The Penta-EF-Hand ALG-2 Protein Interacts with the Cytosolic Domain of the SOCE Regulator SARAF and Interferes with Ubiquitination
Abstract
1. Introduction
2. Results
2.1. Screening for Novel ALG-2-Interacting Proteins
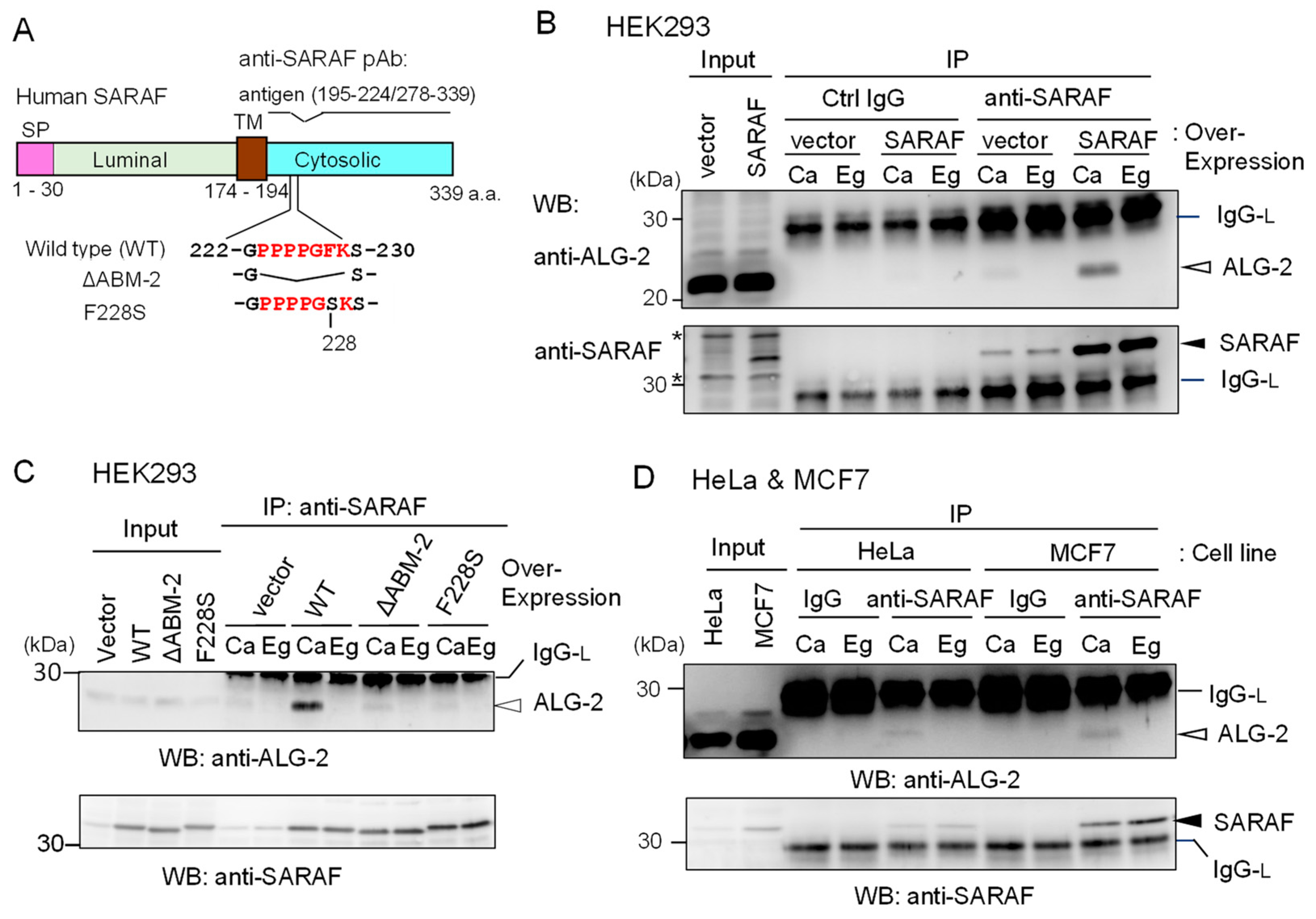
2.2. Co-Immunoprecipitation (co-IP) of Endogenous ALG-2 and SARAF
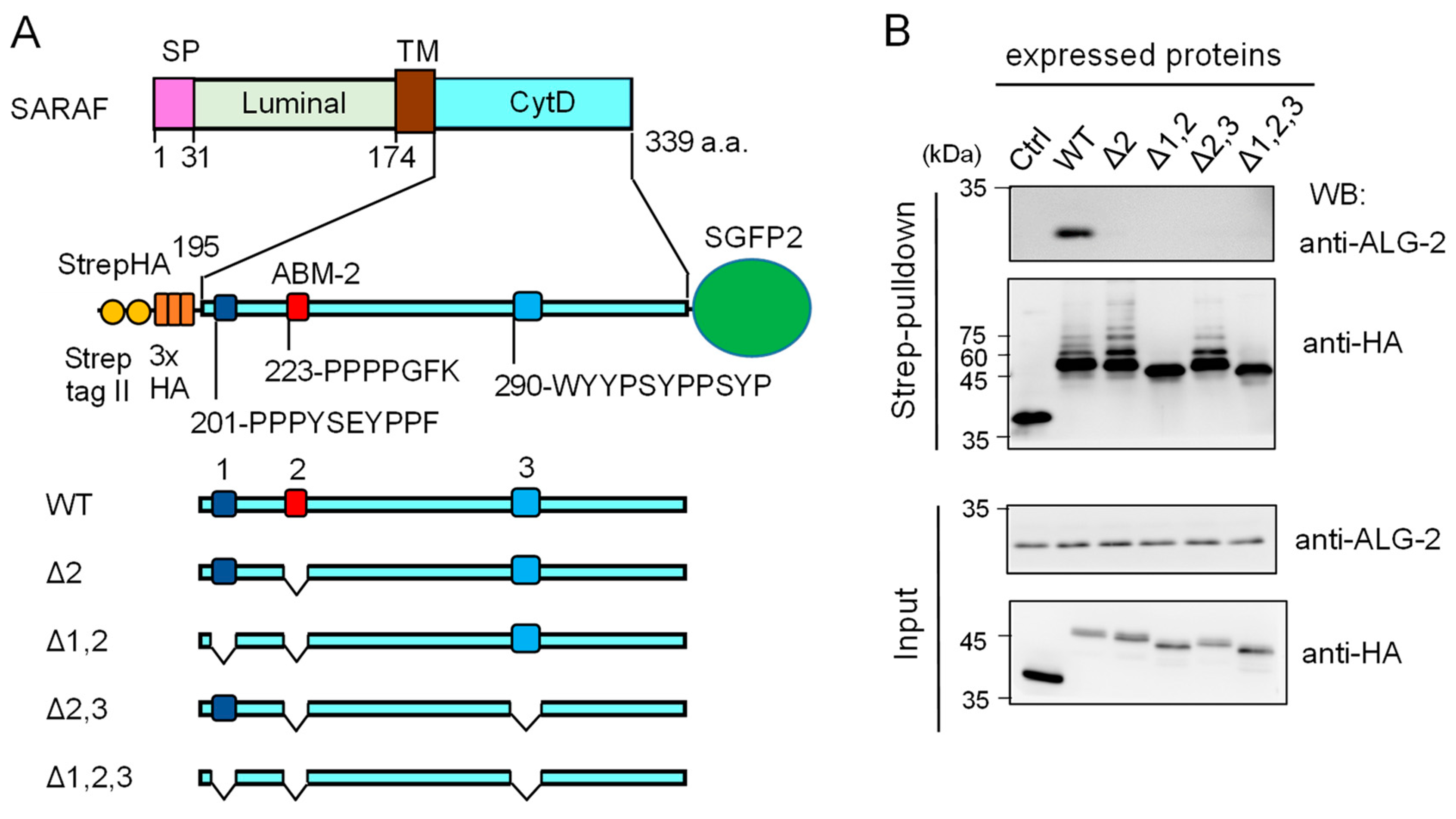
2.3. Essential Region in SARAF for Interaction with ALG-2
2.4. Ubiquitination of SARAF
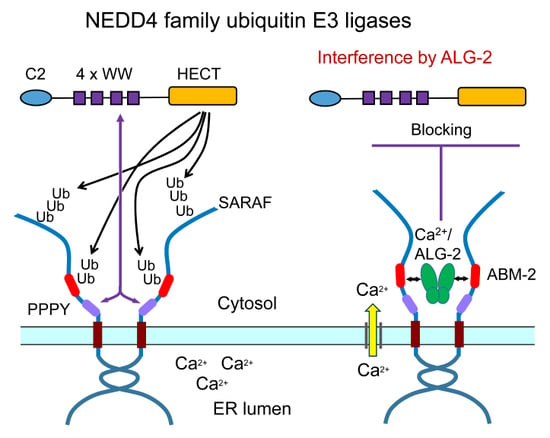
2.5. Suppression of Ubiquitination by Overexpression of ALG-2
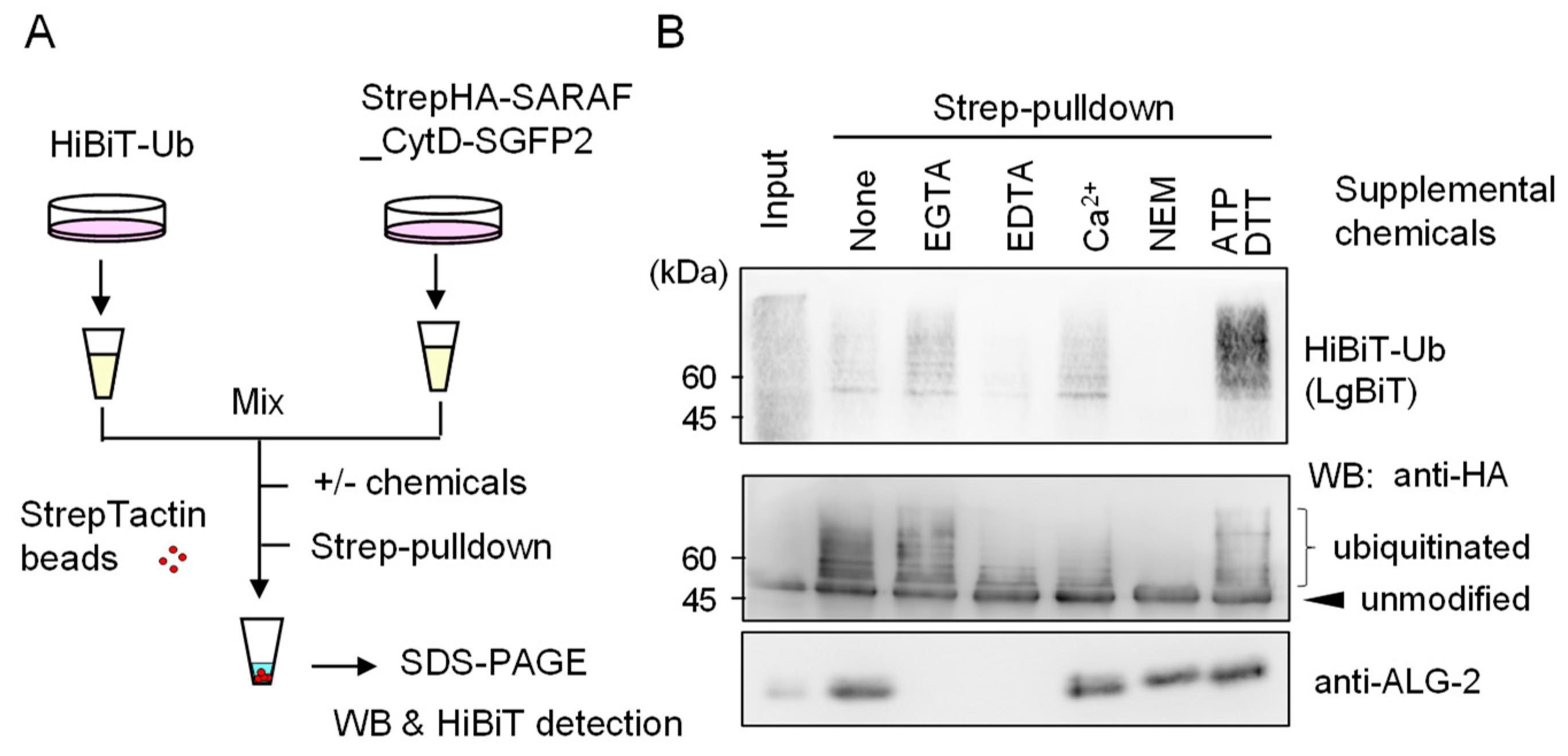
2.6. Evidence of In Vitro Ubiquitination of Expressed SARAF Constructs after Cell Lysis
2.7. In-Cell Ubiquitination of SARAF by NEDD4 Family E3 Ligases
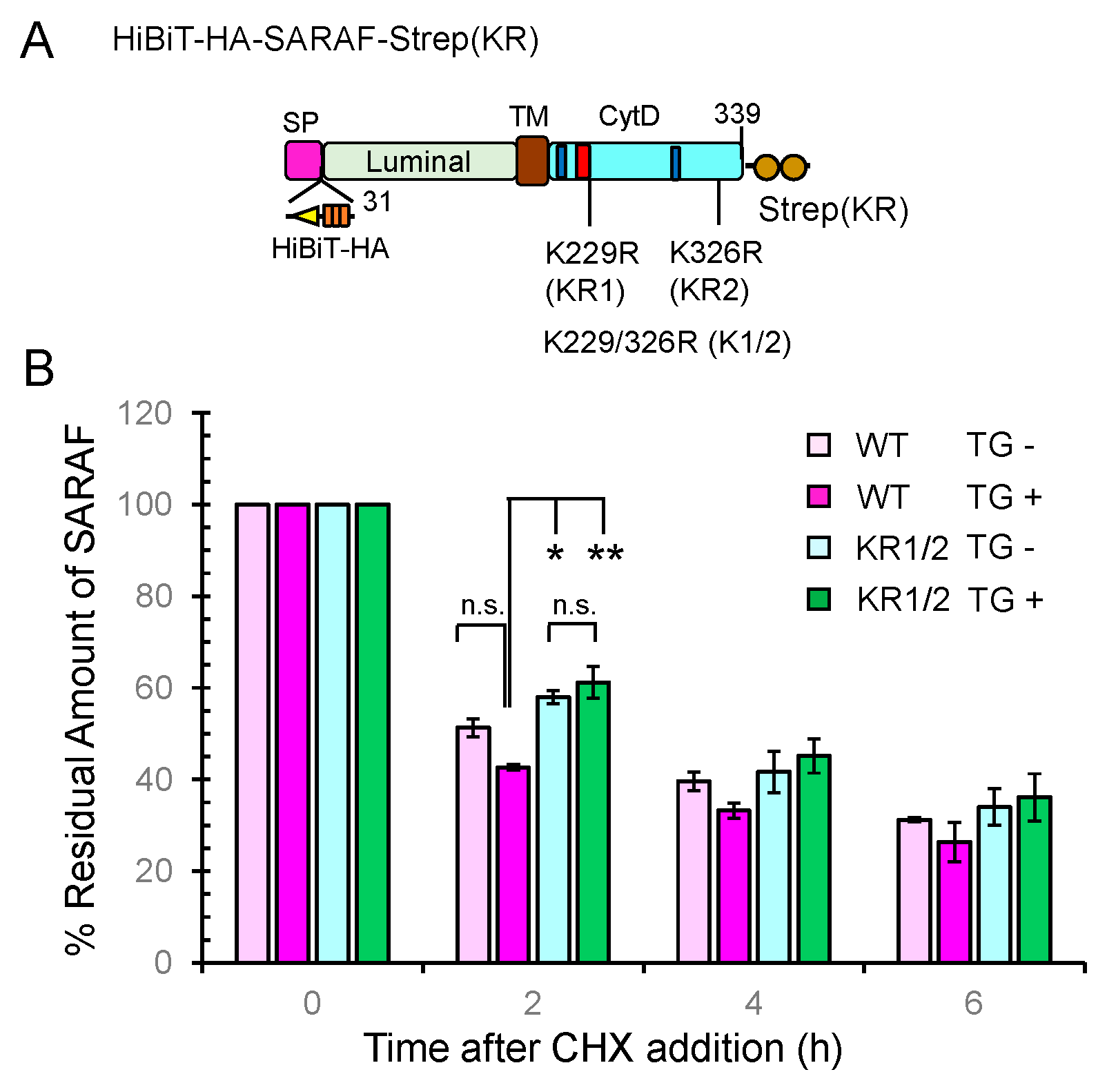
2.8. Slower Degradation Rate of Ubiquitination-Resistant SARAF Mutant
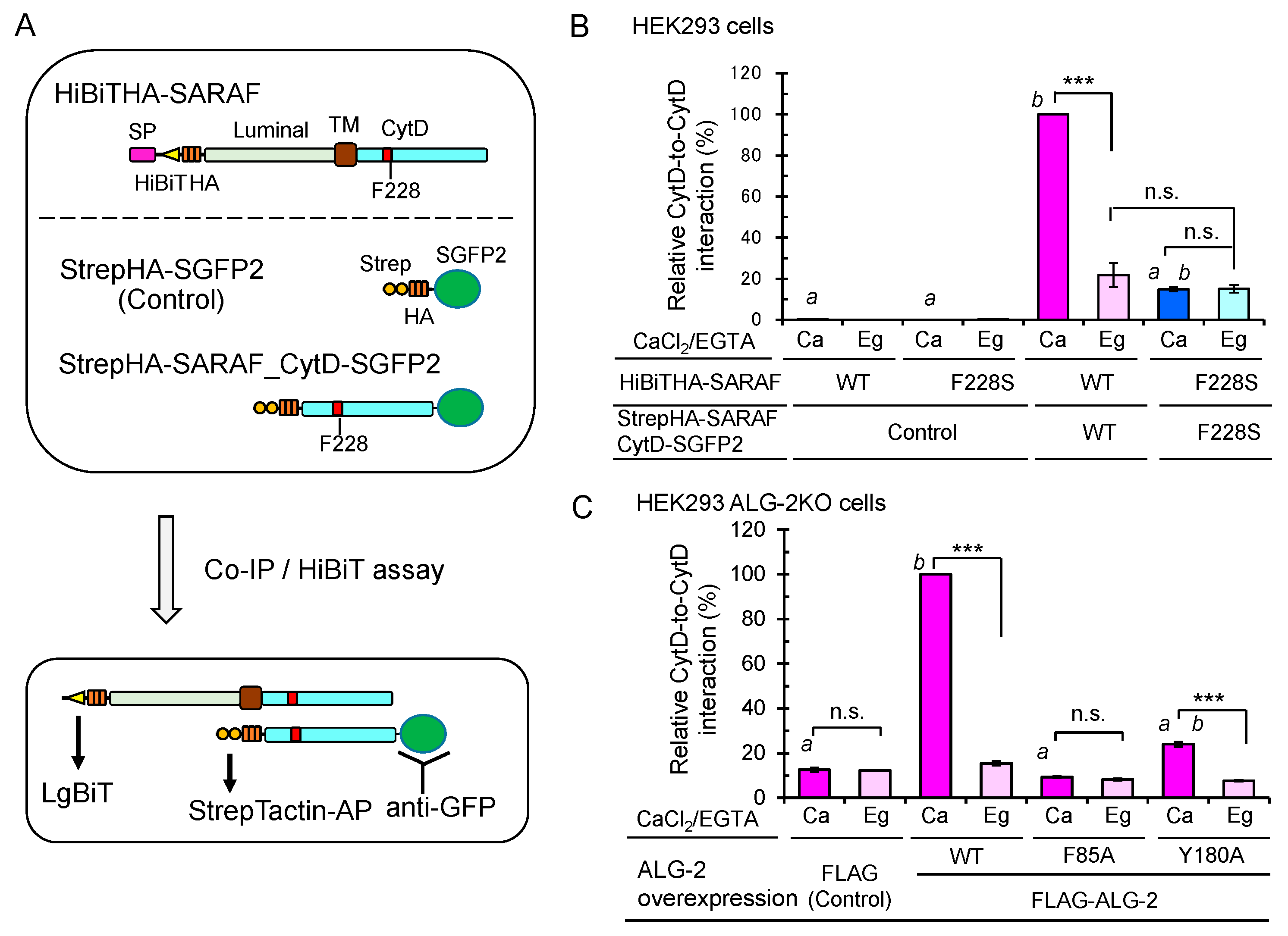
2.9. Enhanced Ca2+-Dependent CytD-to-CytD Interaction by ALG-2
3. Discussion
4. Materials and Methods
4.1. Antibodies and Reagents
4.2. In Silico Screening
4.3. Plasmid Construction
4.4. Cell Culture and DNA Transfection
4.5. Preparation of Nluc-ALG-2
4.6. Binding Assays with Nluc-ALG-2
4.7. Co-Immunoprecipitation (Co-IP) Assays
4.8. Strep-Pulldown Assays
4.9. Detection of HiBiT-Tagged Proteins
4.10. Half-Life Analysis
4.11. Statistical Analysis
4.12. Research Ethics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABM | ALG-2-binding motif |
| AP | alkaline phosphatase |
| CHX | cycloheximide |
| co-IP | co-immunoprecipitation |
| CytD | cytosolic domain |
| FW | Far-Western |
| HECT | homologous to the E6-AP carboxyl terminus |
| IP | Immunoprecipitation |
| NEM | N-Ethylmaleimide |
| Nluc | nanoluciferase |
| PRR | Pro-rich region |
| SOCE | store-operated calcium entry |
| TG | thapsigargin |
| TM | transmembrane |
| WB | Western blot |
| WT | wild type |
References
- Maki, M.; Maemoto, Y.; Osako, Y.; Shibata, H. Evolutionary and physical linkage between calpains and penta-EF-hand Ca2+-binding proteins. FEBS J. 2012, 279, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Vito, P.; Lacanà, E.; D′Adamio, L. Interfering with apoptosis: Ca2+-binding protein ALG-2 and Alzheimer’s disease gene ALG-3. Science 1996, 271, 521–525. [Google Scholar] [CrossRef] [PubMed]
- la Cour, J.M.; Hoj, B.R.; Mollerup, J.; Simon, R.; Sauter, G.; Berchtold, M.W. The apoptosis linked gene ALG-2 is dysregulated in tumors of various origin and contributes to cancer cell viability. Mol. Oncol. 2008, 1, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Yang, Y.; Gao, S.; Liu, Y.; Yu, F.; Zhou, Y.; Lyu, R.; Liu, M.; Liu, X.; Li, D.; et al. Deregulated ALG-2/HEBP2 axis alters microtubule dynamics and mitotic spindle behavior to stimulate cancer development. J. Cell Physiol. 2017, 232, 3067–3076. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Li, S.; Zhou, Q.; Wang, D.; Zou, D.; Shu, J.; Huang, Y. MiR-124 inhibits invasion and induces apoptosis of ovarian cancer cells by targeting programmed cell death 6. Oncol. Lett. 2017, 14, 7311–7317. [Google Scholar] [CrossRef][Green Version]
- Zhang, L.; Chen, X.; Liu, B.; Han, J. MicroRNA-124-3p directly targets PDCD6 to inhibit metastasis in breast cancer. Oncol. Lett. 2018, 15, 984–990. [Google Scholar] [CrossRef]
- Yamada, Y.; Arao, T.; Gotoda, T.; Taniguchi, H.; Oda, I.; Shirao, K.; Shimada, Y.; Hamaguchi, T.; Kato, K.; Hamano, T.; et al. Identification of prognostic biomarkers in gastric cancer using endoscopic biopsy samples. Cancer Sci. 2008, 99, 2193–2199. [Google Scholar] [CrossRef]
- Vito, P.; Pellegrini, L.; Guiet, C.; D′Adamio, L. Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J. Biol. Chem. 1999, 274, 1533–1540. [Google Scholar] [CrossRef]
- Missotten, M.; Nichols, A.; Rieger, K.; Sadoul, R. Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death Differ. 1999, 6, 124–129. [Google Scholar] [CrossRef]
- Strack, B.; Calistri, A.; Craig, S.; Popova, E.; Gottlinger, H.G. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 2003, 114, 689–699. [Google Scholar] [CrossRef]
- von Schwedler, U.K.; Stuchell, M.; Muller, B.; Ward, D.M.; Chung, H.Y.; Morita, E.; Wang, H.E.; Davis, T.; He, G.P.; Cimbora, D.M.; et al. The protein network of HIV budding. Cell 2003, 114, 701–713. [Google Scholar] [CrossRef]
- Katoh, K.; Shibata, H.; Suzuki, H.; Nara, A.; Ishidoh, K.; Kominami, E.; Yoshimori, T.; Maki, M. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J. Biol. Chem. 2003, 278, 39104–39113. [Google Scholar] [CrossRef]
- Rho, S.B.; Song, Y.J.; Lim, M.C.; Lee, S.H.; Kim, B.R.; Park, S.Y. Programmed cell death 6 (PDCD6) inhibits angiogenesis through PI3K/mTOR/p70S6K pathway by interacting of VEGFR-2. Cell Signal 2012, 24, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.V.; Poksay, K.S.; Castro-Obregon, S.; Schilling, B.; Row, R.H.; del Rio, G.; Gibson, B.W.; Ellerby, H.M.; Bredesen, D.E. Molecular components of a cell death pathway activated by endoplasmic reticulum stress. J. Biol. Chem. 2004, 279, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Scheffer, L.L.; Sreetama, S.C.; Sharma, N.; Medikayala, S.; Brown, K.J.; Defour, A.; Jaiswal, J.K. Mechanism of Ca2+-triggered ESCRT assembly and regulation of cell membrane repair. Nat. Commun. 2014, 5, 5646. [Google Scholar] [CrossRef] [PubMed]
- Maki, M.; Takahara, T.; Shibata, H. Multifaceted Roles of ALG-2 in Ca2+-Regulated Membrane Trafficking. Int. J. Mol. Sci. 2016, 17, 1401. [Google Scholar] [CrossRef]
- Maki, M. Structures and functions of penta-EF-hand calcium-binding proteins and their interacting partners: Enigmatic relationships between ALG-2 and calpain-7. Biosci. Biotechnol. Biochem. 2020, 84, 651–660. [Google Scholar] [CrossRef]
- Osugi, K.; Suzuki, H.; Nomura, T.; Ariumi, Y.; Shibata, H.; Maki, M. Identification of the P-body component PATL1 as a novel ALG-2-interacting protein by in silico and far-Western screening of proline-rich proteins. J. Biochem. 2012, 151, 657–666. [Google Scholar] [CrossRef]
- Sasaki-Osugi, K.; Imoto, C.; Takahara, T.; Shibata, H.; Maki, M. Nuclear ALG-2 protein interacts with Ca2+ homeostasis endoplasmic reticulum protein (CHERP) Ca2+-dependently and participates in regulation of alternative splicing of inositol trisphosphate receptor type 1 (IP3R1) pre-mRNA. J. Biol. Chem. 2013, 288, 33361–33375. [Google Scholar] [CrossRef]
- Shibata, H.; Suzuki, H.; Kakiuchi, T.; Inuzuka, T.; Yoshida, H.; Mizuno, T.; Maki, M. Identification of Alix-type and Non-Alix-type ALG-2-binding sites in human phospholipid scramblase 3: Differential binding to an alternatively spliced isoform and amino acid-substituted mutants. J. Biol. Chem. 2008, 283, 9623–9632. [Google Scholar] [CrossRef]
- Palty, R.; Raveh, A.; Kaminsky, I.; Meller, R.; Reuveny, E. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell 2012, 149, 425–438. [Google Scholar] [CrossRef] [PubMed]
- Schwinn, M.K.; Machleidt, T.; Zimmerman, K.; Eggers, C.T.; Dixon, A.S.; Hurst, R.; Hall, M.P.; Encell, L.P.; Binkowski, B.F.; Wood, K.V. CRISPR-Mediated Tagging of Endogenous Proteins with a Luminescent Peptide. ACS Chem. Biol. 2018, 13, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Ingham, R.J.; Gish, G.; Pawson, T. The Nedd4 family of E3 ubiquitin ligases: Functional diversity within a common modular architecture. Oncogene 2004, 23, 1972–1984. [Google Scholar] [CrossRef] [PubMed]
- Rotin, D.; Kumar, S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409. [Google Scholar] [CrossRef]
- Kimberlin, C.R.; Meshcheriakova, A.; Palty, R.; Raveh, A.; Karbat, I.; Reuveny, E.; Minor, D.L., Jr. SARAF Luminal Domain Structure Reveals a Novel Domain-Swapped β-Sandwich Fold Important for SOCE Modulation. J. Mol. Biol. 2019, 431, 2869–2883. [Google Scholar] [CrossRef]
- Takahashi, T.; Kojima, K.; Zhang, W.; Sasaki, K.; Ito, M.; Suzuki, H.; Kawasaki, M.; Wakatsuki, S.; Takahara, T.; Shibata, H.; et al. Structural analysis of the complex between penta-EF-hand ALG-2 protein and Sec31A peptide reveals a novel target recognition mechanism of ALG-2. Int. J. Mol. Sci. 2015, 16, 3677–3699. [Google Scholar] [CrossRef]
- Takahara, T.; Inoue, K.; Arai, Y.; Kuwata, K.; Shibata, H.; Maki, M. The calcium-binding protein ALG-2 regulates protein secretion and trafficking via interactions with MISSL and MAP1B proteins. J. Biol. Chem. 2017, 292, 17057–17072. [Google Scholar] [CrossRef]
- Lo, K.W.; Zhang, Q.; Li, M.; Zhang, M. Apoptosis-linked gene product ALG-2 is a new member of the calpain small subunit subfamily of Ca2+-binding proteins. Biochemistry 1999, 38, 7498–7508. [Google Scholar] [CrossRef]
- Suzuki, H.; Kawasaki, M.; Inuzuka, T.; Okumura, M.; Kakiuchi, T.; Shibata, H.; Wakatsuki, S.; Maki, M. Structural basis for Ca2+-dependent formation of ALG-2/Alix peptide complex: Ca2+/EF3-driven arginine switch mechanism. Structure 2008, 16, 1562–1573. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef]
- Bernassola, F.; Karin, M.; Ciechanover, A.; Melino, G. The HECT family of E3 ubiquitin ligases: Multiple players in cancer development. Cancer Cell 2008, 14, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Grabbe, C.; Husnjak, K.; Dikic, I. The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 2011, 12, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Schwertman, P.; Bekker-Jensen, S.; Mailand, N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 2016, 17, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Livneh, I.; Kravtsova-Ivantsiv, Y.; Braten, O.; Kwon, Y.T.; Ciechanover, A. Monoubiquitination joins polyubiquitination as an esteemed proteasomal targeting signal. Bioessays 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H. Adaptor functions of the Ca2+-binding protein ALG-2 in protein transport from the endoplasmic reticulum. Biosci. Biotechnol. Biochem. 2019, 83, 20–32. [Google Scholar] [CrossRef]
- Okumura, M.; Ichioka, F.; Kobayashi, R.; Suzuki, H.; Yoshida, H.; Shibata, H.; Maki, M. Penta-EF-hand protein ALG-2 functions as a Ca2+-dependent adaptor that bridges Alix and TSG101. Biochem. Biophys. Res. Commun. 2009, 386, 237–241. [Google Scholar] [CrossRef]
- Soboloff, J.; Rothberg, B.S.; Madesh, M.; Gill, D.L. STIM proteins: Dynamic calcium signal transducers. Nat. Rev. Mol. Cell Biol. 2012, 13, 549–565. [Google Scholar] [CrossRef]
- Yeung, P.S.; Yamashita, M.; Prakriya, M. Molecular basis of allosteric Orai1 channel activation by STIM1. J. Physiol. 2020, 598, 1707–1723. [Google Scholar] [CrossRef]
- Lopez, J.J.; Albarran, L.; Gomez, L.J.; Smani, T.; Salido, G.M.; Rosado, J.A. Molecular modulators of store-operated calcium entry. Biochim. Biophys. Acta 2016, 1863, 2037–2043. [Google Scholar] [CrossRef]
- Singh, B.B.; Liu, X.; Tang, J.; Zhu, M.X.; Ambudkar, I.S. Calmodulin regulates Ca2+-dependent feedback inhibition of store-operated Ca2+ influx by interaction with a site in the C terminus of TrpC1. Mol. Cell 2002, 9, 739–750. [Google Scholar] [CrossRef]
- Mullins, F.M.; Park, C.Y.; Dolmetsch, R.E.; Lewis, R.S. STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc. Natl. Acad. Sci. USA 2009, 106, 15495–15500. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, X.; Mueller, G.A.; Sobhany, M.; DeRose, E.F.; Zhang, Y.; London, R.E.; Birnbaumer, L. Crystal structure of calmodulin binding domain of orai1 in complex with Ca2+ calmodulin displays a unique binding mode. J. Biol. Chem. 2012, 287, 43030–43041. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, G.; Yang, Y.; Fu, S.; Liu, X.; Kang, H.; Yang, X.; Su, X.C.; Shen, Y. Calmodulin dissociates the STIM1-Orai1 complex and STIM1 oligomers. Nat. Commun. 2017, 8, 1042. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, S.; Jung, H.J.; Kim, K.D.; Souda, P.; Whitelegge, J.; Gwack, Y. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat. Cell Biol. 2010, 12, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Albarran, L.; Lopez, J.J.; Jardin, I.; Sanchez-Collado, J.; Berna-Erro, A.; Smani, T.; Camello, P.J.; Salido, G.M.; Rosado, J.A. EFHB is a Novel Cytosolic Ca2+ Sensor That Modulates STIM1-SARAF Interaction. Cell Physiol. Biochem. 2018, 51, 1164–1178. [Google Scholar] [CrossRef] [PubMed]
- Albarran, L.; Lopez, J.J.; Amor, N.B.; Martin-Cano, F.E.; Berna-Erro, A.; Smani, T.; Salido, G.M.; Rosado, J.A. Dynamic interaction of SARAF with STIM1 and Orai1 to modulate store-operated calcium entry. Sci. Rep. 2016, 6, 24452. [Google Scholar] [CrossRef] [PubMed]
- Jardin, I.; Albarran, L.; Salido, G.M.; Lopez, J.J.; Sage, S.O.; Rosado, J.A. Fine-tuning of store-operated calcium entry by fast and slow Ca2+-dependent inactivation: Involvement of SARAF. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Albarran, L.; Lopez, J.J.; Gomez, L.J.; Salido, G.M.; Rosado, J.A. SARAF modulates TRPC1, but not TRPC6, channel function in a STIM1-independent manner. Biochem. J. 2016, 473, 3581–3595. [Google Scholar] [CrossRef]
- Shibata, H.; Suzuki, H.; Yoshida, H.; Maki, M. ALG-2 directly binds Sec31A and localizes at endoplasmic reticulum exit sites in a Ca2+-dependent manner. Biochem. Biophys. Res. Commun. 2007, 353, 756–763. [Google Scholar] [CrossRef]
- Kremers, G.J.; Goedhart, J.; van den Heuvel, D.J.; Gerritsen, H.C.; Gadella, T.W., Jr. Improved green and blue fluorescent proteins for expression in bacteria and mammalian cells. Biochemistry 2007, 46, 3775–3783. [Google Scholar] [CrossRef]
- Okegawa, Y.; Motohashi, K. A simple and ultra-low cost homemade seamless ligation cloning extract (SLiCE) as an alternative to a commercially available seamless DNA cloning kit. Biochem. Biophys. Rep. 2015, 4, 148–151. [Google Scholar] [CrossRef] [PubMed]
- Takahara, T.; Arai, Y.; Kono, Y.; Shibata, H.; Maki, M. A microtubule-associated protein MAP1B binds to and regulates localization of a calcium-binding protein ALG-2. Biochem. Biophys. Res. Commun. 2018, 497, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Matsuo, R.; Takahara, T.; Shibata, H.; Maki, M. High Sensitive Quantitative Binding Assays Using a Nanoluciferase-Fused Probe for Analysis of ALG-2-Interacting Proteins. Methods Mol. Biol. 2019, 1929, 501–516. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Muramatsu, A.; Matsuo, R.; Teranishi, N.; Kahara, Y.; Takahara, T.; Shibata, H.; Maki, M. The Penta-EF-Hand ALG-2 Protein Interacts with the Cytosolic Domain of the SOCE Regulator SARAF and Interferes with Ubiquitination. Int. J. Mol. Sci. 2020, 21, 6315. https://doi.org/10.3390/ijms21176315
Zhang W, Muramatsu A, Matsuo R, Teranishi N, Kahara Y, Takahara T, Shibata H, Maki M. The Penta-EF-Hand ALG-2 Protein Interacts with the Cytosolic Domain of the SOCE Regulator SARAF and Interferes with Ubiquitination. International Journal of Molecular Sciences. 2020; 21(17):6315. https://doi.org/10.3390/ijms21176315
Chicago/Turabian StyleZhang, Wei, Ayaka Muramatsu, Rina Matsuo, Naoki Teranishi, Yui Kahara, Terunao Takahara, Hideki Shibata, and Masatoshi Maki. 2020. "The Penta-EF-Hand ALG-2 Protein Interacts with the Cytosolic Domain of the SOCE Regulator SARAF and Interferes with Ubiquitination" International Journal of Molecular Sciences 21, no. 17: 6315. https://doi.org/10.3390/ijms21176315
APA StyleZhang, W., Muramatsu, A., Matsuo, R., Teranishi, N., Kahara, Y., Takahara, T., Shibata, H., & Maki, M. (2020). The Penta-EF-Hand ALG-2 Protein Interacts with the Cytosolic Domain of the SOCE Regulator SARAF and Interferes with Ubiquitination. International Journal of Molecular Sciences, 21(17), 6315. https://doi.org/10.3390/ijms21176315