Focal Accumulation of ROS Can Block Pyricularia oryzae Effector BAS4-Expression and Prevent Infection in Rice
Abstract
:1. Introduction
2. Results
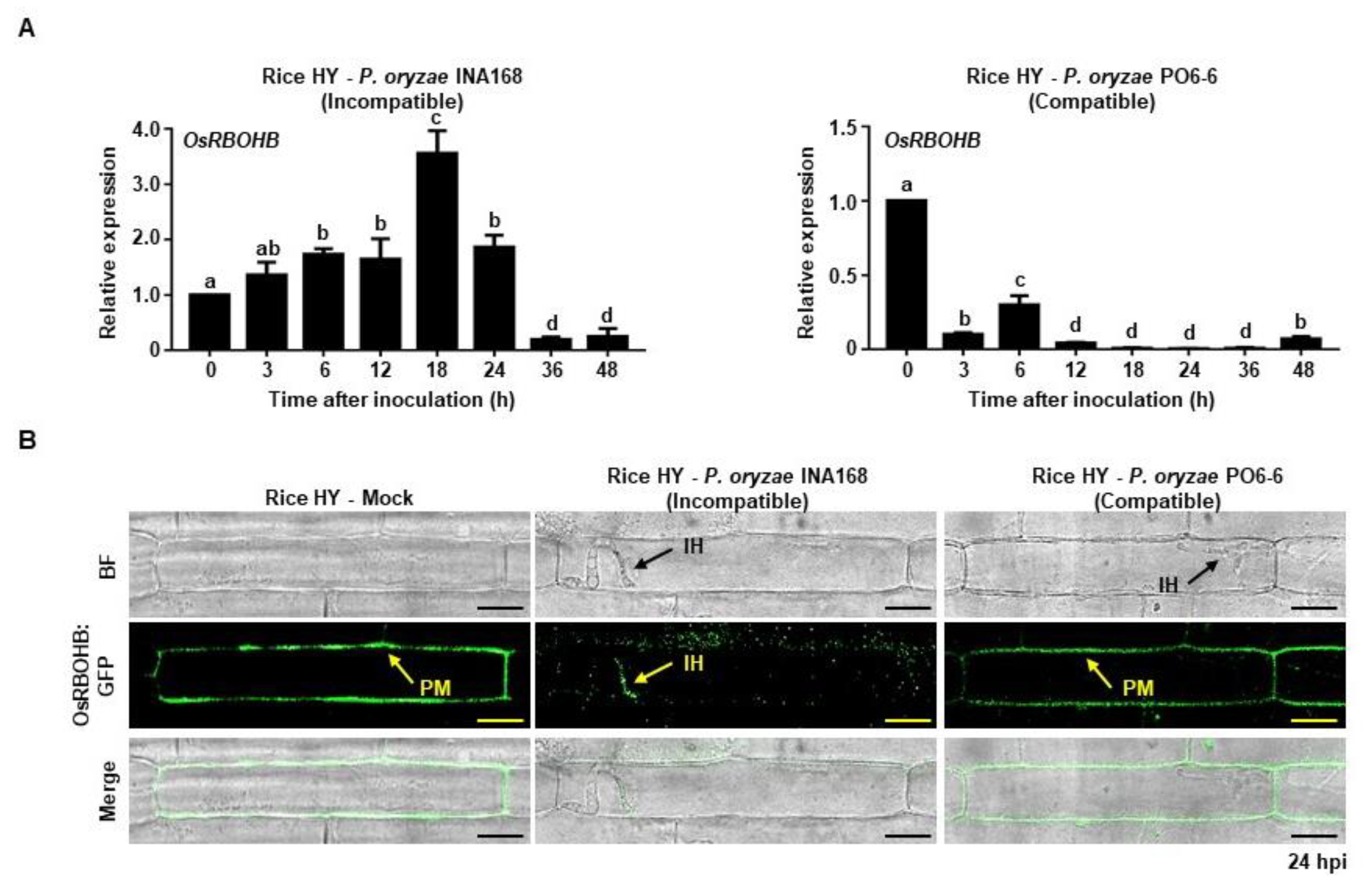
2.1. Avirulent P. oryzae Infection Induced OsRBOHB Expression and Altered OsRBOHB Localization in Rice Cells
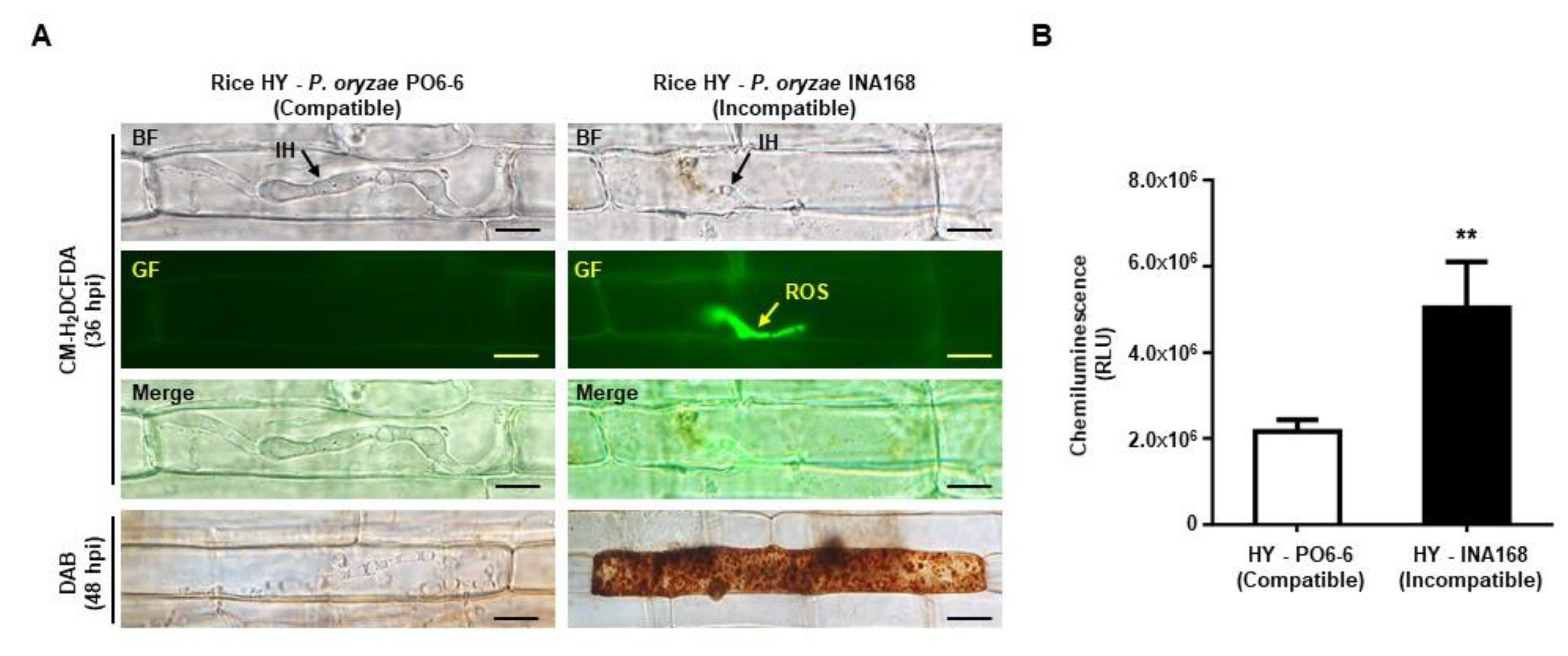
2.2. Avirulent P. oryzae Infection Induced ROS Focal Accumulation
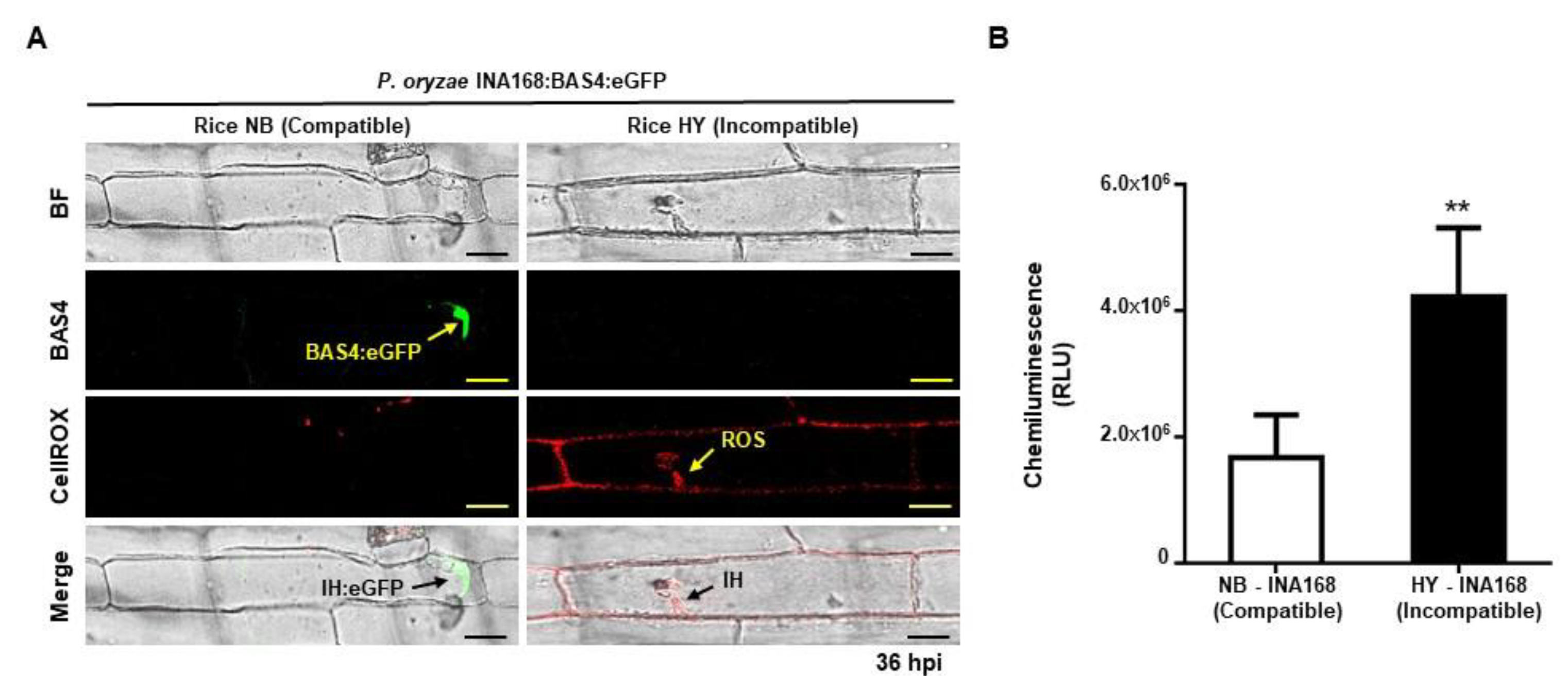
2.3. ROS Focal Accumulation around Invasive Hyphae Suppressed Effector BAS4 Expression
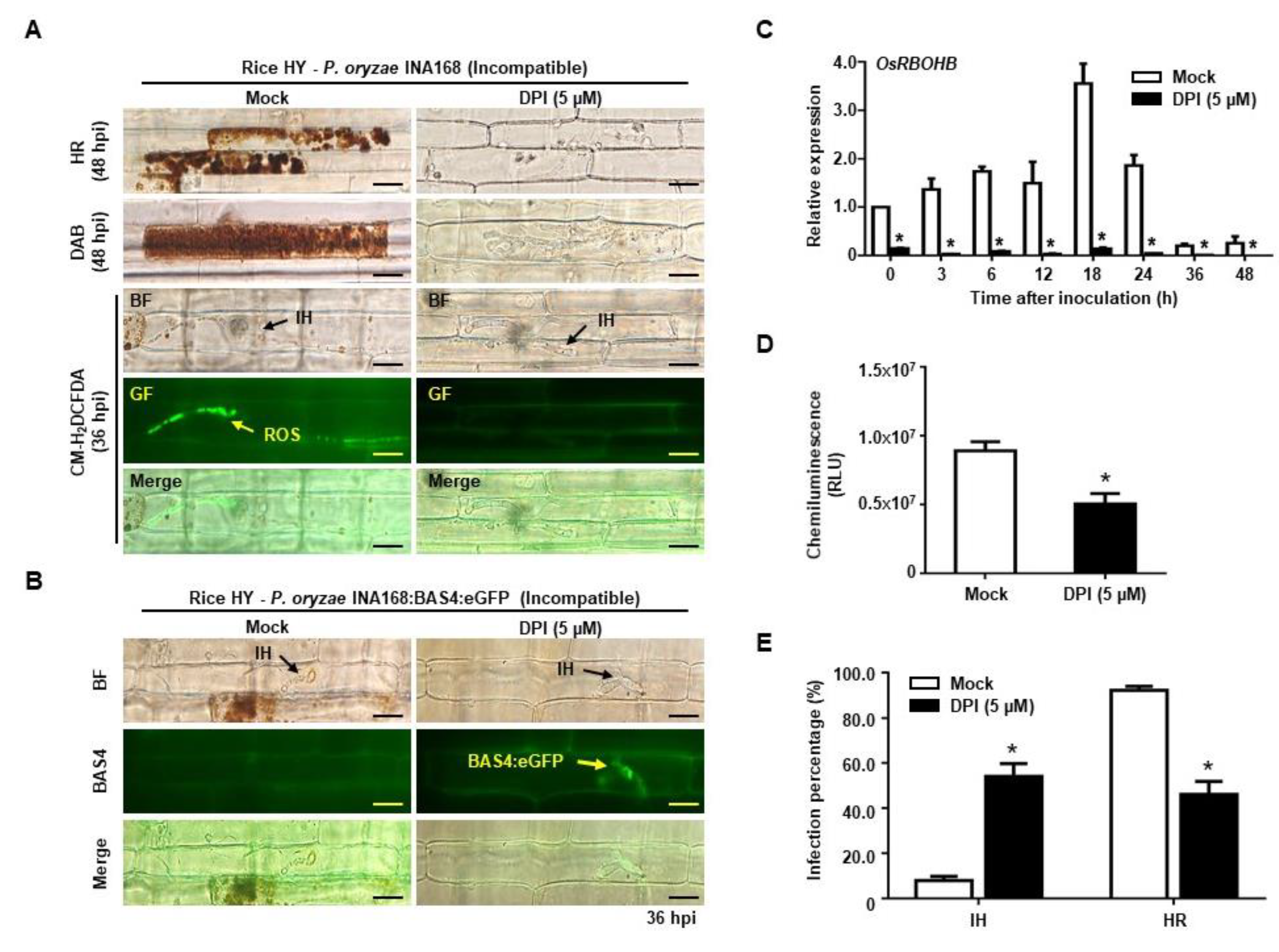
2.4. DPI Suppressed ROS Production but Restored Effector BAS4 Expression
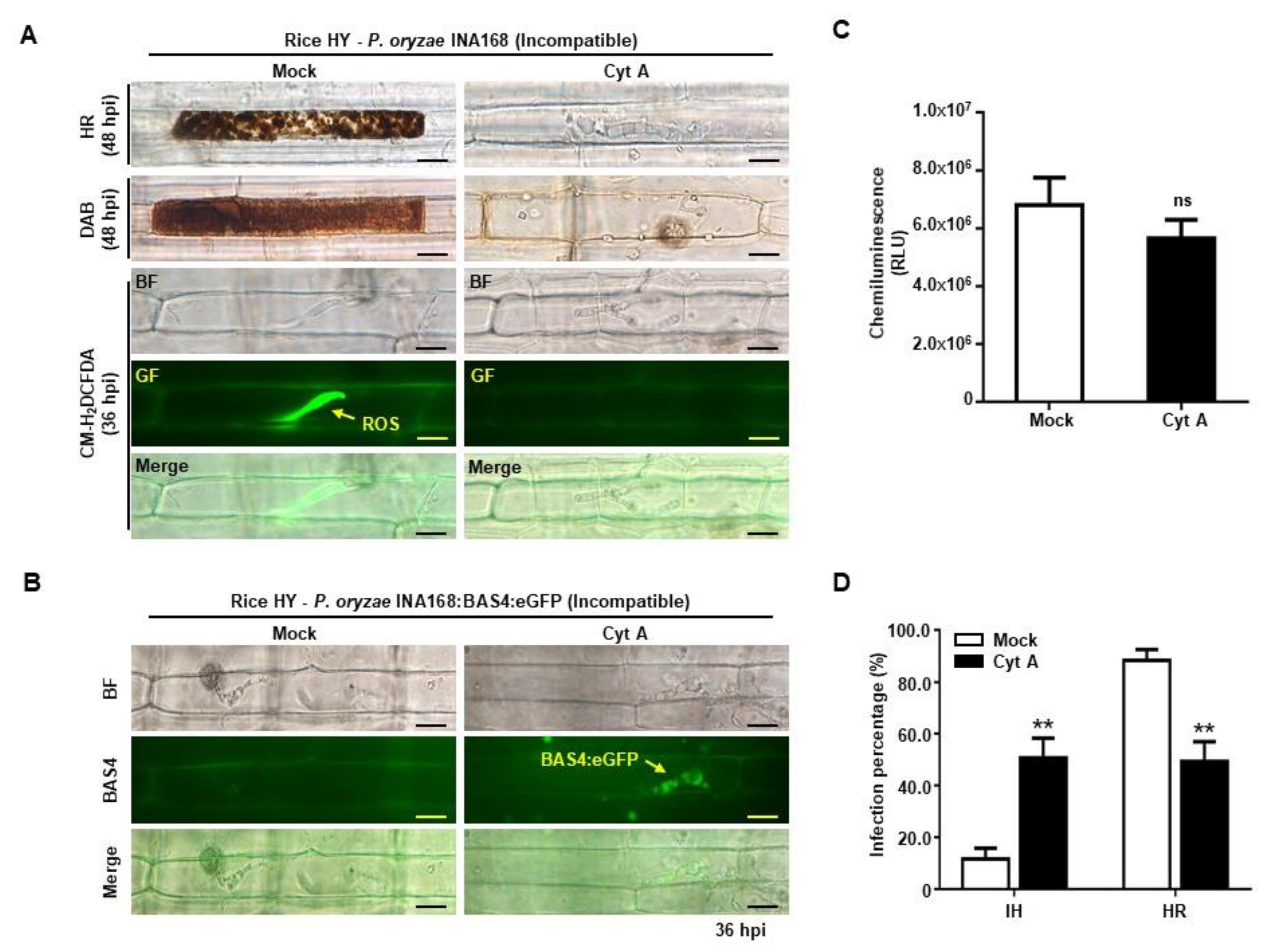
2.5. Cytochalasin a Suppressed ROS Focal Accumulation and Restored Effector BAS4 Expression
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Fungal Culture Conditions
4.3. Fungal Transformation
4.4. Plasmid Construction and Subcellular Localization
4.5. Pathogenicity Test
4.6. Cytochalasin A and Diphenyleneiodonium Treatment
4.7. ROS Detection
4.8. ROS Quantification
4.9. Gene Expression Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BAS4 | Biotrophy-Associated Secreted 4 |
| BIC | Biotrophic Interfacial Complexes |
| Cyt A | Cytochalasin A |
| Cyt E | Cytochalasin E |
| DAB | 3,3′-diaminobenzidine |
| DPI | Diphenyleneiodonium |
| EHM | Extrahaustorial Membrane |
| EIHM | Extra-invasive Hyphal Membrane |
| ETI | Effector-triggered Immunity |
| HR | Hypersensitive Response |
| NADPH | reduced Nicotinamide Adenine Dinucleotide Phosphate |
| NOX | NADPH Oxidase |
| PCD | Programmed Cell Death |
| PAMP | Pathogen-Associated Molecular Pattern |
| PTI | PAMP-triggered Immunity |
| RBOH | Respiratory Burst Oxidase Homologue |
| ROS | Reactive Oxygen Species |
References
- Win, J.; Chaparro-garcia, A.; Belhaj, K.; Saunders, D.G.O.; Yoshida, K.; Dong, S.; Schornack, S.; Zipfel, C.; Robatzek, S.; Hogenhout, S.A.; et al. Effector biology of plant-associated organisms: Concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Moreno, L.; Ebert, M.K.; Bolton, M.D.; Thomma, B.P.H.J. Tools of the crook-infection strategies of fungal plant pathogens. Plant. J. 2018, 93, 664–674. [Google Scholar] [CrossRef] [Green Version]
- Thomma, B.P.H.J.; Nurnberger, T.; Joostena, M.H.A.J. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef] [Green Version]
- Thomas, C.M.; Jones, D.A.; Parniske, M.; Harrison, K.; Balint-Kurti, P.J.; Hatzixanthis, K.; Jones, J.D.G. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 1997, 9, 2209–2224. [Google Scholar] [PubMed] [Green Version]
- Felix, G.; Duran, J.D.; Volko, S.; Boller, T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999, 18, 265–276. [Google Scholar] [CrossRef]
- Khatib, M.; Lafitte, C.; Esquerré-Tugayé, M.-T.; Bottin, A.; Rickauer, M. The CBEL elicitor of Phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signaling pathways. New Phytol. 2004, 162, 501–510. [Google Scholar] [CrossRef]
- Postma, J.; Liebrand, T.W.H.; Bi, G.Z.; Evrard, A.; Bye, R.R.; Mbengue, M.; Kuhn, H.; Joosten, M.H.A.; Robatzek, S. Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol. 2016, 210, 627–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Kadota, Y.; Liebrand, T.W.H.; Goto, Y.; Sklenar, J.; Derbyshire, P.; Menke, F.L.H.; Torres, M.A.; Molina, A.; Zipfel, C.; Coaker, G.; et al. Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytol. 2019, 221, 2160–2175. [Google Scholar] [CrossRef]
- Grant, J.; Loake, G.J. Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol. 2000, 124, 21–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradley, D.J.; Kjellbom, P.; Lamb, C.J. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: A novel, rapid defense response. Cell 1992, 70, 21–30. [Google Scholar] [CrossRef]
- O’Brien, J.A.; Daudi, A.; Butt, V.S.; Bolwell, G.P. Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 2012, 236, 765–779. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A. ROS in biotic interactions. Physiol. Plant. 2010, 138, 414–429. [Google Scholar] [CrossRef] [PubMed]
- Kou, Y.J.; Qiu, J.H.; Tao, Z. Every coin has two sides: Reactive oxygen species during rice-Magnaporthe oryzae interaction. Int. J. Mol. Sci. 2019, 20, 1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morales, J.; Kadota, Y.; Zipfel, C.; Molina, A.; Torres, M.A. The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot. 2016, 67, 1663–1676. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.F.; Li, W.Q.; Li, W.Y.; Wu, G.L.; Zhou, C.Y.; Chen, K.M. Characterization of rice NADPH oxidase genes and their expression under various environmental conditions. Int. J. Mol. Sci. 2013, 14, 9440–9458. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, M.M.; Wang, Y.J.; Gao, Y.T.; Li, R.; Wang, G.F.; Li, W.Q.; Liu, W.T.; Chen, K.M. The plasma membrane NADPH oxidase OsRbohA plays a crucial role in developmental regulation and drought-stress response in rice. Physiol. Plant. 2016, 156, 421–443. [Google Scholar] [CrossRef]
- Yamauchi, T.; Yoshioka, M.; Fukazawa, A.; Mori, H.; Nishizawa, N.K.; Tsutsumi, N.; Yoshioka, H.; Nakazono, M. An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 2017, 29, 775–790. [Google Scholar] [CrossRef] [Green Version]
- Dangol, S.; Chen, Y.F.; Hwang, B.K.; Jwa, N.S. Iron- and reactive oxygen species-dependent ferroptotic cell death in rice-Magnaporthe oryzae interactions. Plant Cell 2019, 31, 189–209. [Google Scholar] [CrossRef] [Green Version]
- Terman, J.R.; Kashina, A. Post-translational modification and regulation of actin. Curr. Opin. Cell Biol. 2013, 25, 30–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdivia, A.; Duran, C.; San, A. The role of Nox-mediated oxidation in the regulation of cytoskeletal dynamics. Curr. Pharm. Des. 2015, 21, 6009–6022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, C.; González-Billault, C. Regulation of cytoskeletal dynamics by redox signaling and oxidative stress: Implications for neuronal development and trafficking. Front. Cell Neurosci. 2015, 9, 381. [Google Scholar] [CrossRef] [Green Version]
- Jwa, N.S.; Hwang, B.K. Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front. Plant Sci. 2017, 8, 1687. [Google Scholar]
- Garre, V.; Tenberge, K.B.; Eising, R. Secretion of a fungal extracellular catalase by Claviceps purpurea during infection of rye: Putative role in pathogenicity and suppression of host defense. Phytopathology 1998, 88, 744–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kankanala, P.; Czymmek, K.; Valent, B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 2007, 19, 706–724. [Google Scholar] [CrossRef] [Green Version]
- Mosquera, G.; Giraldo, M.C.; Khang, C.H.; Coughlan, S.; Valent, B. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-asociated secreted proteins in rice blast disease. Plant Cell 2009, 21, 1273–1290. [Google Scholar] [CrossRef] [Green Version]
- Khang, C.H.; Berruyer, R.; Giraldo, M.C.; Kankanala, P.; Park, S.Y.; Czymmek, K.; Kang, S.C.; Valent, B. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 2010, 22, 1388–1403. [Google Scholar] [CrossRef] [Green Version]
- Selin, C.; Kievit, T.R.; Belmonte, M.F.; Fernando, W.G.D. Elucidating the role of effectors in plant-fungal interactions: Progress and challenges. Front. Microbiol. 2016, 7, 600. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Dangol, S.; Chen, Y.F.; Choi, J.H.; Cho, Y.S.; Lee, J.E.; Choi, M.O.; Jwa, N.S. Magnaporthe oryzae effector AVR-Pii helps to establish compatibility by inhibition of the rice NADP-malic enzyme resulting in disruption of oxidative burst and host innate immunity. Mol. Cells 2016, 39, 426–438. [Google Scholar]
- Caseys, C. Ferroptosis: A companion of ROS in Figurehting Magnaporthe in rice. Plant Cell 2019, 31, 13–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foreman, J.; Demidchik, V.; Bothwell, J.H.F.; Mylona, P.; Miedema, H.; Torresk, M.A.; Linstead, P.; Costa, S.; Brownlee, C.; Jonesk, J.D.G.; et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 2003, 422, 442–446. [Google Scholar] [CrossRef] [PubMed]
- Sagi, M.; Fluhr, R. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 2006, 141, 336–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, S.; Kimura, S.; Kaya, H.; Iizuka, A.; Wong, H.L.; Shimamoto, K. Reactive oxygen species production and activation mechanism of the rice NADPH oxidase OsRbohB. J. Biochem. 2012, 152, 37–43. [Google Scholar] [CrossRef]
- Nagano, M.; Ishikawa, T.; Fujiwara, M.; Fukao, Y.; Kawano, Y.; Kawai-Yamada, M.; Shimamoto, K. Plasma Membrane Microdomains are Essential for Rac1-RbohB/H-Mediated Immunity in Rice. Plant Cell 2016, 28, 1966–1983. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Talbot, N.J. Investigating the cell biology of plant infection by the rice blast fungus Magnaporthe oryzae. Curr. Opin. Microbiol. 2016, 34, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Pfeifer, M.A.; Khang, C.H. A nuclear contortionist: The mitotic migration of Magnaporthe oryzae nuclei during plant infection. Mycology 2018, 9, 202–210. [Google Scholar] [CrossRef]
- Park, C.H.; Chen, S.; Shirsekar, G.; Zhou, B.; Khang, C.H.; Songkumarn, P.; Afzal, A.J.; Ning, Y.; Wang, R.; Bellizzi, M.; et al. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen associated molecular pattern-triggered immunity in rice. Plant Cell 2012, 24, 4748–4762. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.J.; Xu, J.R. Effectors and effector delivery in Magnaporthe oryzae. PLoS Pathog. 2014, 10, e1003826. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.B.; Trush, M.A. Diphenyleneiodonium, an NAD(P)H oxidase inhibitor, also potently inhibits mitochondrial reactive oxygen species production. Biochem. Biophys. Res. Commun. 1998, 253, 295–299. [Google Scholar] [CrossRef]
- Jiang, M.Y.; Zhang, J.H. Involvement of plasma-membrane NADPH oxidase in abscisic acid and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 2002, 215, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Overmyer, K.; Brosché, M.; Kangasjärvi, J. Reactive oxygen species and hormonal control of cell death. Trends Plant Sci. 2003, 8, 335–342. [Google Scholar] [CrossRef]
- Cooper, J.A. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 1987, 105, 1473–1478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, I.; Hakuno, H. Actin-related defense mechanism to reject penetration attempt by a non-pathogen is maintained in tobacco BY-2 cells. Planta 2003, 217, 340–345. [Google Scholar] [CrossRef]
- Wang, W.M.; Wen, Y.Q.; Berkey, R.; Xiao, S.Y. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 2009, 21, 2898–2913. [Google Scholar] [CrossRef] [Green Version]
- Berkey, R.; Zhang, Y.; Ma, X.; King, H.; Zhang, Q.; Wang, W.; Xiao, S. Homologues of the RPW8 resistance protein are localized to the extrahaustorial membrane that is likely synthesized De Novo. Plant Physiol. 2017, 173, 600–613. [Google Scholar] [CrossRef] [Green Version]
- Galletti, R.; Denoux, C.; Gambetta, S.; Dewdney, J.; Ausubel, F.M.; Lorenzo, G.; Ferrari, S. The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol. 2008, 148, 1695–1706. [Google Scholar] [CrossRef] [Green Version]
- Noirot, E.; Der, C.; Lherminier, J.; Robert, F.; Moricova, P.; Kiêu, K.; Leborgne-Castel, N.; Simon-Plas, F.; Bouhidel, K. Dynamic changes in the subcellular distribution of the tobacco ROS-producing enzyme RBOHD in response to the oomycete elicitor cryptogein. J. Exp. Bot. 2014, 65, 5011–5022. [Google Scholar] [CrossRef]
- Li, X.H.; Zhang, H.J.; Tian, L.M.; Huang, L.; Liu, S.X.; Li, D.Y.; Song, F.M. Tomato SlRbohB, a member of the NADPH oxidase family, is required for disease resistance against Botrytis cinerea and tolerance to drought stress. Front. Plant Sci. 2015, 6, 463. [Google Scholar] [CrossRef] [Green Version]
- Wong, H.L.; Pinontoan, R.; Hayashi, K.; Tabata, R.; Yaeno, T.; Hasegawa, K.; Kojima, C.; Yoshioka, H.; Iba, K.; Kawasaki, T.; et al. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 2007, 9, 4022–4034. [Google Scholar] [CrossRef] [Green Version]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2009, 2, 45. [Google Scholar] [CrossRef] [Green Version]
- Daudi, A.; O’Brien, J.A. Detection of Hydrogen Peroxide by DAB Staining in Arabidopsis Leaves. Bio Protoc 2012, 2, e263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, J.F.; Torres, M.S.; Somu, M.P.; Johnson, H.; Irizarry, I.; Chen, Q.; Zhang, N.; Walsh, E.; Tadych, M.; Bergen, M. Hydrogen peroxide staining to visualize intracellular bacterial infections of seedling root cells. Microsc. Res. Tech. 2014, 77, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Bestwick, C.S.; Brown, I.R.; Bennett, M.H.; Mansfield, J.W. Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseolicola. Plant Cell 1997, 9, 209–221. [Google Scholar]
- Liu, G.S.; Zhu, D.Z.; Chen, G.H.; Gao, X.Q.; Zhang, X.S. Disrupted actin dynamics trigger an increment in the reactive oxygen species levels in the Arabidopsis root under salt stress. Plant Cell Rep. 2012, 31, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Kaya, H.; Nakajima, R.; Iwano, M.; Kanaoka, M.M.; Kimura, S.; Takeda, S.; Kawarazaki, T.; Senzaki, E.; Hamamura, Y.; Higashiyama, T.; et al. Ca2+-activated reactive oxygen species production by Arabidopsis RbohH and RbohJ is essential for proper pollen tube tip growth. Plant Cell 2014, 26, 1069–1080. [Google Scholar] [CrossRef] [Green Version]
- Vazquez, L.A.B.; Sanchez, R.; Hernandez-Barrera, A.; Zepeda-Jazo, I.; Sanchez, F.; Quinto, C.; Torres, L.C. Actin polymerization drives polar growth in Arabidopsis root hair cells. Plant Signal. Behav. 2014, 9, e29401. [Google Scholar] [CrossRef]
- Schuh, M. An actin-dependent mechanism for long range vesicle transport. Nat. Cell Biol. 2013, 13, 1431–1436. [Google Scholar] [CrossRef]
- Baral, A.; Shruthi, K.S.; Mathew, M.K. Vesicular Trafficking and Salinity Responses in Plants. IUBMB Life 2015, 67, 677–686. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Yamada, M.; Kobayashi, I.; Kunoh, H. Actin microfilaments are required for the expression of nonhost resistance in higher plants. Plant Cell Physiol. 1997, 38, 725–733. [Google Scholar] [CrossRef]
- Opalski, K.S.; Schultheiss, H.; Kogel, K.H.; Hückelhoven, R. The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. Plant J. 2005, 41, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, D.; Jones, D.A.; Hardham, A.R. Re-organization of the cytoskeleton and endoplasmic reticulum in the Arabidopsis pen1-1 mutant inoculated with the non-adapted powdery mildew pathogen, Blumeria graminis f. sp. hordei. Mol. Plant Pathol. 2006, 7, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Hückelhoven, R.; Panstruga, R. Cell biology of the plant–powdery mildew interaction. Curr. Opin. Plant Biol. 2011, 14, 738–746. [Google Scholar] [CrossRef]
- Crivello, J.F.; Jefcoate, C.R. Intracellular movement of cholesterol in rat adrenal cells. Kinetics and effects of inhibitors. J. Biol. Chem. 1980, 255, 8144–8151. [Google Scholar]
- Bonder, E.M. Cytochalasin B slows but does not prevent monomer addition at the barbed end of the actin filament. Int. J. Cell Biol. 1986, 102, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Yun, B.W.; Atkinson, H.A.; Gaborit, C.; Greenland, A.; Read, N.D.; Pallas, J.A.; Loake, G.J. Loss of actin cytoskeletal function and EDS1 activity, in combination, severely compromises non-host resistance in Arabidopsis against wheat powdery mildew. Plant J. 2003, 34, 768–777. [Google Scholar] [CrossRef]
- Hussey, P.J.; Ketelaar, T.; Deeks, M.J. Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 2006, 57, 109–125. [Google Scholar] [CrossRef] [Green Version]
- Shimada, C.; Lipka, V.; O’Connel, R.; Okuno, T.; Schulze-Lefert, P.; Takano, Y. Nonhost resistance in Arabidopsis-Colletotrichum interactions acts at the cell periphery and requires actin filament function. Mol. Plant Microbe Interact. 2006, 19, 270–279. [Google Scholar] [CrossRef] [Green Version]
- Foissner, I.; Wasteneys, G.O. Wide-ranging effects of eight Cytochalasins and Latrunculin A and B on intracellular motility and actin filament reorganization in characean internodal cells. Plant Cell Physiol. 2007, 48, 585–597. [Google Scholar] [CrossRef]
- Holzinger, A.; Blaas, K. Actin-Dynamics in Plant Cells: The function of actin perturbing substances Jasplakinolide, Chondramides, Phalloidin, Cytochalasins, and Latrunculins. Methods Mol. Biol. 2016, 1365, 243–261. [Google Scholar]
- Munnamalai, V.; Weaver, C.J.; Weisheit, C.E.; Venkatraman, P.; Agim, Z.S.; Quinn, M.T.; Suter, D.M. Bidirectional interactions between NOX2-type NADPH oxidase and the F-actin cytoskeleton in neuronal growth cones. J. Neurochem. 2014, 130, 526–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rampal, A.L.; Pinkofsky, H.B.; Jung, C.Y. Structure of cytochalasins and cytochalasin B binding sites in human erythrocyte membranes. Biochemistry 1980, 19, 679–683. [Google Scholar] [CrossRef]
- Scherlach, K.; Boettger, D.; Remmea, N.; Hertweck, C. The chemistry and biology of cytochalasans. Nat. Prod. Rep. 2010, 27, 869–886. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.S.; Greenshields, D.L.; Sammynaiken, R.; Hirji, R.N.; Selvaraj, G.; Wei, Y.D. Targeted alterations in iron homeostasis underlie plant defense responses. J. Cell Sci. 2007, 120, 596–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kováčik, J.; Babula, P.; Hedbavny, J.; Švec, P. Manganese-induced oxidative stress in two ontogenetic stages of chamomile and amelioration by nitric oxide. Plant Sci. 2014, 215–216, 1–10. [Google Scholar]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Dangol, S.; Wang, J.; Jwa, N.-S. Focal Accumulation of ROS Can Block Pyricularia oryzae Effector BAS4-Expression and Prevent Infection in Rice. Int. J. Mol. Sci. 2020, 21, 6196. https://doi.org/10.3390/ijms21176196
Chen Y, Dangol S, Wang J, Jwa N-S. Focal Accumulation of ROS Can Block Pyricularia oryzae Effector BAS4-Expression and Prevent Infection in Rice. International Journal of Molecular Sciences. 2020; 21(17):6196. https://doi.org/10.3390/ijms21176196
Chicago/Turabian StyleChen, Yafei, Sarmina Dangol, Juan Wang, and Nam-Soo Jwa. 2020. "Focal Accumulation of ROS Can Block Pyricularia oryzae Effector BAS4-Expression and Prevent Infection in Rice" International Journal of Molecular Sciences 21, no. 17: 6196. https://doi.org/10.3390/ijms21176196
APA StyleChen, Y., Dangol, S., Wang, J., & Jwa, N. -S. (2020). Focal Accumulation of ROS Can Block Pyricularia oryzae Effector BAS4-Expression and Prevent Infection in Rice. International Journal of Molecular Sciences, 21(17), 6196. https://doi.org/10.3390/ijms21176196





