit ripening is unknown and whether it is involved in ethylene biosynthesis and signaling pathway is unclear. It has been shown that overexpressing OsARD1 enhanced the endogenous ethylene production in rice by upregulating ACC synthase genes including OsACS2, OsACS4, OsACS5, and OsACS6 in OsARD1-overexpressed plants [46]. Similarly, this study, we found that overexpression of MdARD4 in tomato promoted ripening by activating the expression of four ethylene biosynthetic genes ACS2, ACO3, ACO1, and RIN. Furthermore, the expression of ethylene-responsive genes E4, E8, and ERF1 was more in transgenic fruits than in WT fruits compared at the same stage of maturity (Figure 4). Thus, overexpression of MdARD4 induced the expression of genes involved in ethylene biosynthesis and response, improved ethylene concentration and shortened fruit maturation time. Further, higher concentration of carotenoids and enhanced expression of carotenoid biosynthetic genes were detected in transgenic fruits when compared with WT plants at the same stage of maturity (Figure 5). Previous studies have shown that overexpression of apple RNA binding protein MhYTP2, which interacts with MhARD4, accelerated fruit ripening in tomato [47]. This finding indicated that MdARD4 might regulate fruit ripening by improving ethylene and carotenoid concentrations.
Cold, a common abiotic stress, influences the geographical distribution and negatively impacts productivity and quality of many important crops, such as an apple. Cold stress could reduce cell turgor and destabilize the plasma membrane of plants via ROS burst and oxidative damage, which result in cell damage and even plant death [48,49]. Electrolyte leakage and MDA concentration are typically used to evaluate the peroxidation of plasma membrane. In this study, the electrolyte leakage and MDA concentration were significantly lower in MdARD4 transgenic tomato plants under cold stress (Figure 6). The antioxidative defense system of higher plants is one of the primary physiological response mechanisms under cold stress [50]. CAT, POD, and SOD, which have the ability to eliminate reactive oxygen species and protect the membrane, are essential indicators used to evaluate plant redox status. High CAT, POD, and SOD activities suggested high antioxidative ability and high cold resistance in Arabidopsis [29]. Numerous studies have shown that cold hardiness in various plants is affected by the levels of these antioxidant enzymes [51,52,53]. In this study, transgenic plants showed significantly decreased H2O2 concentration than WT plants after 5 days of 4 °C low temperature, and CAT, POD, and SOD activities were significantly higher in MdARD4 transgenic tomato plants under cold conditions. Decreased H2O2 concentration is attributed to higher POD and CAT abilities. These findings indicated that MdARD4 could increase the antioxidative ability, decrease the membrane lipid peroxidation, and improve the plasma membrane stability in plants in response to cold stress. On the one hand, overexpression of MdARD4 improved ethylene concentration and shortened fruit maturation time, which may reduce the damage from cold stress due to the shorted growth stages. It may be an adaptation mechanism of the different biotic and abiotic stresses [54].
In conclusion, we identified three acireductone dioxygenase genes involved in both fruit ripening and cold hardiness: MdARD1, MdARD2, and MdARD4. MdARD4 showed higher expression under cold stress and during fruit ripening. Overexpression of MdARD4 accelerated fruit ripening in tomato via the regulation of ethylene and carotenoid signaling pathway and increase in ethylene and carotenoid concentrations. Furthermore, MdARD4 transgenic tomato plants demonstrated better growth and cold tolerance compared with the WT plants partially due to the increase in antioxidative ability. This study provides new insights into the regulatory mechanism of ARD in fruit ripening and the cold stress response and proposes MdARD4 as an ideal target candidate for breeding to regulate fruit ripening and cold tolerance in plants.
4. Material and Methods
4.1. Gene Cloning and Plasmid Construction
Leaves were collected from healthy ‘Golden delicious’ (Malus domestica) apple trees at the Horticultural Experimental Station of Northwest A & F University, Yangling (34°20′ N, 108°24′ E), China. These leaves were immediately frozen in liquid nitrogen and stored at −80 °C.
The open reading frames of MdARD4 were obtained through expressed sequence tags (EST) assembly. RNA was extracted from the leaves using the CTAB method [55]. First-strand cDNA was synthesized using RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, Burlington, ON, Canada) according to the manufacturer’s protocol. Primers for cloning were designed using Primer Premier 6.0 (PREMIER Biosoft Int., San Francisco, CA, USA). The sequences of primers used for cDNA cloning are listed in Supplementary Table S1. The amplified MdARD4 product was inserted into the pCambia2300 plant expression vector under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The plasmid was transformed into Agrobacterium tumefaciens EHA105 by the heat shock method [56] and was confirmed by sequencing.
4.2. Plant Materials and Treatments
Fruits of the ‘Honey crisp’ apple (M. domestica) were sampled for gene expression analysis at 0, 15, 30, 47, 61, 75, 92, 104, and 120 days after pollination between 3:00 PM and 4:00 PM. On each collection day, six apples per replicate were harvested from three trees, with a total of five replicates. The fruits were immediately weighed, cut into small pieces after removing the core and frozen on-site in liquid nitrogen. All frozen samples were stored at −80 °C.
The ‘Golden delicious’ apple grafted on the rootstock of Malus hupehensis and grown in pots at the Horticultural Experimental Station of Northwest A & F University, Yangling, China were used for expression analysis of MdARD under cold stress. To induce cold stress, the potted plants were placed at 4 °C and a 16 h photoperiod in the growth chamber. Mature leaves were sampled at 0, 2, 4, 8, and 12 h. Three biological replicates were collected for each treatment with five seedlings combined as one replicate. All collected tissues were immediately frozen in liquid nitrogen and stored at −80 °C.
4.3. Sequence Retrieval and Phylogenetic Tree Construction
The following databases were used to retrieve and compare the putative ARDs from the apple and Arabidopsis: Genome Database for Rosaceae (GDR; https://www.rosaceae.org/species/malus/malus_x_domestica) and Arabidopsis Information Resource (TAIR) database (http://www.Arabidopsis.org). Arabidopsis ARD sequences were used as queries to perform TBLASTN (Basic Local Alignment Search Tool; http://blast.ncbi.nlm.nih.gov) against the apple genome. We searched for ARD homologous protein sequences in other species using BLASTP. A phylogenetic tree was constructed using MEGA 5 software (http://www.megasoftware.net/) [57].
4.4. Subcellular Localization Analysis
MdARD4 was cloned into the pGWB405 vector with a C-terminal green fluorescence protein (GFP). The fused vector and the empty vector were transformed into the Agrobacterium tumefaciens strain EHA105. Further, 5-week-old leaves of tobacco (Nicotiana tabacum) plants were transformed with these vectors. The infiltrated tobacco plants were grown for an additional 3 days under a 16 h:8 h, light:dark photoperiod at 21 °C in a growth chamber. The GFP signal in tobacco leaves was then detected by the Nikon A1R/A1 confocal microscope (Nikon, Tokyo, Japan).
4.5. Production of Transgenic Tomato Plants
The ORF of MdARD4 was inserted into the pCambia2300 vector to construct the MdARD4 overexpression vector. This construct was introduced into Agrobacterium tumefaciens and used to transform ‘Micro-Tom’ tomato at Wuhan Double helix Biology Science and Technology Co., Ltd. (http://www.bioslx.xom/; http://www.biogo.net/com/whslxgs/). Putative transgenic tomato plants were selected on the MS medium containing 50 mg·L−1 kanamycin. Further, 3:1 segregating lines were selected, and T3 homozygous transgenic lines were further confirmed by quantitative real-time PCR.
4.6. Wild-Type (WT) and Transgenic Plant Treatment
To analyze fruit ripening, fruits were sampled at the green fruit stage (G) and red fruit stage (R). The concentration of carotenoids and the expression levels of ethylene biosynthetic genes (ACS2, ACO3, ACO1, and RIN), ethylene-responsive genes (E4, E8, and ERF1), carotenoid biosynthetic genes (PSY1, PDS, and ZDS), and the internal control gene (CAC) were determined in the fruits at different developmental stages of wild type (WT) and transgenic (overexpressing MdARD4) tomato plants.
To further test the response of tomato plants to chilling, seeds of WT and transgenic tomato plants were sown in 250 cm3 plastic pots in a growth chamber under controlled conditions of 50% relative humidity, 28 °C and a long-day photoperiod (16 h:8 h, light:dark). The seedlings were watered regularly and supplied with half-strength Hoagland’s nutrient solution (pH 6.0) twice a week for 40 days to maintain growth. Seedlings of similar size were selected and divided into two groups. One group was placed in a growth chamber at 25 °C and the other group was placed in a growth chamber at 4 °C. After 5 days of treatment under the same photoperiod, H2O2 concentration, electrolyte leakage, MDA, and SOD, CAT, and POD activities were determined in the samples collected. The survival rates were determined as the number of visibly green plants 3 days after being under the normal conditions.
4.7. Ethylene Measurement
Ethylene production was measured according to the method by Han et al. with some modifications [58]. Five fruits were enclosed in a 3.6 L airtight vessel for 1 h at 25 °C, and 1 mL gas sample was collected with a syringe. This gas sample was injected into GC-14A flame ionization detector gas chromatograph (Shimadzu, Kyoto, Japan) to determine ethylene concentration. The ethylene production rate was expressed as µL·kg−1·h−1. Three biological replicates were maintained.
4.8. Analysis of MDA, Electrolyte Leakage and H2O2 Concentrations and Antioxidant Enzyme Activities
The concentration of MDA in the leaf extracts was determined by the thiobarbituric acid (TBA) method [59]. Electrolyte leakage was determined from leaves as described by [60,61] with an electrical conductivity meter (DSS-307; SPSIC, Shanghai, China). The level of H2O2 was measured as described by [62]. Leaf samples (0.1 g) were ground in a chilled mortar with 1% (w/v) polyvinylpolypyrrolidone and homogenized with 1.2 mL of 50 mM potassium phosphate buffer (pH 7.8) containing 1 mM EDTA-Na2 and 0.3% Triton X-100. Each homogenate was centrifuged at 13,000× g for 10 min at 4 °C. The supernatant was used to determine the activities of SOD, CAT, and POD following the established protocols [63,64,65,66,67].
4.9. RNA Extraction and Gene Expression Analysis
Total RNA from snap-frozen tissues was extracted by the CTAB method [55]. The total RNA was reverse transcribed into cDNA using PrimeScript ® RT reagent Kit with gDNA Eraser (Perfect Real Time, Takara). Real-time quantitative PCR (Q-RT-PCR) was conducted on a LightCycler® (Roche Ltd., Basel, Switzerland) 96 real-time PCR detection system (Roche) using the ChamQ SYBR qPCR Master Mixture (Vazyme biotech co., Ltd., Nanjing, China). Apple ELONGATION FACTOR1α (DQ341381)/MdActin (XM_008344381) and tomato CAC (NM_001324017)/SlActin (NM_001308447) were used as the internal control genes to compensate variation in cDNA concentrations and PCR efficiency between samples. The relative quantity of target gene transcript was determined by the 2−ΔΔCt method [68,69]. The internal control genes were amplified in each sample and used for normalization. The results are presented in arbitrary units, as the ratio of the target gene expression to the expression of the reference gene in the indicated group was calculated as 1. All experiments had three biological replicates. Primers used for Q-RT-PCR are listed in Supplementary Table S1.
4.10. Statistical Analysis
Data were subjected to one-way analysis of variance (ANOVA), and mean differences were assessed by a Tukey test (p < 0.05).
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/17/6182/s1, Table S1. Primers used in our study.
Author Contributions
Experimental design and implementation and manuscript composition: T.G.; Experimental implementation and assistance: X.Z., Y.L., C.L. (Chenlu Liu), N.W., Q.J. and J.W.; Experimental design and implementation, manuscript composition and review, financial support: C.L. (Changhai Liu) and F.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
This work was supported by the Young Scientist’s Fund of the National Natural Science Foundation of China (31701897) and the Natural Science Basic Research Plan in Shaanxi Province (2018JQ3008).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, K.L.; Li, H.; Ecker, J.R. Ethylene biosynthesis and signaling networks. Plant Cell. 2002, 14, S131–S151. [Google Scholar] [CrossRef] [PubMed]
- Hiwasa, K.; Kinugasa, Y.; Amano, S.; Hashimoto, A.; Nakano, R.; Inaba, A.; Kubo, Y. Ethylene is required for both the initiation and progression of softening in pear (Pyrus communis L.) fruit. J. Exp. Bot. 2003, 54, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Hirschberg, J. Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant. Biol. 2001, 4, 210–218. [Google Scholar]
- Naeem, M.; Muqarab, R.; Waseem, M. The Solanum melongena COP1 delays fruit ripening and influences Ethylene signaling in tomato. J. Plant Physiol. 2019, 240, 152997. [Google Scholar] [CrossRef]
- Liu, M.C.; Gomes, B.L.; Mila, I.; Purgatto, E.; Peres, L.E.P.; Frasse, P.; Maza, E.; Zouine, M.; Roustan, J.P.; Bouzayen, M.; et al. Comprehensive Profiling of Ethylene Response Factor Expression Identifies Ripening-Associated ERF Genes and Their Link to Key Regulators of Fruit Ripening in Tomato. Plant Physiol. 2016, 170, 1732–1744. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, Y.; Liu, C.; Chen, L.; Ma, J.; Sheng, J.; Shen, L. Inhibition of nitricoxide synthesis delayed mature-green tomato fruits ripening induced by inhibition of ethylene. Sci. Hortic. 2016, 211, 95–101. [Google Scholar] [CrossRef]
- Hao, P.P.; Wang, G.M.; Cheng, H.Y.; Ke, Y.Q.; Qi, K.J.; Gu, C.; Zhang, S.L. Transcriptome analysis unravels an ethylene response factor involved in regulating fruit ripening in pear. Physiol. Plant. 2018, 163, 124–135. [Google Scholar] [CrossRef]
- Li, S.; Zhu, B.Z.; Pirrello, J.; Xu, C.J.; Zhang, B.; Bouzayen, M.; Chen, K.S.; Grierson, D. Roles of RIN and ethylene in tomato fruit ripening and ripening-associated traits. New Phytol. 2020, 226, 460–475. [Google Scholar] [CrossRef]
- Cook, D.; Fowler, S.; Fiehn, O.; Thomashow, M.F. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl Acad. Sci USA 2004, 101, 15243–15248. [Google Scholar] [CrossRef]
- Qin, K.; Shinozaki, K. Yamaguchi-Shinozaki, Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef]
- Shi, H.G.; Wang, X.; Ye, T.T.; Chen, F.F.; Deng, J.; Yang, P.F.; Zhang, Y.S.; Chan, Z.L. The Cysteine2/Histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA6 modulates biotic and abiotic stress responses by activating salicylic acid-related genes and C-REPEAT-BINDING FACTOR genes in Arabidopsis. Plant Physiol. 2014, 165, 1367–1379. [Google Scholar] [CrossRef] [PubMed]
- Zarka, D.G.; Vogel, J.T.; Cook, D. Thomashow MF: Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol. 2003, 133, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Chen, X.; Hong, Y.Y.; Wang, Y.; Xu, P.; Ke, S.D.; Liu, H.Y.; Zhu, J.K.; Oliver, D.J.; Xiang, C.B. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell. 2008, 20, 1134–1151. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Harb, A.; Krishnan, A.; Ambavaram, M.M.; Pereira, A. Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol. 2010, 154, 1254–1271. [Google Scholar] [CrossRef]
- Zhu, J.; Dong, C.H.; Zhu, J.K. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 2007, 10, 290–295. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 2015, 56, 7–15. [Google Scholar] [CrossRef]
- Morgan, P.W.; Drew, M.C. Ethylene and plant responses to stress. Physiol. Plant. 1997, 100, 620–630. [Google Scholar] [CrossRef]
- Wang, C.Y.; Adams, D.O. Chilling-induced ethylene production in cucumbers (Cucumis sativus L.). Plant Physiol. 1982, 69, 424–427. [Google Scholar] [CrossRef]
- Field, R.J. The role of 1-aminocyclopropane-1-carboxylic acid in the control of low temperature induced ethylene production in leaf tissue of Phaseolus vulgaris L. Ann. Bot. 1984, 54, 61–67. [Google Scholar] [CrossRef]
- Chu, C.; Lee, T.M. The relationship between ethylene biosynthesis and chilling tolerance in seedlings of rice (Oryza sativa L.). Bot. Bull. Acad. Sin. 1989, 30, 263–273. [Google Scholar]
- Macháčcková, I.; Hanišová, A.; Krekule, J. Levels of ethylene, ACC, MACC, ABA and proline as indicators of cold hardening and frost resistance in winter wheat. Physiol. Plant. 1989, 76, 603–607. [Google Scholar] [CrossRef]
- Shi, Y.; Tian, S.W.; Hou, L.H.; Huang, X.Z.; Zhang, X.Y.; Guo, H.W. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell. 2012, 24, 2578–2595. [Google Scholar] [CrossRef]
- Catalá, R.; López-Cobollo, R.; Castellano, M.M.; Angosto, T.; Alonso, J.M.; Ecker, J.R.; Salinas, E. The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell. 2014, 26, 3326–3342. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Liu, W.J.; Xia, X.Z.; Wang, T.Z.; Zhang, W.H. Cold acclimation-induced freezing tolerance of Medicago truncatula seedlings is negatively regulated by ethylene. Physiol. Plant. 2014, 152, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Shen, L.; Fan, B.; Yu, M.M.; Zheng, Y.; Lv, S.N.; Sheng, J.P. Ethylene and cold participate in the regulation of LeCBF1 gene expression in postharvest tomato fruits. FEBS Lett. 2009, 583, 3329–3334. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, R. Enhanced tolerance to freezing in tobacco and tomato over-expressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Biol. 2010, 73, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.M.; Zhu, Z.F.; Zhang, L.L.; Fang, L.C.; Zhang, J.S.; Wang, Q.F.; Li, S.H.; Liang, Z.C.; Xin, H.P. Overexpression of ethylene response factors VaERF080 and VaERF087 from Vitis amurensis enhances cold tolerance in Arabidopsis. Sci. Hortic. 2019, 243, 320–326. [Google Scholar] [CrossRef]
- Hu, Z.G.; Huang, X.B.; Amombo, E.; Liu, A.; Fan, J.B.; Bi, A.; Ji, K.; Xin, H.P.; Chen, L.; Fu, J. The ethylene responsive factor CdERF1 from bermudagrass (Cynodon dactylon) positively regulates cold tolerance. Plant Sci. 2020, 294, 110432. [Google Scholar] [CrossRef]
- Bürstenbinder, K.; Rzewuski, G.; Wirtz, M.; Sauter, M. The role of methionine recycling for ethylene synthesis in Arabidopsis. Plant J. 2007, 49, 238–249. [Google Scholar] [CrossRef]
- Sauter, M.; Lorbiecke, R.; OuYang, B.; Pochapsky, T.C.; Rzewuski, G. The immediate early ethylene response gene OsARD1 encodes an acireductone dioxygenase involved in recycling of the ethylene precursor S-adenosylmethionine. Plant J. 2005, 44, 718–729. [Google Scholar] [CrossRef]
- Ramanathan, V.; Rahman, H.; Subramanian, S.; Nallathambi, J.; Kaliyaperumal, A.; Manickam, S.; Ranganathan, C.; Muthurajan, R. OsARD4 encoding an acireductone dioxygenase improves root architecture in rice by promoting development of secondary roots. Sci. Rep. 2018, 8, 15713. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.S.; Jia, J.G.; Lv, J.; Liang, X.F.; Han, D.J.; Huang, L.L.; Kang, Z.S. Characterization of the expression profile of a wheat aci-reductone-dioxygenase-like gene in response to stripe rust pathogen infection and abiotic stresses. Plant Physiol. Biochem. 2010, 48, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Lelievre, J.M.; Latche, A.; Jones, B.; Bouzayen, M.; Pech, J.C. Ethylene and fruit ripening. Physiol. Plant. 1997, 101, 727–739. [Google Scholar] [CrossRef]
- Zarembinski, T.I.; Theologis, A. Ethylene biosynthesis and action: A case of conservation. Plant Mol. Biol. 1994, 26, 1579–1597. [Google Scholar] [CrossRef] [PubMed]
- Oetiker, J.H.; Olson, D.C.; Shiu, O.Y.; Yang, S.F. Differential induction of seven 1-aminocyclopropane-1-carboxylate synthase genes by elicitor in suspension cultures of tomato (Lycopersicon esculentum). Plant Mol. Biol. 1997, 34, 275–286. [Google Scholar] [CrossRef]
- Lee, J.M.; Joung, J.G.; McQuinn, R.; Chung, M.Y.; Fei, Z.; Tieman, D.; Klee, H.; Giovannoni, J. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 2012, 70, 191–204. [Google Scholar] [CrossRef]
- Alexander, L.; Grierson, D. Ethylene biosynthesis and action in tomato: A model for climacteric fruit ripening. J. Exp. Bot. 2002, 53, 2039–2055. [Google Scholar] [CrossRef]
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998, 12, 3703–3714. [Google Scholar] [CrossRef]
- Yang, Z.; Tian, L.; Latoszek-Green, M.; Brown, D.; Wu, K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 2005, 58, 585–596. [Google Scholar] [CrossRef] [PubMed]
- 42 Xiao, Y.Y.; Chen, J.Y.; Kuang, J.F.; Shan, W.; Xie, H.; Jiang, Y.M.; Lu, W.J. Banana ethylene response factors are involved in fruit ripening through their interactions with ethylene biosynthesis genes. J. Exp. Bot. 2013, 64, 2499–2510. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.C.; Han, Y.C.; Qi, X.Y.; Shan, W.; Chen, J.Y.; Lu, W.J.; Kuang, J.F. Papaya CpERF9 acts as a transcriptional repressor of cell-wall-modifying genes CpPME1/2 and CpPG5 involved in fruit ripening. Plant Cell Rep. 2016, 35, 2341–2352. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Jiang, Z.Y.; Zhang, L.C.; Tan, D.M.; Wei, Y.; Yuan, H.; Li, T.L.; Wang, A.D. Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J. 2016, 88, 735–748. [Google Scholar] [CrossRef]
- Lincoln, J.E.; Fischer, R.L. Diverse mechanisms for the regulation of ethylene-inducible gene expression. Mol. Gen. Genet. 1988, 212, 71–75. [Google Scholar] [CrossRef]
- Liang, S.S.; Xiong, W.; Yin, C.C.; Xie, X.D.; Jin, Y.J.; Zhang, S.J.; Yang, B.; Ye, G.Y.; Chen, S.Y.; Luan, W.J. Overexpression of OsARD1 improves submergence, drought, and salt tolerances of seedling through the enhancement of ethylene synthesis in rice. Front. Plant Sci. 2019, 10, 1088. [Google Scholar] [CrossRef]
- Wang, N.; Guo, T.L.; Wang, P.; Sun, X.; Shao, Y.; Liang, B.W.; Jia, X.; Gong, X.Q.; Ma, F.W. Functional analysis of apple MhYTP1 and MhYTP2 genes in leaf senescence and fruit ripening. Sci. Hortic. 2017, 221, 23–32. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Lee, D.H.; Lee, C.B. Chilling stress-induced changes of antioxidant enzymes in the leaves of cucumber: In gel enzyme activity assays. Plant Sci. 2000, 159, 75–85. [Google Scholar] [CrossRef]
- Kuk, Y.I.; Shin, J.S.; Burgos, N.R.; Hwang, T.E.; Han, O.; Cho, B.H.; Jung, S.; Guh, J.O. Antioxidative enzymes offer protection from chilling damage in rice plants. Crop. Sci. 2003, 43, 2109–2117. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Shi, K.; Xia, X.J.; Zhou, Y.H.; Yu, J.Q. Hydrogen peroxide is involved in the cold acclimation-induced chilling tolerance of tomato plants. Plant Physiol. Biochem. 2012, 60, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.J.; Chen, H.W.; Ma, B.; Zhang, W.K.; Chen, S.Y.; Zhang, J.S. The role of ethylene in plants under salinity stress. Front. Plant Sci. 2015, 6, 1059. [Google Scholar] [CrossRef]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2010, 19, 520–525. [Google Scholar] [CrossRef]
- Hood, E.E.; Jen, G.; Kayes, L.; Kramer, J.; Fraley, R.T.; Chilton, M.D. Restriction endonuclease map of pTi Bo542, a potential Ti plasmid vector for genetic engineering of plants. Nat. Biotechnol. 1984, 2, 702–709. [Google Scholar] [CrossRef]
- Jiang, Q.; Yang, J.; Wang, Q.; Zhou, K.; Mao, K.; Ma, F.W. Overexpression of MdEPF2 improves water use efficiency and reduces oxidative stress in tomato. Env. Exp. Bot. 2019, 162, 321–332. [Google Scholar] [CrossRef]
- Han, Y.; Zhu, Q.; Zhang, Z.; Meng, K.; Hou, Y.; Ban, Q.; Suo, J.; Rao, J. Analysis of xyloglucan endotransglycosylase/hydrolase (XTH) genes and diverse roles of isoenzymes during persimmon fruit development and postharvest softening. PLoS ONE 2015, 10, e0123668. [Google Scholar] [CrossRef]
- Hodges, D.M.; Delong, J.M.; Forney, C.F.; Prange, R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Guo, T.L.; Wang, N.; Xue, Y.C.; Guan, Q.M.; van Nocker, S.; Ma, F.W.; Liu, C.H. Overexpression of the rna binding protein MhYTP1 in transgenic apple enhances drought tolerance and WUE by improving ABA level under drought condition. Plant Sci. 2019, 280, 397–407. [Google Scholar] [CrossRef]
- Liu, C.H.; Guo, T.L.; Wang, N.; Wang, Q.; Xue, Y.C.; Zhan, M.H.; Guan, Q.M.; Ma, F.W. Overexpression of MhYTP2 enhances apple water-use efficiency by activating ABA and ethylene signaling. Env. Exp. Bot. 2019, 157, 260–268. [Google Scholar] [CrossRef]
- Patterson, B.D.; Macrae, E.A.; Ferguson, I.B. Estimation of hydrogen peroxide in plantextracts using titanium (IV). Anal. Biochem. 1984, 139, 487–492. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutases: I. Occurrence in higher plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yin, L.; Liang, D.; Li, C.; Ma, F.; Yue, Z. Delayed senescence of apple leaves by exogenous melatonin treatment: Toward regulating the ascorbate-glutathione cycle. J. Pineal Res. 2012, 53, 11–20. [Google Scholar] [CrossRef]
- Zhang, J.L.; Niu, J.P.; Duan, Y.; Zhang, M.X.; Liu, J.Y.; Li, P.M.; Ma, F.W. Photoprotection mechanism in the ‘Fuji’ apple peel at different levels of photooxidative sunburn. Physiol. Plant. 2015, 154, 54–65. [Google Scholar] [CrossRef]
- Li, Y.X.; Liu, B.Y.; Peng, Y.X.; Liu, C.L.; Zhang, X.Z.; Zhang, Z.J.; Liang, W.; Ma, F.W.; Li, C.Y. Exogenous GABA alleviates alkaline stress in Malus hupehensis by regulating the accumulation of organic acids. Sci. Hortic. 2020, 261, 108982. [Google Scholar] [CrossRef]
- Li, Y.X.; Liu, C.L.; Sun, X.; Liu, B.Y.; Zhang, X.Z.; Liang, W.; Huo, L.Q.; Wang, P.; Ma, F.W.; Li, C.Y. Overexpression of MdATG18a enhances alkaline tolerance and GABA shunt in apple through increased autophagy under alkaline conditions. Tree Physiol. 2020, in press. [Google Scholar] [CrossRef]
- Kim, K.M.; Cho, H.; Choi, K.; Kim, J.; Kim, B.W.; Ko, Y.G.; Jang, S.K.; Kim, Y.K. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes Dev. 2009, 23, 2033–2045. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).