Role of miR-30a-3p Regulation of Oncogenic Targets in Pancreatic Ductal Adenocarcinoma Pathogenesis
Abstract
1. Introduction
2. Results
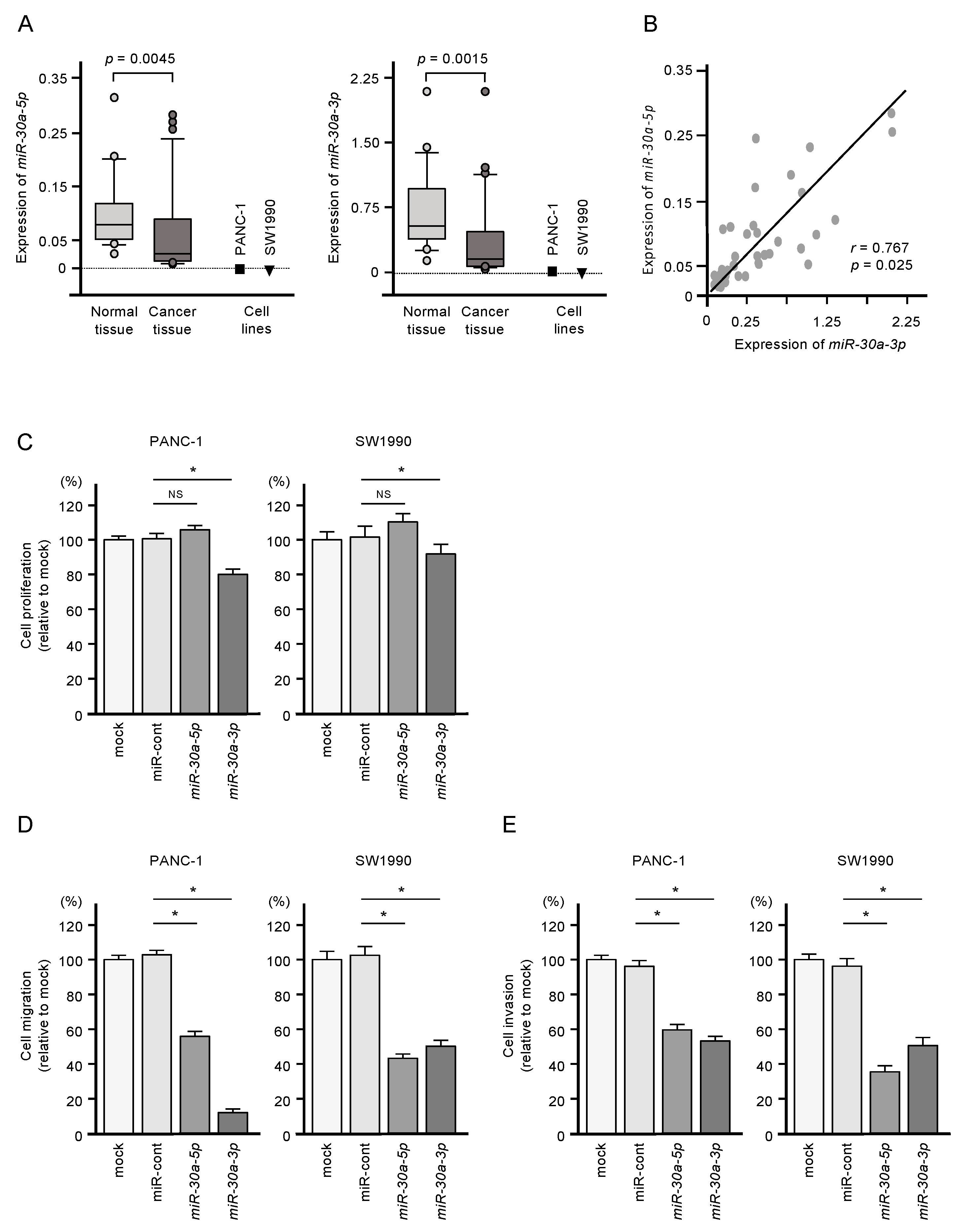
2.1. Downregulation of miR-30a-5p and miR-30a-3p in PDAC Cells and Their Tumor-Suppressive Roles in PDAC Cell Lines
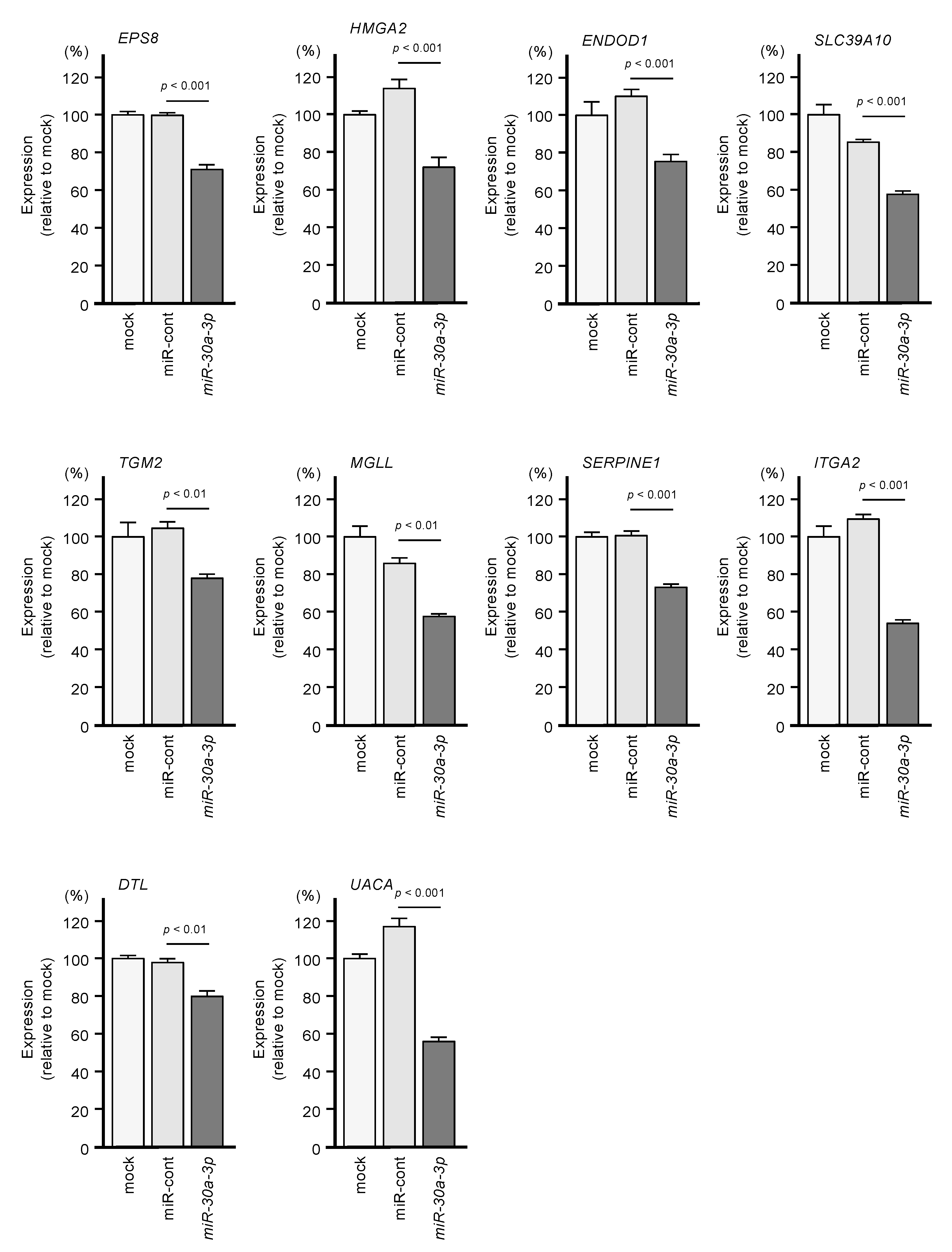
2.2. Identification of Genes Targeted by miR-30a-3p in PDAC Cells
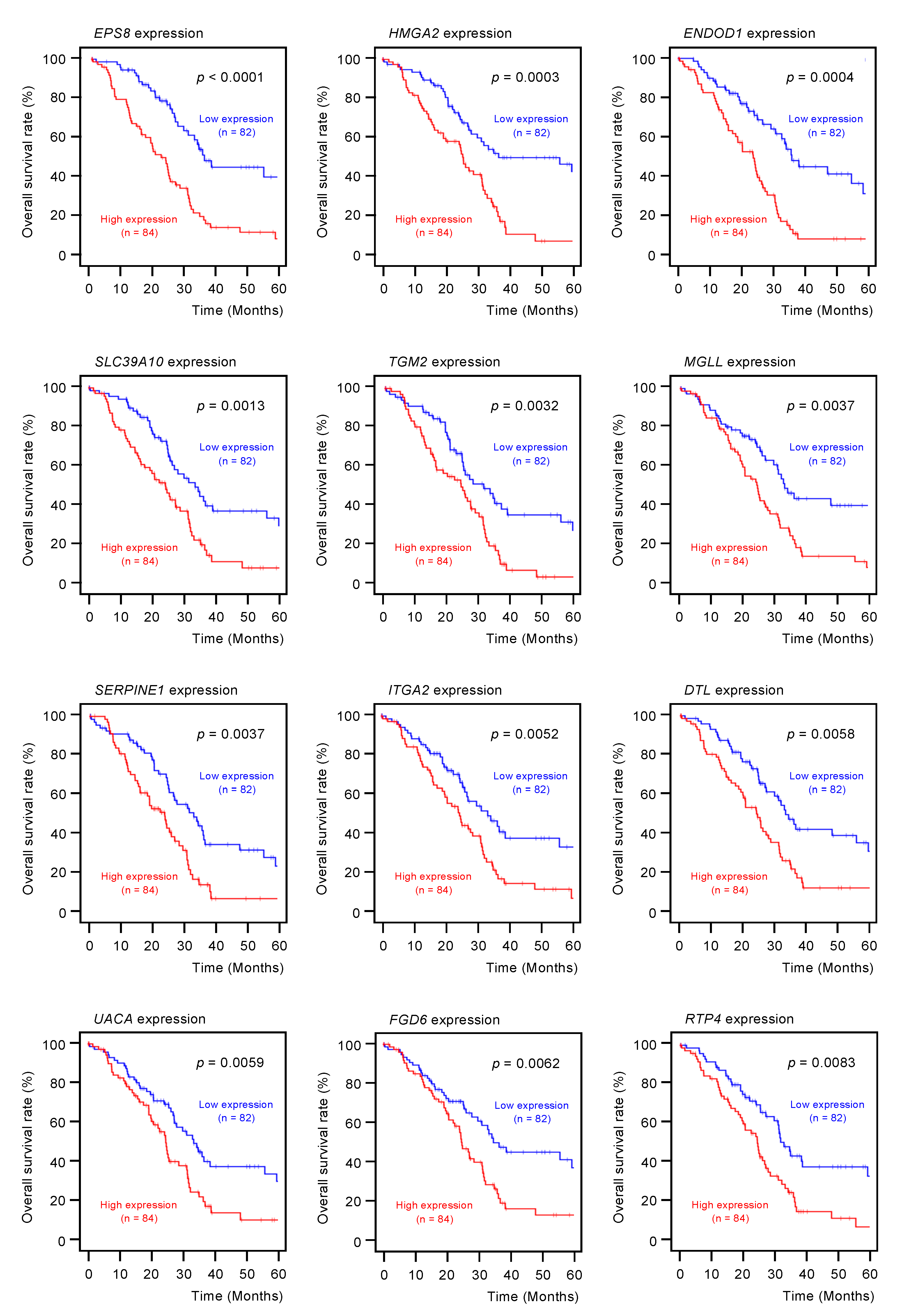
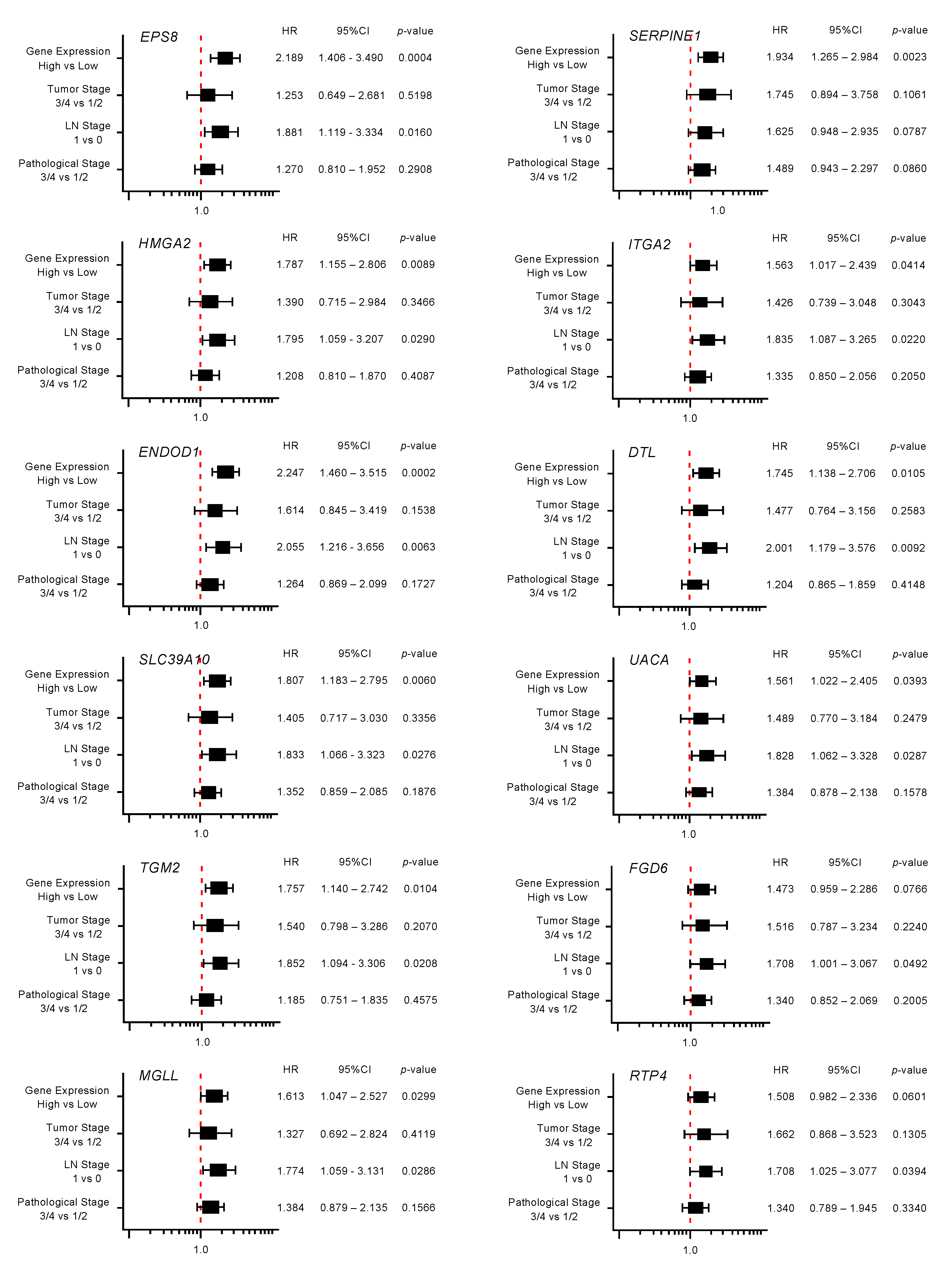
2.3. Clinical Significance of miR-30a-3p Target Genes in PDAC
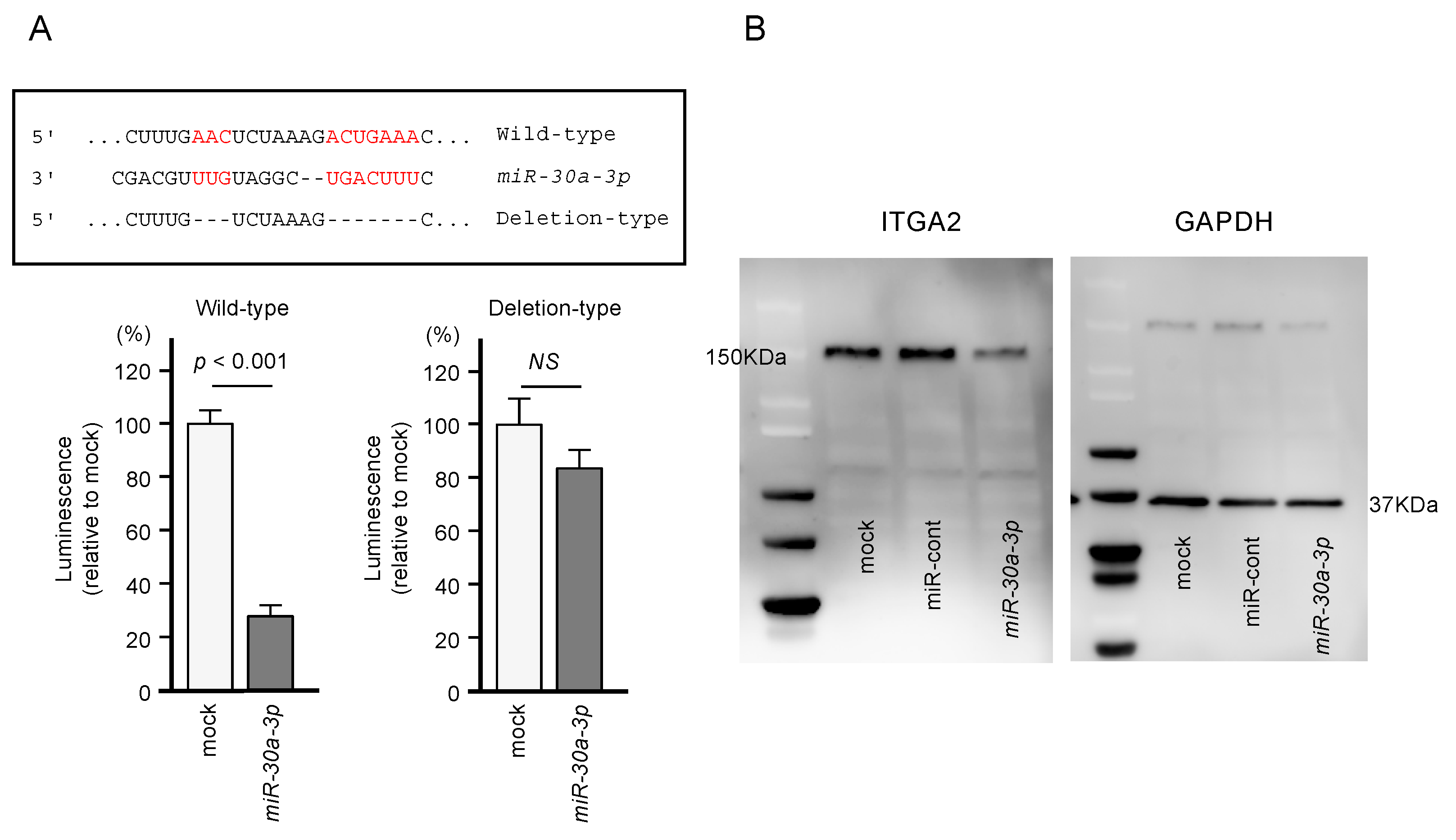
2.4. Direct Regulation of ITGA2 Expression by miR-30a-3p in PDAC Cells
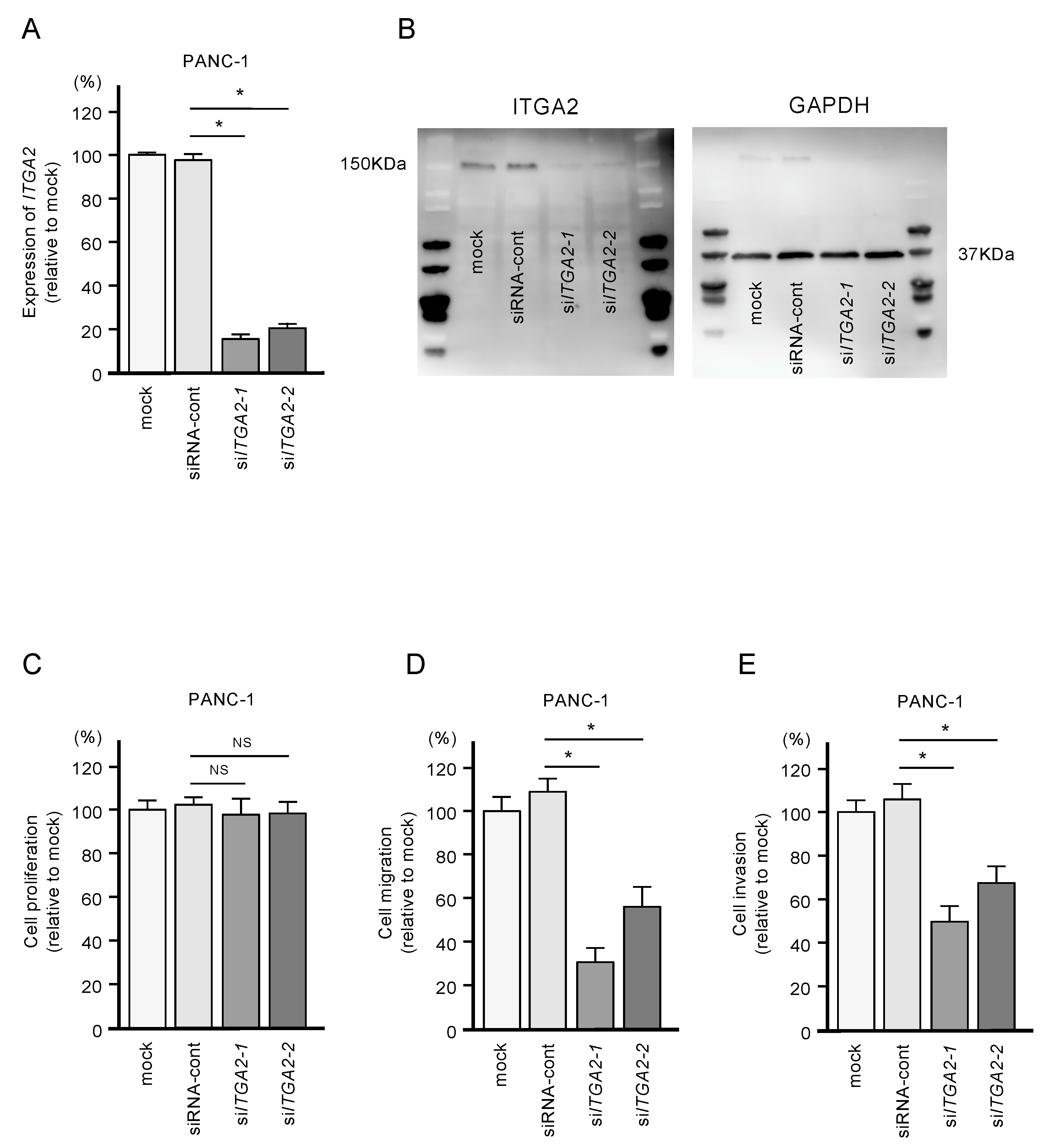
2.5. Functional Significance of ITGA2 in PDAC Cells
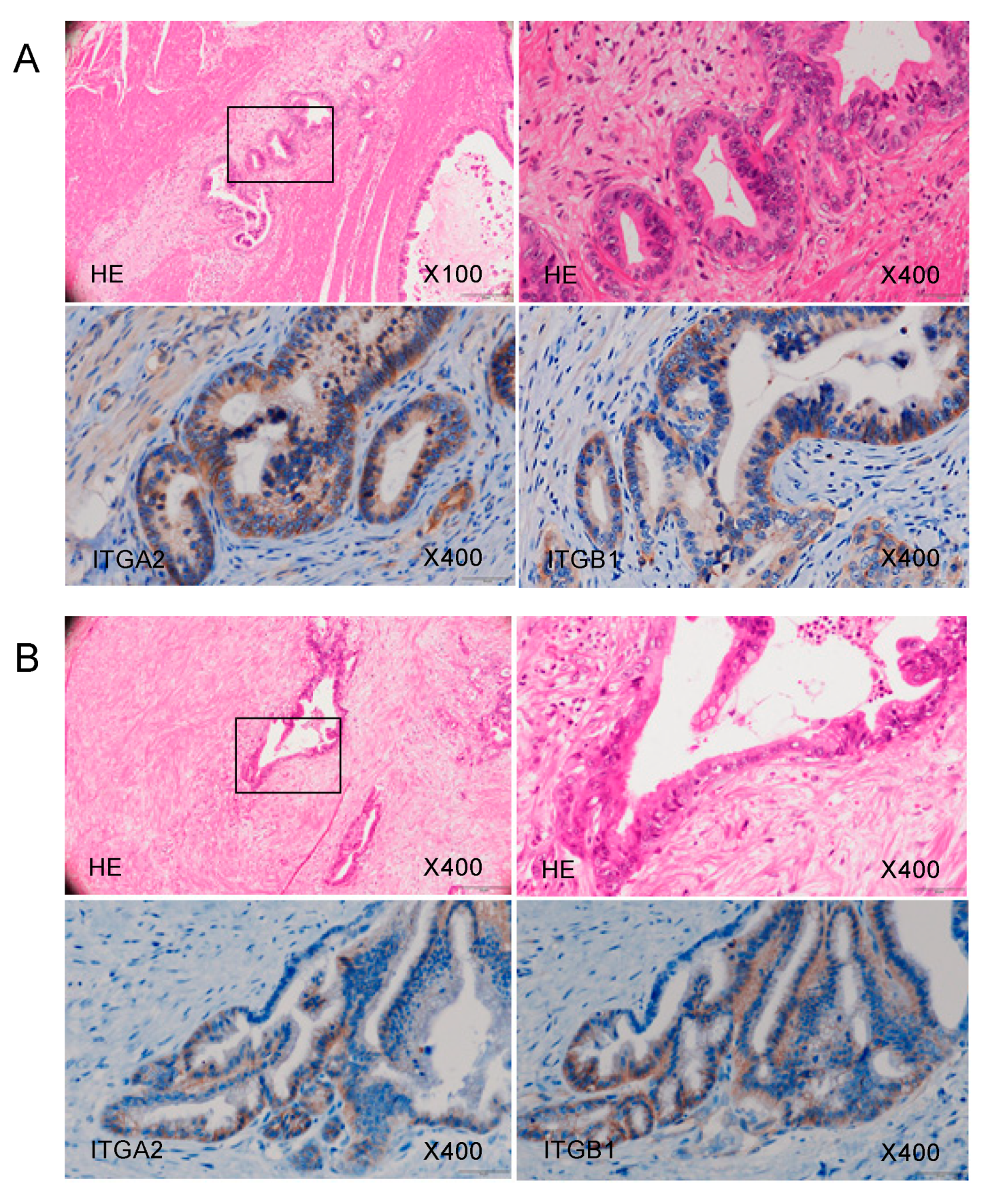
2.6. Overexpression of ITGA2 in Immunohistochemical Staining of PDAC Clinical Specimens
3. Discussion
4. Materials and Methods
4.1. Collection of Clinical Human PDAC Specimens, Pancreas Tissue Specimens, and PDAC Cell Lines
4.2. RNA Extraction and Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
4.3. Transfection of PDAC Cells with miRNAs, siRNAs, and Plasmid Vectors
4.4. Functional Assays in PDAC Cells (Cell Proliferation, Migration, and Invasion Assays)
4.5. Identification of the miR-30a-5p and miR-30a-3p Targets in PDAC Cells
4.6. Clinical Database Analysis of miRNA Target Genes in PDAC Clinical Specimens
4.7. Plasmid Construction and Dual-Luciferase Reporter Assays
4.8. Western Blotting and Immunohistochemistry
4.9. Downregulation of miR-30a-5p and miR-30a-3p in PDAC Cells and Their Tumor-Suppressive Roles in PDAC Cell Lines
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- Adamska, A.; Domenichini, A.; Falasca, M. Pancreatic ductal adenocarcinoma: Current and evolving therapies. Int. J. Mol. Sci. 2017, 18, 1338. [Google Scholar] [CrossRef] [PubMed]
- Ansari, D.; Gustafsson, A.; Andersson, R. Update on the management of pancreatic cancer: Surgery is not enough. World J. Gastroenterol. 2015, 21, 3157–3165. [Google Scholar] [CrossRef] [PubMed]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Gillen, S.; Schuster, T.; Meyer Zum Buschenfelde, C.; Friess, H.; Kleeff, J. Preoperative/neoadjuvant therapy in pancreatic cancer: A systematic review and meta-analysis of response and resection percentages. PLoS Med. 2010, 7, e1000267. [Google Scholar] [CrossRef]
- Du, J.; Gu, J.; Li, J. Mechanisms of drug resistance of pancreatic ductal adenocarcinoma at different levels. Biosci. Rep. 2020. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs–An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Tarang, S.; Weston, M.D. Macros in microRNA target identification: A comparative analysis of in silico, in vitro, and in vivo approaches to microRNA target identification. RNA Biol. 2014, 11, 324–333. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Chen, C.Z. MicroRNAs as oncogenes and tumor suppressors. N. Engl. J. Med. 2005, 353, 1768–1771. [Google Scholar] [CrossRef] [PubMed]
- Yonemori, K.; Seki, N.; Idichi, T.; Kurahara, H.; Osako, Y.; Koshizuka, K.; Arai, T.; Okato, A.; Kita, Y.; Arigami, T.; et al. The microRNA expression signature of pancreatic ductal adenocarcinoma by RNA sequencing: Anti-tumour functions of the microRNA-216 cluster. Oncotarget 2017, 8, 70097–70115. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 2018, 9, 402. [Google Scholar] [CrossRef]
- Idichi, T.; Seki, N.; Kurahara, H.; Fukuhisa, H.; Toda, H.; Shimonosono, M.; Okato, A.; Arai, T.; Kita, Y.; Mataki, Y.; et al. Molecular pathogenesis of pancreatic ductal adenocarcinoma: Impact of passenger strand of pre-miR-148a on gene regulation. Cancer Sci. 2018, 109, 2013–2026. [Google Scholar] [CrossRef]
- Fukuhisa, H.; Seki, N.; Idichi, T.; Kurahara, H.; Yamada, Y.; Toda, H.; Kita, Y.; Kawasaki, Y.; Tanoue, K.; Mataki, Y.; et al. Gene regulation by antitumor miR-130b-5p in pancreatic ductal adenocarcinoma: The clinical significance of oncogenic EPS8. J. Hum. Genet. 2019, 64, 521–534. [Google Scholar] [CrossRef]
- Kumar, M.S.; Lu, J.; Mercer, K.L.; Golub, T.R.; Jacks, T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007, 39, 673–677. [Google Scholar] [CrossRef]
- Chevalier, D.; Borchert, G.M. Genome-wide analysis of MicroRNA-regulated transcripts. Methods Mol. Biol. 2017, 1617, 93–107. [Google Scholar]
- Tafrihi, M.; Hasheminasab, E. MiRNAs: Biology, biogenesis, their web-based tools, and databases. Microrna 2019, 8, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Yonemori, K.; Kurahara, H.; Maemura, K.; Natsugoe, S. MicroRNA in pancreatic cancer. J. Hum. Genet. 2017, 62, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Rawat, M.; Kadian, K.; Gupta, Y.; Kumar, A.; Chain, P.S.G.; Kovbasnjuk, O.; Kumar, S.; Parasher, G. MicroRNA in Pancreatic Cancer: From Biology to Therapeutic Potential. Genes (Basel) 2019, 10, 752. [Google Scholar] [CrossRef] [PubMed]
- Daoud, A.Z.; Mulholland, E.J.; Cole, G.; McCarthy, H.O. MicroRNAs in Pancreatic Cancer: Biomarkers, prognostic, and therapeutic modulators. BMC Cancer 2019, 19, 1130. [Google Scholar] [CrossRef]
- Toda, H.; Seki, N.; Kurozumi, S.; Shinden, Y.; Yamada, Y.; Nohata, N.; Moriya, S.; Idichi, T.; Maemura, K.; Fujii, T.; et al. RNA-sequence-based microRNA expression signature in breast cancer: Tumor-suppressive miR-101-5p regulates molecular pathogenesis. Mol. Oncol. 2020, 14, 426–446. [Google Scholar] [CrossRef]
- Toda, H.; Kurozumi, S.; Kijima, Y.; Idichi, T.; Shinden, Y.; Yamada, Y.; Arai, T.; Maemura, K.; Fujii, T.; Horiguchi, J.; et al. Molecular pathogenesis of triple-negative breast cancer based on microRNA expression signatures: Antitumor miR-204-5p targets AP1S3. J. Hum. Genet. 2018, 63, 1197–1210. [Google Scholar] [CrossRef]
- Wada, M.; Goto, Y.; Tanaka, T.; Okada, R.; Moriya, S.; Idichi, T.; Noda, M.; Sasaki, K.; Kita, Y.; Kurahara, H.; et al. RNA sequencing-based microRNA expression signature in esophageal squamous cell carcinoma: Oncogenic targets by antitumor miR-143-5p and miR-143-3p regulation. J. Hum. Genet. 2020. [Google Scholar] [CrossRef]
- Yonemori, M.; Seki, N.; Yoshino, H.; Matsushita, R.; Miyamoto, K.; Nakagawa, M.; Enokida, H. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci. 2016, 107, 1233–1242. [Google Scholar] [CrossRef]
- Matsushita, R.; Seki, N.; Chiyomaru, T.; Inoguchi, S.; Ishihara, T.; Goto, Y.; Nishikawa, R.; Mataki, H.; Tatarano, S.; Itesako, T.; et al. Tumour-suppressive microRNA-144-5p directly targets CCNE1/2 as potential prognostic markers in bladder cancer. Br. J. Cancer 2015, 113, 282–289. [Google Scholar] [CrossRef]
- Goto, Y.; Kurozumi, A.; Arai, T.; Nohata, N.; Kojima, S.; Okato, A.; Kato, M.; Yamazaki, K.; Ishida, Y.; Naya, Y.; et al. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br. J. Cancer 2017, 117, 409–420. [Google Scholar] [CrossRef]
- Mitra, R.; Adams, C.M.; Jiang, W.; Greenawalt, E.; Eischen, C.M. Pan-cancer analysis reveals cooperativity of both strands of microRNA that regulate tumorigenesis and patient survival. Nat. Commun. 2020, 11, 968. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Cui, H.; Liang, J.; Su, X. Role of MicroRNA-30c in cancer progression. J. Cancer 2020, 11, 2593–2601. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Liu, S.; Hu, L.; Jia, L.; Wang, H.; Guo, M.; Chen, C.; Liu, Y.; Xu, L. miR-30 Family: A promising regulator in development and disease. Biomed Res. Int. 2018, 2018, 9623412. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.H.; Zhang, H.D.; Tang, J.H. MiR-30a: A novel biomarker and potential therapeutic target for cancer. J. Oncol. 2018, 2018, 5167829. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, F.; He, J.; Du, J.; Zhang, H.; Shi, H.; Chen, Y.; Wei, Y.; Xue, W.; Yan, J.; et al. miR-30a-3p targets MAD2L1 and regulates proliferation of gastric cancer cells. Onco Targets Ther. 2019, 12, 11313–11324. [Google Scholar] [CrossRef]
- Wang, H.; Kanmangne, D.; Li, R.; Qian, Z.; Xia, X.; Wang, X.; Wang, T. miR-30a-3p suppresses the proliferation and migration of lung adenocarcinoma cells by downregulating CNPY2. Oncol. Rep. 2020, 43, 646–654. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, J.; Wu, X.; Huang, J.; Chen, W.; Liu, D.; Zhang, J.; Huang, Y.; Xue, W. miR-30a-3p inhibits renal cancer cell invasion and metastasis through targeting ATG12. Transl. Urol. 2020, 9, 646–653. [Google Scholar] [CrossRef]
- Li, D.; Yang, M.; Liao, A.; Zeng, B.; Liu, D.; Yao, Y.; Hu, G.; Chen, X.; Feng, Z.; Du, Y.; et al. Linc00483 as ceRNA regulates proliferation and apoptosis through activating MAPKs in gastric cancer. J. Cell Mol. Med. 2018, 22, 3875–3886. [Google Scholar] [CrossRef]
- Hu, X.; Liu, W.; Jiang, X.; Wang, B.; Li, L.; Wang, J.; Ma, J. Long non-coding RNA LINC00460 aggravates invasion and metastasis by targeting miR-30a-3p/Rap1A in nasopharyngeal carcinoma. Hum. Cell 2019, 32, 465–476. [Google Scholar] [CrossRef]
- Yuan, S.; Xiang, Y.; Wang, G.; Zhou, M.; Meng, G.; Liu, Q.; Hu, Z.; Li, C.; Xie, W.; Wu, N.; et al. Hypoxia-sensitive LINC01436 is regulated by E2F6 and acts as an oncogene by targeting miR-30a-3p in non-small cell lung cancer. Mol. Oncol. 2019, 13, 840–856. [Google Scholar] [CrossRef]
- Qi, B.; Wang, Y.; Chen, Z.-J.; Li, X.-N.; Qi, Y.; Yang, Y.; Cui, G.-H.; Guo, H.-Z.; Li, W.-H.; Zhao, S. Down-regulation of miR-30a-3p/5p promotes esophageal squamous cell carcinoma cell proliferation by activating the Wnt signaling pathway. World J. Gastroenterol. 2017, 23, 7965–7977. [Google Scholar] [CrossRef] [PubMed]
- Argentiero, A.; De Summa, S.; Di Fonte, R.; Iacobazzi, R.M.; Porcelli, L.; Da Vià, M.; Brunetti, O.; Azzariti, A.; Silvestris, N.; Solimando, A.G. Gene expression comparison between the lymph node-positive and -negative reveals a peculiar immune microenvironment signature and a theranostic role for WNT targeting in pancreatic ductal adenocarcinoma: A pilot study. Cancers (Basel) 2019, 11, 942. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Jia, S.; Ding, G.; Zhang, M.; Yu, W.; Wu, Z.; Cao, L. Down-regulation of miR-30a-5p is associated with poor prognosis and promotes chemoresistance of gemcitabine in pancreatic ductal adenocarcinoma. J. Cancer 2019, 10, 5031–5040. [Google Scholar] [CrossRef] [PubMed]
- Young, A.R.; Narita, M. Oncogenic HMGA2: Short or small? Genes. Dev. 2007, 21, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Pfannkuche, K.; Summer, H.; Li, O.; Hescheler, J.; Dröge, P. The high mobility group protein HMGA2: A co-regulator of chromatin structure and pluripotency in stem cells? Stem. Cell Rev. Rep. 2009, 5, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Mo, Q.; Wang, X. Oncological role of HMGA2 (Review). Int. J. Oncol. 2019, 55, 775–788. [Google Scholar] [CrossRef]
- Takada, Y.; Ye, X.; Simon, S. The integrins. Genome Biol. 2007, 8, 215. [Google Scholar] [CrossRef]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef]
- Seguin, L.; Desgrosellier, J.S.; Weis, S.M.; Cheresh, D.A. Integrins and cancer: Regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015, 25, 234–240. [Google Scholar] [CrossRef]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef]
- Cruz da Silva, E.; Dontenwill, M.; Choulier, L.; Lehmann, M. Role of integrins in resistance to therapies targeting growth factor receptors in cancer. Cancers (Basel) 2019, 11, 692. [Google Scholar] [CrossRef]
- Wong, K.F.; Liu, A.M.; Hong, W.; Xu, Z.; Luk, J.M. Integrin α2β1 inhibits MST1 kinase phosphorylation and activates Yes-associated protein oncogenic signaling in hepatocellular carcinoma. Oncotarget 2016, 7, 77683–77695. [Google Scholar] [CrossRef] [PubMed]
- Adorno-Cruz, V.; Liu, H. Regulation and functions of integrin α2 in cell adhesion and disease. Genes Dis. 2019, 6, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Sun, Y.; Li, D.; Li, H.; Jin, X.; Ren, D. Overexpressed ITGA2 contributes to paclitaxel resistance by ovarian cancer cells through the activation of the AKT/FoxO1 pathway. Aging (Albany NY) 2020, 12, 5336–5351. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cao, T.; Guo, K.; Zhou, Y.; Liu, H.; Pan, Y.; Hou, Q.; Nie, Y.; Fan, D.; Lu, Y.; et al. Regulation of integrin subunit alpha 2 by miR-135b-5p modulates chemoresistance in gastric cancer. Front. Oncol. 2020, 10, 308. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Zhao, J.; Sun, Y.; Li, D.; Meng, Z.; Wang, B.; Fan, P.; Liu, Z.; Jin, X.; Wu, H. Overexpressed ITGA2 promotes malignant tumor aggression by up-regulating PD-L1 expression through the activation of the STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 485. [Google Scholar] [CrossRef]
- Idichi, T.; Seki, N.; Kurahara, H.; Fukuhisa, H.; Toda, H.; Shimonosono, M.; Yamada, Y.; Arai, T.; Kita, Y.; Kijima, Y.; et al. Involvement of anti-tumor miR-124-3p and its targets in the pathogenesis of pancreatic ductal adenocarcinoma: Direct regulation of ITGA3 and ITGB1 by miR-124-3p. Oncotarget 2018, 9, 28849–28865. [Google Scholar] [CrossRef]


| Entrez Gene ID | Gene Symbol | Gene Name | Expression in PANC-1 miR-30a-3p Transfectants (FClog2 < −1.0) | GSE15471 (FClog2 > 1.0) | TCGA OncoLnc OS p-Value (In 5 Years) |
|---|---|---|---|---|---|
| 2059 | EPS8 | epidermal growth factor receptor pathway substrate 8 | −1.4210253 | 1.262866923 | 0.0001 |
| 8091 | HMGA2 | high mobility group AT-hook 2 | −1.3260773 | 1.319899625 | 0.0003 |
| 23052 | ENDOD1 | endonuclease domain containing 1 | −1.023117 | 1.177174988 | 0.0004 |
| 57181 | SLC39A10 | solute carrier family 39 (zinc transporter), member 10 | −1.305532 | 1.770031909 | 0.0013 |
| 7052 | TGM2 | transglutaminase 2 | −1.1769093 | 2.232351312 | 0.0032 |
| 11343 | MGLL | monoglyceride lipase | −2.0041718 | 1.656630451 | 0.0037 |
| 5054 | SERPINE1 | serpin peptidase inhibitor, clade E (nexin, plasminogen ativator inhibitor type 1), member 1 | −1.8854839 | 1.966089688 | 0.0037 |
| 3673 | ITGA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | −2.235423 | 2.619190426 | 0.0052 |
| 51514 | DTL | denticleless E3 ubiquitin protein ligase homolog (Drosophila) | −1.4902039 | 1.119465213 | 0.0058 |
| 55075 | UACA | uveal autoantigen with coiled−coil domains and ankyrin repeats | −2.600379 | 1.2785210 | 0.0059 |
| 55785 | FGD6 | FYVE, RhoGEF and PH domain containing 6 | −1.1297837 | 1.472426111 | 0.0062 |
| 64108 | RTP4 | receptor (chemosensory) transporter protein 4 | −1.6157867 | 1.179594488 | 0.0083 |
| 118429 | ANTXR2 | anthrax toxin receptor 2 | −1.5205879 | 1.927129062 | 0.0101 |
| 55704 | CCDC88A | coiled-coil domain containing 88A | −1.9858294 | 1.104710907 | 0.0112 |
| 5358 | PLS3 | plastin 3 | −1.2563933 | 1.111472402 | 0.0144 |
| 25907 | TMEM158 | transmembrane protein 158 (gene/pseudogene) | −1.7876587 | 2.088651268 | 0.0178 |
| 79026 | AHNAK | AHNAK nucleoprotein | −1.2770338 | 1.085859303 | 0.0179 |
| 54739 | XAF1 | XIAP associated factor 1 | −1.237152 | 1.868846867 | 0.018 |
| 26509 | MYOF | myoferlin | −1.5663022 | 2.424595363 | 0.019 |
| 5782 | PTPN12 | protein tyrosine phosphatase, non-receptor type 12 | −1.2349758 | 1.501494024 | 0.0205 |
| 55450 | CAMK2N1 | calcium/calmodulin-dependent protein kinase II inhibitor 1 | -1.5350429 | 1.530841589 | 0.0265 |
| 22822 | PHLDA1 | pleckstrin homology-like domain, family A, member 1 | −2.3576307 | 1.629925658 | 0.031 |
| 9976 | CLEC2B | C-type lectin domain family 2, member B | −1.2099471 | 1.787926875 | 0.0393 |
| 3217 | HOXB7 | homeobox B7 | −1.9851137 | 1.546718079 | 0.0423 |
| 301 | ANXA1 | annexin A1 | −1.7945881 | 2.109602404 | 0.0441 |
| 285761 | DCBLD1 | discoidin, CUB and LCCL domain containing 1 | −1.3696324 | 1.736051328 | 0.0452 |
| 3656 | IRAK2 | interleukin-1 receptor-associated kinase 2 | −2.023258 | 1.139261425 | 0.0527 |
| 7074 | TIAM1 | T−cell lymphoma invasion and metastasis 1 | −1.0873771 | 1.191799754 | 0.0567 |
| 91404 | SESTD1 | SEC14 and spectrin domains 1 | −1.0042286 | 1.389203339 | 0.0615 |
| 9770 | RASSF2 | Ras association (RalGDS/AF-6) domain family member 2 | −1.1753412 | 1.512689587 | 0.0623 |
| 2113 | ETS1 | v-ets avian erythroblastosis virus E26 oncogene homolog 1 | −1.5893964 | 1.15605574 | 0.0733 |
| 6566 | SLC16A1 | solute carrier family 16 (monocarboxylate transporter), member 1 | −1.7025123 | 1.289199126 | 0.0748 |
| 7421 | VDR | vitamin D (1,25- dihydroxyvitamin D3) receptor | −1.0838604 | 1.641355784 | 0.0829 |
| 54810 | GIPC2 | GIPC PDZ domain containing family, member 2 | −1.1281186 | 1.357104573 | 0.0863 |
| 56937 | PMEPA1 | prostate transmembrane protein, androgen induced 1 | −1.1079406 | 2.165168297 | 0.0901 |
| 55686 | MREG | melanoregulin | −2.0134192 | 1.110134893 | 0.0916 |
| 7046 | TGFBR1 | transforming growth factor, beta receptor 1 | −1.5660658 | 1.567228059 | 0.101 |
| 57713 | SFMBT2 | Scm-like with four mbt domains 2 | −2.1900904 | 1.273632017 | 0.107 |
| 4642 | MYO1D | myosin ID | −1.0740082 | 1.117494381 | 0.113 |
| 4131 | MAP1B | microtubule-associated protein 1B | −1.5398299 | 1.867512419 | 0.122 |
| 3397 | ID1 | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | −1.0427207 | 1.289751677 | 0.134 |
| 25927 | CNRIP1 | cannabinoid receptor interacting protein 1 | −1.5424297 | 1.231971168 | 0.147 |
| 54855 | FAM46C | family with sequence similarity 46, member C | −1.192339 | 1.326045782 | 0.147 |
| 90459 | ERI1 | exoribonuclease 1 | −1.0246572 | 1.087664329 | 0.149 |
| 7035 | TFPI | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | −2.0193892 | 1.701846836 | 0.151 |
| 3433 | IFIT2 | interferon-induced protein with tetratricopeptide repeats 2 | −1.7064413 | 1.460924272 | 0.169 |
| 10100 | TSPAN2 | tetraspanin 2 | −1.0831516 | 1.41156579 | 0.169 |
| 18 | ABAT | 4-aminobutyrate aminotransferase | −1.3639479 | 1.797952852 | 0.174 |
| 27286 | SRPX2 | sushi-repeat containing protein, X−linked 2 | −1.1989174 | 2.472950218 | 0.182 |
| 9456 | HOMER1 | homer homolog 1 (Drosophila) | −1.0073853 | 1.299511365 | 0.191 |
| 3696 | ITGB8 | integrin, beta 8 | −1.4881207 | 1.573789775 | 0.203 |
| 2633 | GBP1 | guanylate binding protein 1, interferon-inducible | −1.7881731 | 1.911867379 | 0.215 |
| 83700 | JAM3 | junctional adhesion molecule 3 | −1.1177526 | 1.065465122 | 0.217 |
| 7498 | XDH | xanthine dehydrogenase | −2.073526 | 1.186465821 | 0.22 |
| 10846 | PDE10A | phosphodiesterase 10A | −1.1002035 | 1.167788231 | 0.223 |
| 11031 | RAB31 | RAB31, member RAS oncogene family | −1.2654961 | 2.855210292 | 0.225 |
| 135228 | CD109 | CD109 molecule | −2.4484148 | 2.363606904 | 0.231 |
| 64105 | CENPK | centromere protein K | −1.186185 | 1.179594934 | 0.233 |
| 57045 | TWSG1 | twisted gastrulation BMP signaling modulator 1 | −2.5573916 | 1.19989424 | 0.234 |
| 22891 | ZNF365 | zinc finger protein 365 | −1.1564493 | 1.11269613 | 0.256 |
| 2635 | GBP3 | guanylate binding protein 3 | −1.5946016 | 1.672916793 | 0.26 |
| 56913 | C1GALT1 | core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 | −1.5604228 | 1.322049565 | 0.262 |
| 2697 | GJA1 | gap junction protein, alpha 1, 43kDa | −2.5297747 | 1.75081194 | 0.264 |
| 2004 | ELK3 | ELK3, ETS-domain protein (SRF accessory protein 2) | −1.2669431 | 1.489197891 | 0.264 |
| 5125 | PCSK5 | proprotein convertase subtilisin/kexin type 5 | −1.0345638 | 1.411697675 | 0.286 |
| 57157 | PHTF2 | putative homeodomain transcription factor 2 | −1.0247078 | 1.303249973 | 0.287 |
| 51762 | RAB8B | RAB8B, member RAS oncogene family | −1.4737062 | 1.194672398 | 0.299 |
| 3176 | HNMT | histamine N-methyltransferase | −1.017499 | 1.193174738 | 0.327 |
| 9644 | SH3PXD2A | SH3 and PX domains 2A | −1.789658 | 1.631376261 | 0.362 |
| 143903 | LAYN | layilin | −1.4523163 | 1.461949991 | 0.368 |
| 8876 | VNN1 | vanin 1 | −2.3857656 | 2.077747375 | 0.395 |
| 11329 | STK38 | serine/threonine kinase 38 | −1.2525054 | 1.275932579 | 0.403 |
| 9890 | LPPR4 | Lipid phosphate phosphatase-related protein type 4 | −2.102301 | 1.232443047 | 0.412 |
| 2313 | FLI1 | Fli-1 proto-oncogene, ETS transcription factor | −1.902698 | 1.132852853 | 0.428 |
| 6925 | TCF4 | transcription factor 4 | −1.1343423 | 1.6263018 | 0.43 |
| 10487 | CAP1 | CAP, adenylate cyclase-associated protein 1 (yeast) | −1.3670303 | 1.280092932 | 0.456 |
| 10687 | PNMA2 | paraneoplastic Ma antigen 2 | −1.3189737 | 2.060441736 | 0.471 |
| 55790 | CSGALNACT1 | chondroitin sulfate N-acetylgalactosaminyltransferase 1 | −1.5380203 | 1.083904879 | 0.482 |
| 79772 | MCTP1 | multiple C2 domains, transmembrane 1 | −2.0109892 | 1.206431617 | 0.483 |
| 89795 | NAV3 | neuron navigator 3 | −1.9344572 | 1.513845893 | 0.485 |
| 135892 | TRIM50 | tripartite motif containing 50 | −1.2579012 | 1.230273363 | 0.494 |
| 83716 | CRISPLD2 | cysteine-rich secretory protein LCCL domain containing 2 | −1.025413 | 1.461831106 | 0.494 |
| 6558 | SLC12A2 | solute carrier family 12 (sodium/potassium/chloride transporter), member 2 | −2.2465677 | 1.117656881 | 0.524 |
| 26064 | RAI14 | retinoic acid induced 14 | −1.1299313 | 1.438363973 | 0.562 |
| 6443 | SGCB | sarcoglycan, beta (43 kDa dystrophin-associated glycoprotein) | −2.1484566 | 1.119623109 | 0.624 |
| 92241 | RCSD1 | RCSD domain containing 1 | −1.6868691 | 1.19300128 | 0.645 |
| 3106 | HLA-B | major histocompatibility complex, class I, B | −1.2878256 | 1.410905751 | 0.658 |
| 5592 | PRKG1 | protein kinase, cGMP-dependent, type I | −1.2409348 | 1.102178721 | 0.668 |
| 9590 | AKAP12 | A kinase (PRKA) anchor protein 12 | −1.5021276 | 1.672466982 | 0.689 |
| 493869 | GPX8 | glutathione peroxidase 8 (putative) | −1.1502694 | 2.136870238 | 0.699 |
| 285203 | EOGT | EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase | −1.5562044 | 1.219417735 | 0.711 |
| 11010 | GLIPR1 | GLI pathogenesis-related 1 | −1.0720639 | 1.936762677 | 0.733 |
| 5167 | ENPP1 | ectonucleotide pyrophosphatase/phosphodiesterase 1 | −1.0345893 | 1.107172826 | 0.756 |
| 9615 | GDA | guanine deaminase | −1.0771136 | 1.280713291 | 0.803 |
| 1687 | DFNA5 | deafness, autosomal dominant 5 | −1.2998511 | 1.348055613 | 0.816 |
| 5552 | SRGN | serglycin | −1.7418242 | 1.491958762 | 0.867 |
| 9444 | QKI | QKI, KH domain containing, RNA binding | −1.3413125 | 1.553606699 | 0.871 |
| 84056 | KATNAL1 | katanin p60 subunit A-like 1 | −1.6962346 | 1.164463984 | 0.877 |
| 151887 | CCDC80 | coiled-coil domain containing 80 | −1.1713166 | 2.301304933 | 0.901 |
| 133418 | EMB | embigin | −1.0146027 | 1.447293274 | 0.963 |
| 2124 | EVI2B | ecotropic viral integration site 2B | −1.6717868 | 1.54469176 | 0.97 |
| 79625 | NDNF | neuron-derived neurotrophic factor | −1.3546426 | 1.114794439 | 0.983 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimomura, H.; Okada, R.; Tanaka, T.; Hozaka, Y.; Wada, M.; Moriya, S.; Idichi, T.; Kita, Y.; Kurahara, H.; Ohtsuka, T.; et al. Role of miR-30a-3p Regulation of Oncogenic Targets in Pancreatic Ductal Adenocarcinoma Pathogenesis. Int. J. Mol. Sci. 2020, 21, 6459. https://doi.org/10.3390/ijms21186459
Shimomura H, Okada R, Tanaka T, Hozaka Y, Wada M, Moriya S, Idichi T, Kita Y, Kurahara H, Ohtsuka T, et al. Role of miR-30a-3p Regulation of Oncogenic Targets in Pancreatic Ductal Adenocarcinoma Pathogenesis. International Journal of Molecular Sciences. 2020; 21(18):6459. https://doi.org/10.3390/ijms21186459
Chicago/Turabian StyleShimomura, Hiroki, Reona Okada, Takako Tanaka, Yuto Hozaka, Masumi Wada, Shogo Moriya, Tetsuya Idichi, Yoshiaki Kita, Hiroshi Kurahara, Takao Ohtsuka, and et al. 2020. "Role of miR-30a-3p Regulation of Oncogenic Targets in Pancreatic Ductal Adenocarcinoma Pathogenesis" International Journal of Molecular Sciences 21, no. 18: 6459. https://doi.org/10.3390/ijms21186459
APA StyleShimomura, H., Okada, R., Tanaka, T., Hozaka, Y., Wada, M., Moriya, S., Idichi, T., Kita, Y., Kurahara, H., Ohtsuka, T., & Seki, N. (2020). Role of miR-30a-3p Regulation of Oncogenic Targets in Pancreatic Ductal Adenocarcinoma Pathogenesis. International Journal of Molecular Sciences, 21(18), 6459. https://doi.org/10.3390/ijms21186459





