Markers of Angiogenesis, Lymphangiogenesis, and Epithelial–Mesenchymal Transition (Plasticity) in CIN and Early Invasive Carcinoma of the Cervix: Exploring Putative Molecular Mechanisms Involved in Early Tumor Invasion
Abstract
:1. Introduction
2. Results
2.1. Differential Gene Expression Analysis
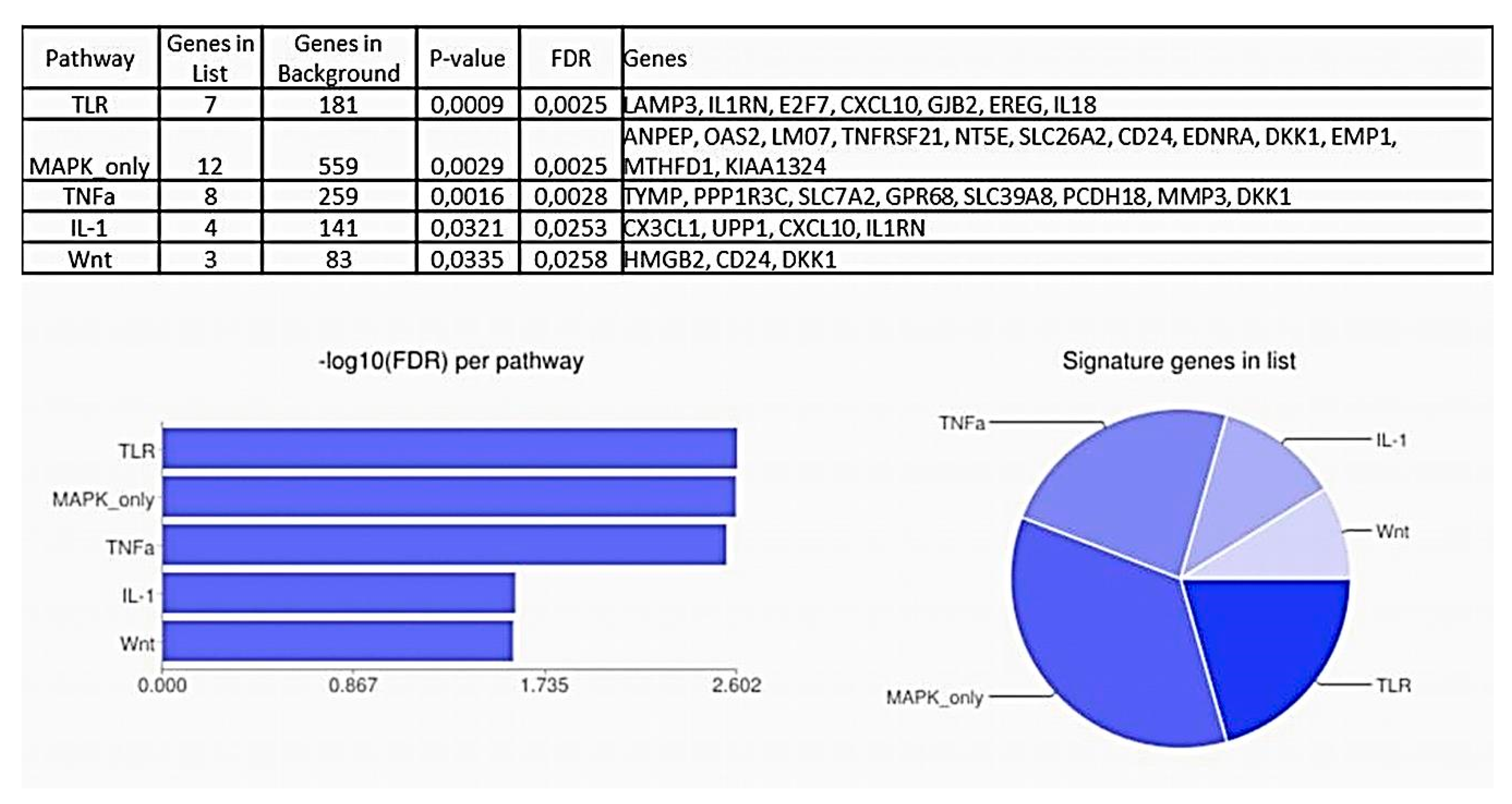
2.2. Pathway Analysis
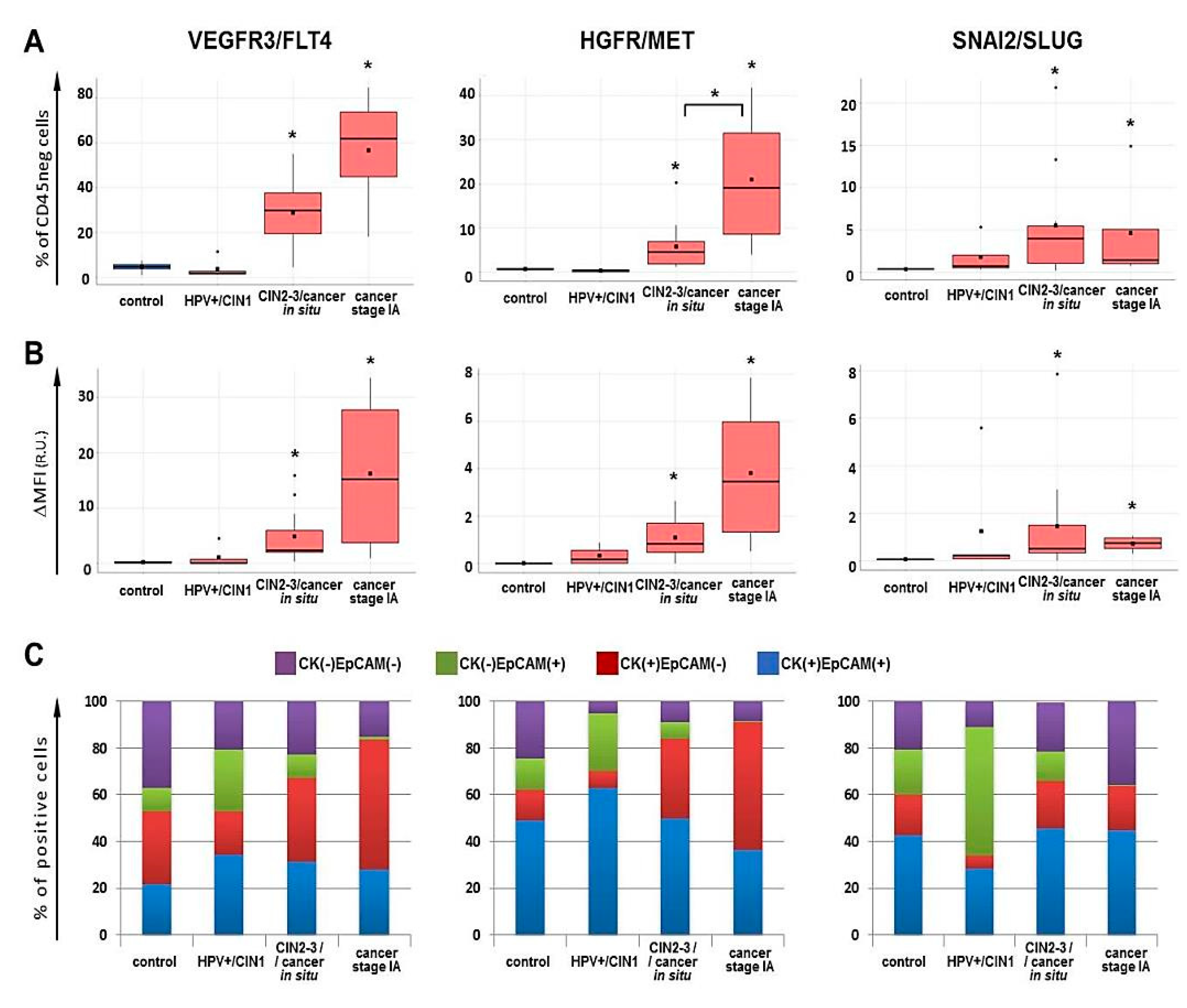
2.3. Flow Cytometry for FLT4/VEGFR3, MET/HGFR and SLUG/SNAI2 Expression
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. RNA Sequencing (RNA-Seq)
4.3. Real-Time Polymerase Chain Reaction (RT-PCR)
4.4. Western Blot
4.5. Tissue Dissociation and Flow Cytometry
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CIN | cervical intraepithelial neoplasia |
| CIS | carcinoma in situ |
| CK | cytokeratin |
| CPM | Counts Per Million reads mapped |
| CR | invasive cervical cancer specimens |
| DEGs | differentially expressed genes |
| ECM | extracellular matrix |
| EMT | epithelial-to-mesenchymal transition |
| EMP | epithelial-mesenchymal plasticity |
| FC | fold change |
| FDR | false discovery rate |
| FFPE | formalin-fixed paraffin-embedded |
| FIGO | International Federation of Gynecology and Obstetrics |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| HPV | human papillomavirus |
| IFN | interferon |
| ISG | interferon-stimulated genes |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MFI | Median Fluorescence Intensity |
| qPCR | quantitative polymerase chain reaction |
| RNA-Seq | RNA sequencing |
| TILs | tumor-infiltrating lymphocytes |
References
- Paduch, R. The role of lymphangiogenesis and angiogenesis in tumor metastasis. Cell Oncol. 2016, 39, 397–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomida, C.; Yamagishi, N.; Nagano, H.; Uchida, T.; Ohno, A.; Hirasaka, K.; Nikawa, T.; Teshima-Kondo, S. Antiangiogenic agent sunitinib induces epithelial to mesenchymal transition and accelerates motility of colorectal cancer cells. J. Med. Investig. 2017, 64, 250–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufies, M.; Giuliano, S.; Ambrosetti, D.; Claren, A.; Ndiaye, P.D.; Mastri, M.; Moghrabi, W.; Cooley, L.S.; Ettaiche, M.; Chamorey, E.; et al. Sunitinib Stimulates Expression of VEGFC by Tumor Cells and Promotes Lymphangiogenesis in Clear Cell Renal Cell Carcinomas. Cancer Res. 2017, 77, 1212–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Xu, C.; Hong, S.; Xia, X.; Cao, Y.; McDermott, J.; Mu, Y.; Han, J.J. Immune Cell Types and Secreted Factors Contributing to Inflammation-to-Cancer Transition and Immune Therapy Response. Cell Rep. 2019, 26, 1965–1977.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anschau, F.; da Silva, M.C.; Torrens, M.C.; Guimarães Gonçalves, M.A. Microinvasive Carcinoma of the Cervix. In Book Topics on Cervical Cancer with an Advocacy for Prevention; Rajkumar, R., Ed.; InTech: London, UK, 2012; pp. 131–138. [Google Scholar] [CrossRef] [Green Version]
- Masterson, L.; Sorgeloos, F.; Winder, D.; Lechner, M.; Marker, A.; Malhotra, S.; Sudhoff, H.; Jani, P.; Goon, P.; Sterling, J. Deregulation of SYCP2 predicts early stage human papillomavirus-positive oropharyngeal carcinoma: A prospective whole transcriptome analysis. Cancer Sci. 2015, 106, 1568–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, S.H.; Choi, Y.J.; Kim, M.S.; Baek, I.P.; Lee, S.H.; Lee, A.W.; Hur, S.Y.; Kim, T.M.; Lee, S.H.; Chung, Y.J. Progression of naive intraepithelial neoplasia genome to aggressive squamous cell carcinoma genome of uterine cervix. Oncotarget 2015, 6, 4385–4393. [Google Scholar] [CrossRef]
- Royse, K.E.; Zhi, D.; Conner, M.G.; Clodfelder-Miller, B.; Srinivasasainagendra, V.; Vaughan, L.K.; Skibola, C.F.; Crossman, D.K.; Levy, S.; Shrestha, S. Differential Gene Expression Landscape of Co-Existing Cervical Pre-Cancer Lesions Using RNA-seq. Front. Oncol. 2014, 4, 339. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Feng, M.; Li, X.; Zhong, P.; Guo, A.; Chen, G.; Xu, Q.; Ye, Y. Transcriptome profiling of cancer and normal tissues from cervical squamous cancer patients by deep sequencing. Mol. Med. Rep. 2017, 16, 2075–2088. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Feng, M.; Chen, G.; Zhou, Z.; Li, J.; Ye, Y. Characterization of the microRNA profile in early-stage cervical squamous cell carcinoma by next-generation sequencing. Oncol. Rep. 2017, 37, 1477–1486. [Google Scholar] [CrossRef]
- Rofstad, E.K.; Huang, R.; Galappathi, K.; Andersen, L.M.; Wegner, C.S.; Hauge, A.; Gaustad, J.V.; Simonsen, T.G. Functional intratumoral lymphatics in patient-derived xenograft models of squamous cell carcinoma of the uterine cervix: Implications for lymph node metastasis. Oncotarget 2016, 7, 56986–56997. [Google Scholar] [CrossRef] [Green Version]
- Ou, J.; Guan, D.; Yang, Y. Non-contact co-culture with human vascular endothelial cells promotes epithelial-to-mesenchymal transition of cervical cancer SiHa cells by activating the NOTCH1/LOX/SNAIL pathway. Cell Mol. Biol. Lett. 2019, 24, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gius, D.; Funk, M.C.; Chuang, E.Y.; Feng, S.; Huettner, P.C.; Nguyen, L.; Bradbury, C.M.; Mishra, M.; Gao, S.; Buttin, B.M.; et al. Profiling microdissected epithelium and stroma to model genomic signatures for cervical carcinogenesis accommodating for covariates. Cancer Res. 2007, 67, 7113–7123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatla, N.; Berek, J.S.; Cuello Fredes, M.; Denny, L.A.; Grenman, S.; Karunaratne, K.; Kehoe, S.T.; Konishi, I.; Olawaiye, A.B.; Prat, J.; et al. Revised FIGO staging for carcinoma of the cervix uteri. Int. J. Gynaecol. Obstet. 2019, 145, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Klymenko, T.; Gu, Q.; Herbert, I.; Stevenson, A.; Iliev, V.; Watkins, G.; Pollock, C.; Bhatia, R.; Cuschieri, K.; Herzyk, P.; et al. RNA-Seq Analysis of Differentiated Keratinocytes Reveals a Massive Response to Late Events during Human Papillomavirus 16 Infection, Including Loss of Epithelial Barrier Function. J. Virol. 2017, 91, e01001-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, X.; Huang, O.; Jiang, M.; Xie, Z.; Chen, D.; Zhang, X. Prognostic value of ephrin B receptors in breast cancer: An online survival analysis using the microarray data of 3,554 patients. Oncol. Lett. 2019, 18, 742–750. [Google Scholar] [CrossRef] [Green Version]
- Sato, S.; Vasaikar, S.; Eskaros, A.; Kim, Y.; Lewis, J.S.; Zhang, B.; Zijlstra, A.; Weaver, A.M. EPHB2 carried on small extracellular vesicles induces tumor angiogenesis via activation of ephrin reverse signaling. JCI Insight 2019, 4, e132447. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Liu, W.; Cai, J.; Li, M.; Gao, Y.; Lin, W.; Li, Z. EphB2 promotes cervical cancer progression by inducing epithelial-mesenchymal transition. Hum. Pathol. 2014, 45, 372–381. [Google Scholar] [CrossRef]
- Wang, Y.; Xuan, Z.; Wang, B.; Zhang, D.; Zhang, C.; Wang, J.; Sun, Y. EphA3 Downregulation by Hypermethylation Associated with Lymph Node Metastasis and TNM Stage in Colorectal Cancer. Dig. Dis. Sci. 2019, 64, 1514–1522. [Google Scholar] [CrossRef]
- Chen, X.; Lu, B.; Ma, Q.; Ji, C.D.; Li, J.Z. EphA3 inhibits migration and invasion of esophageal cancer cells by activating the mesenchymal-epithelial transition process. Int. J. Oncol. 2019, 54, 722–732. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Yu, C.L.; Zheng, Y. NSD2 inhibition suppresses metastasis in cervical cancer by promoting TGF-β/TGF-βRI/SMADs signaling. Biochem. Biophys. Res. Commun. 2019, 519, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, H.; Liang, Z. circ-MYBL2 Serves as A Sponge For miR-361-3p Promoting Cervical Cancer Cells Proliferation and Invasion. Onco Targets Ther. 2019, 12, 9957–9964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Yang, S.; Xu, J.; Lu, W.; Xie, X. Transcriptome sequencing profiles of cervical cancer tissues and SiHa cells. Funct. Integr. Genom. 2020, 20, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Chen, H.; Qiao, L.; Zhang, W.; Zheng, J.; Zhao, W.; Chen, J.J.; Zhang, W. CIP2A facilitates the G1/S cell cycle transition via B-Myb in human papillomavirus 16 oncoprotein E6-expressing cells. J. Cell. Mol. Med. 2018, 22, 4150–4160. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, A.; Macdonald, A. Stathmin drives virus-induced metastasis. Oncotarget 2015, 6, 32289–32290. [Google Scholar] [CrossRef]
- Irudayam, J.I.; Contreras, D.; Spurka, L.; Subramanian, A.; Allen, J.; Ren, S.; Kanagavel, V.; Nguyen, Q.; Ramaiah, A.; Ramamoorthy, K.; et al. Characterization of type I interferon pathway during hepatic differentiation of human pluripotent stem cells and hepatitis C virus infection. Stem Cell Res. 2015, 15, 354–364. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.Y.; Zhang, H.; Hu, Y.J.; Chen, Y.W.; Zhao, X.N. Identification of key genes associated with cervical cancer by comprehensive analysis of transcriptome microarray and methylation microarray. Oncol. Lett. 2016, 12, 473–478. [Google Scholar] [CrossRef] [Green Version]
- Narayan, G.; Goparaju, C.; Arias-Pulido, H.; Kaufmann, A.M.; Schneider, A.; Dürst, M.; Mansukhani, M.; Pothuri, B.; Murty, V.V. Promoter hypermethylation-mediated inactivation of multiple Slit-Robo pathway genes in cervical cancer progression. Mol. Cancer. 2006, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Yoon, Y.S.; Chung, J.H. Epigenetic silencing of the WNT antagonist DICKKOPF-1 in cervical cancer cell lines. Gynecol. Oncol. 2008, 109, 270–274. [Google Scholar] [CrossRef]
- Brebi, P.; Hoffstetter, R.; Andana, A.; Ili, C.G.; Saavedra, K.; Viscarra, T.; Retamal, J.; Sanchez, R.; Roa, J.C. Evaluation of ZAR1 and SFRP4 methylation status as potentials biomarkers for diagnosis in cervical cancer: Exploratory study phase I. Biomarkers 2014, 19, 181–188. [Google Scholar] [CrossRef]
- Chakraborty, C.; Dutta, S.; Mukherjee, N.; Samadder, S.; Roychowdhury, A.; Roy, A.; Mondal, R.K.; Basu, P.; Roychoudhury, S.; Panda, C.K. Inactivation of PTCH1 is associated with the development of cervical carcinoma: Clinical and prognostic implication. Tumour Biol. 2015, 36, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Beckenkamp, A.; Willig, J.B.; Santana, D.B.; Nascimento, J.; Paccez, J.D.; Zerbini, L.F.; Bruno, A.N.; Pilger, D.A.; Wink, M.R.; Buffon, A. Differential Expression and Enzymatic Activity of DPPIV/CD26 Affects Migration Ability of Cervical Carcinoma Cells. PLoS ONE 2015, 10, e0134305. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, J.; Wang, H.; Jiang, Y.; Wan, Y.; Xia, Y.; Cheng, W. Identification of crucial aberrantly methylated and differentially expressed genes related to cervical cancer using an integrated bioinformatics analysis. Biosci. Rep. 2020, 40, BSR20194365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ge, Y.; Cheng, Q.; Zhang, Q.; Fang, L.; Zheng, J. Decorin is a pivotal effector in the extracellular matrix and tumour microenvironment. Oncotarget 2018, 9, 5480–5491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, T.; Terai, Y.; Ohmichi, M. Association of matrix metalloproteinase-9 and decorin expression with the infiltration of cervical cancer. Oncol. Lett. 2019, 17, 1306–1312. [Google Scholar] [CrossRef] [Green Version]
- Ahmat Amin, M.K.B.; Shimizu, A.; Ogita, H. The Pivotal Roles of the Epithelial Membrane Protein Family in Cancer Invasiveness and Metastasis. Cancers 2019, 11, 1620. [Google Scholar] [CrossRef] [Green Version]
- Sun, G.G.; Lu, Y.F.; Fu, Z.Z.; Cheng, Y.J.; Hu, W.N. EMP1 inhibits nasopharyngeal cancer cell growth and metastasis through induction apoptosis and angiogenesis. Tumour Biol. 2014, 35, 3185–3193. [Google Scholar] [CrossRef]
- Abdalla, Z.; Walsh, T.; Thakker, N.; Ward, C.M. Loss of epithelial markers is an early event in oral dysplasia and is observed within the safety margin of dysplastic and T1 OSCC biopsies. PLoS ONE 2017, 12, e0187449. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y. Identification of hub genes and key pathways associated with the progression of gynecological cancer. Oncol. Lett. 2019, 18, 6516–6524. [Google Scholar] [CrossRef] [Green Version]
- Yun, J.; Song, S.H.; Park, J.; Kim, H.P.; Yoon, Y.K.; Lee, K.H.; Han, S.W.; Oh, D.Y.; Im, S.A.; Bang, Y.J.; et al. Gene silencing of EREG mediated by DNA methylation and histone modification in human gastric cancers. Lab. Investig. 2012, 92, 1033–1044. [Google Scholar] [CrossRef]
- Liao, T.; Kaufmann, A.M.; Qian, X.; Sangvatanakul, V.; Chen, C.; Kube, T.; Zhang, G.; Albers, A.E. Susceptibility to cytotoxic T cell lysis of cancer stem cells derived from cervical and head and neck tumor cell lines. J. Cancer Res. Clin. Oncol. 2013, 139, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Okudela, K.; Woo, T.; Mitsui, H.; Suzuki, T.; Tajiri, M.; Sakuma, Y.; Miyagi, Y.; Tateishi, Y.; Umeda, S.; Masuda, M.; et al. Downregulation of ALDH1A1 expression in non-small cell lung carcinomas--its clinicopathologic and biological significance. Int. J. Clin. Exp. Pathol. 2013, 6, 1–12. [Google Scholar] [PubMed]
- He, Y.; Xiao, M.; Fu, H.; Chen, L.; Qi, L.; Liu, D.; Guo, P.; Chen, L.; Luo, Y.; Xiao, H.; et al. cPLA2α reversibly regulates different subsets of cancer stem cells transformation in cervical cancer. Stem Cells 2020, 38, 487–503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, J.; Zeng, Z.; Fan, J.; Huang, S.; Zhang, L.; Zhang, B.; Wang, X.; Feng, Y.; Ye, Z.; et al. lncRNA Rmst acts as an important mediator of BMP9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) by antagonizing Notch-targeting microRNAs. Aging (Albany NY) 2019, 11, 12476–12496. [Google Scholar] [CrossRef]
- Wang, L.; Liu, D.; Wu, X.; Zeng, Y.; Li, L.; Hou, Y.; Li, W.; Liu, Z. Long non-coding RNA (LncRNA) RMST in triple-negative breast cancer (TNBC): Expression analysis and biological roles research. J. Cell Physiol. 2018, 233, 6603–6612. [Google Scholar] [CrossRef]
- Yan, S.P.; Chu, D.X.; Qiu, H.F.; Xie, Y.; Wang, C.F.; Zhang, J.Y.; Li, W.C.; Guo, R.X. LncRNA LINC01305 silencing inhibits cell epithelial-mesenchymal transition in cervical cancer by inhibiting TNXB-mediated PI3K/Akt signalling pathway. J. Cell Mol. Med. 2019, 23, 2656–2666. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Hong, L.; Yang, W.; Zhao, Y.; Tan, A.; Li, Y. Co-expression network analysis of the lncRNAs and mRNAs associated with cervical cancer progression. Arch. Med. Sci. 2019, 15, 754–764. [Google Scholar] [CrossRef]
- Abhishek, S.; Palamadai Krishnan, S. Epidermal Differentiation Complex: A Review on Its Epigenetic Regulation and Potential Drug Targets. Cell J. 2016, 18, 1–6. [Google Scholar] [CrossRef]
- Siret, C.; Dobric, A.; Martirosyan, A.; Terciolo, C.; Germain, S.; Bonier, R.; Dirami, T.; Dusetti, N.; Tomasini, R.; Rubis, M.; et al. Cadherin-1 and cadherin-3 cooperation determines the aggressiveness of pancreatic ductal adenocarcinoma. Br. J. Cancer 2018, 118, 546–557. [Google Scholar] [CrossRef] [Green Version]
- Elisha, Y.; Kalchenko, V.; Kuznetsov, Y.; Geiger, B. Dual role of E-cadherin in the regulation of invasive collective migration of mammary carcinoma cells. Sci. Rep. 2018, 8, 4986. [Google Scholar] [CrossRef] [Green Version]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Fattet, L.; Jung, H.Y.; Matsumoto, M.W.; Aubol, B.E.; Kumar, A.; Adams, J.A.; Chen, A.C.; Sah, R.L.; Engler, A.J.; Pasquale, E.B.; et al. Matrix Rigidity Controls Epithelial-Mesenchymal Plasticity and Tumor Metastasis via a Mechanoresponsive EPHA2/LYN Complex. Dev. Cell 2020, 54, 302–316.e7. [Google Scholar] [CrossRef] [PubMed]
- Ieguchi, K.; Maru, Y. Roles of EphA1/A2 and ephrin-A1 in cancer. Cancer Sci. 2019, 110, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Jun, T.; Nie, Y.; Hao, J.; Fan, D. The Role of the Slit/Robo Signaling Pathway. J. Cancer 2019, 10, 2694–2705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, P.T.; Zeng, Z.; Salim, N.; Mattarollo, S.; Wells, J.W.; Leggatt, G.R. The Role of CXCR3 and Its Chemokine Ligands in Skin Disease and Cancer. Front. Med. 2018, 5, 271. [Google Scholar] [CrossRef] [PubMed]
- Karki, R.; Kanneganti, T.D. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer 2019, 9, 197–214. [Google Scholar] [CrossRef]
- Hsu, K.S.; Kao, H.Y. PML: Regulation and multifaceted function beyond tumor suppression. Cell Biosci. 2018, 8, 5. [Google Scholar] [CrossRef] [Green Version]
- Parikh, J.R.; Klinger, B.; Xia, Y.; Marto, J.A.; Blüthgen, N. Discovering causal signaling pathways through gene-expression patterns. Nucleic Acids Res. 2010, 38, W109–W117. [Google Scholar] [CrossRef]
- Yi, Y.; Fang, Y.; Wu, K.; Liu, Y.; Zhang, W. Comprehensive gene and pathway analysis of cervical cancer progression. Oncol. Lett. 2020, 19, 3316–3332. [Google Scholar] [CrossRef] [Green Version]
- Rong, C.; Muller, M.; Flechtenmacher, C.; Holzinger, D.; Dyckhoff, G.; Bulut, O.C.; Horn, D.; Plinkert, P.; Hess, J.; Affolter, A. Differential Activation of ERK Signaling in HPV-Related Oropharyngeal Squamous Cell Carcinoma. Cancers 2019, 11, 584. [Google Scholar] [CrossRef] [Green Version]
- Branca, M.; Ciotti, M.; Santini, D.; Bonito, L.D.; Benedetto, A.; Giorgi, C.; Paba, P.; Favalli, C.; Costa, S.; Agarossi, A.; et al. Activation of the ERK/MAP kinase pathway in cervical intraepithelial neoplasia is related to grade of the lesion but not to high-risk human papillomavirus, virus clearance, or prognosis in cervical cancer. Am. J. Clin. Pathol. 2004, 122, 902–911. [Google Scholar] [CrossRef]
- Shi, B.; Bao, J.; Liu, Y.; Shi, J. Death receptor 6 promotes ovarian cancer cell migration through KIF11. FEBS Open Biol. 2018, 8, 1497–1507. [Google Scholar] [CrossRef] [Green Version]
- Ooshio, T.; Irie, K.; Morimoto, K.; Fukuhara, A.; Imai, T.; Takai, Y. Involvement of LMO7 in the association of two cell-cell adhesion molecules, nectin and E-cadherin, through afadin and alpha-actinin in epithelial cells. J. Biol. Chem. 2004, 279, 31365–31373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Zhou, J.; Mei, S.; Wu, D.; Mu, Z.; Chen, B.; Xie, Y.; Ye, Y.; Liu, J. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J. Cell. Mol. Med. 2017, 21, 1228–1236. [Google Scholar] [CrossRef]
- Xie, Y.; Ostriker, A.C.; Jin, Y.; Hu, H.; Sizer, A.J.; Peng, G.; Morris, A.H.; Ryu, C.; Herzog, E.L.; Kyriakides, T.; et al. LMO7 Is a Negative Feedback Regulator of Transforming Growth Factor β Signaling and Fibrosis. Circulation 2019, 39, 679–693. [Google Scholar] [CrossRef] [PubMed]
- Bello, J.O.; Nieva, L.O.; Paredes, A.C.; Gonzalez, A.M.; Zavaleta, L.R.; Lizano, M. Regulation of the Wnt/β-Catenin Signaling Pathway by Human Papillomavirus E6 and E7 Oncoproteins. Viruses 2015, 7, 4734–4755. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traweek, S.T.; Liu, J.; Battifora, H. Keratin gene expression in non-epithelial tissues. Detection with polymerase chain reaction. Am. J. Pathol. 1993, 142, 1111–1118. [Google Scholar]
- Miettinen, M.; Fetsch, J.F. Distribution of keratins in normal endothelial cells and a spectrum of vascular tumors: Implications in tumor diagnosis. Hum. Pathol. 2000, 31, 1062–1067. [Google Scholar] [CrossRef]
- Ikeda, K.; Tate, G.; Suzuki, T.; Mitsuya, T. Coordinate expression of cytokeratin 8 and cytokeratin 17 immunohistochemical staining in cervical intraepithelial neoplasia and cervical squamous cell carcinoma: An immunohistochemical analysis and review of the literature. Gynecol. Oncol. 2008, 108, 598–602. [Google Scholar] [CrossRef]
- Lee, H.; Lee, H.; Cho, Y.K. Cytokeratin7 and cytokeratin19 expression in high grade cervical intraepithelial neoplasm and squamous cell carcinoma and their possible association in cervical carcinogenesis. Diagn. Pathol. 2017, 12, 18. [Google Scholar] [CrossRef] [Green Version]
- Trzpis, M.; McLaughlin, P.M.; de Leij, L.M.; Harmsen, M.C. Epithelial cell adhesion molecule: More than a carcinoma marker and adhesion molecule. Am. J. Pathol. 2007, 171, 386–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Punt, S.; Corver, W.E.; van der Zeeuw, S.A.; Kielbasa, S.M.; Osse, E.M.; Buermans, H.P.; de Kroon, C.D.; Jordanova, E.S.; Gorter, A. Whole-transcriptome analysis of flow-sorted cervical cancer samples reveals that B cell expressed TCL1A is correlated with improved survival. Oncotarget 2015, 6, 38681–38694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, N.; Yang, W.T.; Zheng, P.S. Slug inhibits the proliferation and tumor formation of human cervical cancer cells by up-regulating the p21/p27 proteins and down-regulating the activity of the Wnt/β-catenin signaling pathway via the trans-suppression Akt1/p-Akt1 expression. Oncotarget 2016, 7, 26152–26167. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Albert Einstein College of Medicine; Analytical Biological Services. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Nicol, A.F.; de Andrade, C.V.; Gomes, S.C., Jr.; Brusadelli, M.G.; Lodin, H.M.; Wells, S.I.; Nuovo, G.J. The distribution of novel biomarkers in carcinoma-in-situ, microinvasive, and squamous cell carcinoma of the uterine cervix. Ann. Diagn. Pathol. 2019, 38, 115–122. [Google Scholar] [CrossRef]
- Qiao, X.; Sun, Y.; Zou, Y.; Lin, J.; Xu, W.; Zong, Y.; Guan, X.; Zhang, R.; Wang, X. Immunohistochemical staining of podoplanin is helpful in determining the microinvasion of cervical squamous cell carcinoma. Ann. Diagn. Pathol. 2020, 46, 151493. [Google Scholar] [CrossRef]
- Lessi, F.; Scatena, C.; Aretini, P.; Menicagli, M.; Franceschi, S.; Naccarato, A.G.; Mazzanti, C.M. Molecular profiling of microinvasive breast cancer microenvironment progression. J. Transl. Med. 2019, 17, 187. [Google Scholar] [CrossRef]
- Kang, S.D.; Chatterjee, S.; Alam, S.; Salzberg, A.C.; Milici, J.; van der Burg, S.H.; Meyers, C. Effect of Productive Human Papillomavirus 16 Infection on Global Gene Expression in Cervical Epithelium. J. Virol. 2018, 92, e01261-18. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Klimentová, J.; Göckel-Krzikalla, E.; Ly, R.; Gmelin, N.; Hotz-Wagenblatt, A.; Řehulková, H.; Stulík, J.; Rösl, F.; Niebler, M. Combined Transcriptome and Proteome Analysis of Immortalized Human Keratinocytes Expressing Human Papillomavirus 16 (HPV16) Oncogenes Reveals Novel Key Factors and Networks in HPV-Induced Carcinogenesis. mSphere 2019, 4, e00129-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Peng, L.; Zhang, Y.; Chen, S.; Lei, Q.; Li, G.; Zhang, C. Identification of Key Genes and Pathways in Cervical Cancer by Bioinformatics Analysis. Int. J. Med. Sci. 2019, 16, 800–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kori, M.; Yalcin Arga, K. Potential biomarkers and therapeutic targets in cervical cancer: Insights from the meta-analysis of transcriptomics data within network biomedicine perspective. PLoS ONE 2018, 13, e0200717. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.C.; Korkut, A.; Kanchi, R.S.; Hegde, A.M.; Lenoir, W.; Liu, W.; Liu, Y.; Fan, H.; Shen, H.; Ravikumar, V.; et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell 2018, 33, 690–705.e9. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Ke, J.; Liu, J.; Su, J. DNA methylation data-based molecular subtype classification related to the prognosis of patients with cervical cancer. J. Cell. Biochem. 2020, 121, 2713–2724. [Google Scholar] [CrossRef]
- Beamish, I.V.; Hinck, L.; Kennedy, T.E. Making Connections: Guidance Cues and Receptors at Nonneural Cell-Cell Junctions. Cold Spring Harb. Perspect. Biol. 2018, 10, a029165. [Google Scholar] [CrossRef] [Green Version]
- Rozbesky, D.; Jones, E.Y. Cell Guidance Ligands, Receptors and Complexes—Orchestrating Signalling in Time and Space. Curr. Opin. Struct. Biol. 2020, 61, 79–85. [Google Scholar] [CrossRef]
- Mitra, S.; Mazumder-Indra, D.; Mondal, R.K.; Basu, P.S.; Roy, A.; Roychoudhury, S.; Panda, C.K. Inactivation of SLIT2-ROBO1/2 pathway in premalignant lesions of uterine cervix: Clinical and prognostic significances. PLoS ONE 2012, 7, e38342. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.J.; Schönleben, F.; Banuchi, V.E.; Qiu, W.; Close, L.G.; Assaad, A.M.; Su, G.H. Multiple tumor-suppressor genes on chromosome 3p contribute to head and neck squamous cell carcinoma tumorigenesis. Cancer Biol. Ther. 2010, 10, 689–693. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Xu, H.; Cao, G.; Wu, Z.; Wang, J. Loss of EphA3 Protein Expression Is Associated With Advanced TNM Stage in Clear-Cell Renal Cell Carcinoma. Clin. Genitourin. Cancer. 2017, 15, e169–e173. [Google Scholar] [CrossRef]
- Li, S.; Wu, Z.; Ma, P.; Xu, Y.; Chen, Y.; Wang, H.; He, P.; Kang, Z.; Yin, L.; Zhao, Y.; et al. Ligand-dependent EphA7 signaling inhibits prostate tumor growth and progression. Cell Death Dis. 2017, 8, e3122. [Google Scholar] [CrossRef] [Green Version]
- Di, W.; Weinan, X.; Xin, L.; Zhiwei, Y.; Xinyue, G.; Jinxue, T.; Mingqi, L. Long noncoding RNA SNHG14 facilitates colorectal cancer metastasis through targeting EZH2-regulated EPHA7. Cell Death Dis. 2019, 10, 514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.Q.; Zhang, J.Y.; Bai, C.Y.; Xu, X.E.; Wu, J.Y.; Chen, B.; Wu, Z.Y.; Wang, S.H.; Shen, J.; Shen, J.H.; et al. Low EphA7 Expression Correlated with Lymph Node Metastasis and Poor Prognosis of Patients with Esophageal Squamous Cell Carcinoma. Acta Histochem. Cytochem. 2015, 48, 75–81. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.Y.; Patel, O.; Janes, P.W.; Murphy, J.M.; Lucet, I.S. Eph receptor signalling: From catalytic to non-catalytic functions. Oncogene 2019, 38, 6567–6584. [Google Scholar] [CrossRef] [Green Version]
- Neufeld, G.; Mumblat, Y.; Smolkin, T.; Toledano, S.; Nir-Zvi, I.; Ziv, K.; Kessler, O. The role of the semaphorins in cancer. Cell Adh. Migr. 2016, 10, 652–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toledano, S.; Nir-Zvi, I.; Engelman, R.; Kessler, O.; Neufeld, G. Class-3 Semaphorins and Their Receptors: Potent Multifunctional Modulators of Tumor Progression. Int. J. Mol. Sci. 2019, 20, 556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Klamer, B.; Li, J.; Fernandez, S.; Li, L. A pan-cancer study of class-3 semaphorins as therapeutic targets in cancer. BMC Med. Genomics 2020, 13 (Suppl. 5), 45. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.C.; Yang, J.; Liu, D.; Wu, M.F.; Qiao, L.; Wang, J.N.; Ma, Q.F.; Zeng, Z.; Ye, S.M.; Guo, E.S.; et al. Tumor-associated Lymphatic Endothelial Cells Promote Lymphatic Metastasis By Highly Expressing and Secreting SEMA4C. Clin. Cancer Res. 2017, 23, 214–224. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Yu, Y.; Xiong, Z.; Chen, H.; Tan, B.; Hu, H. Downregulation of SEMA4C Inhibit Epithelial-Mesenchymal Transition (EMT) and the Invasion and Metastasis of Cervical Cancer Cells via Inhibiting Transforming Growth Factor-beta 1 (TGF-β1)-Induced Hela cells p38 Mitogen-Activated Protein Kinase (MAPK) Activation. Med. Sci. Monit. 2020, 26, e918123. [Google Scholar] [CrossRef]
- Jing, L.; Bo, W.; Yourong, F.; Tian, W.; Shixuan, W.; Mingfu, W. Sema4C mediates EMT inducing chemotherapeutic resistance of miR-31-3p in cervical cancer cells. Sci. Rep. 2019, 9, 17727. [Google Scholar] [CrossRef]
- Ding, Y.; He, D.; Florentin, D.; Frolov, A.; Hilsenbeck, S.; Ittmann, M.; Kadmon, D.; Miles, B.; Rowley, D.; Ayala, G. Semaphorin 4F as a critical regulator of neuroepithelial interactions and a biomarker of aggressive prostate cancer. Clin. Cancer Res. 2013, 19, 6101–6111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.J.; Guo, J.L.; Wang, H.K.; Xu, X. Semaphorin-7A contributes to growth, migration and invasion of oral tongue squamous cell carcinoma through TGF-β-mediated EMT signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Elder, A.M.; Tamburini, B.A.J.; Crump, L.S.; Black, S.A.; Wessells, V.M.; Schedin, P.J.; Borges, V.F.; Lyons, T.R. Semaphorin 7A Promotes Macrophage-Mediated Lymphatic Remodeling during Postpartum Mammary Gland Involution and in Breast Cancer. Cancer Res. 2018, 78, 6473–6485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangino, G.; Chiantore, M.V.; Iuliano, M.; Fiorucci, G.; Romeo, G. Inflammatory microenvironment and human papillomavirus-induced carcinogenesis. Cytokine Growth Factor Rev. 2016, 30, 103–111. [Google Scholar] [CrossRef]
- Szewczyk, G.; Maciejewski, T.M.; Szukiewicz, D. Current progress in the inflammatory background of angiogenesis in gynecological cancers. Inflamm. Res. 2019, 68, 247–260. [Google Scholar] [CrossRef] [Green Version]
- Garnier, L.; Gkountidi, A.O.; Hugues, S. Tumor-Associated Lymphatic Vessel Features and Immunomodulatory Functions. Front. Immunol. 2019, 10, 720. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.C.; Hwang, W.T.; Davis, C.; Deshpande, C.; Jeffries, S.; Rajpurohit, Y.; Krishna, V.; Smirnov, D.; Verona, R.; Lorenzi, M.V.; et al. Gene signatures of tumor inflammation and epithelial-to-mesenchymal transition (EMT) predict responses to immune checkpoint blockade in lung cancer with high accuracy. Lung Cancer 2020, 139, 1–8. [Google Scholar] [CrossRef]
- Chen, Y.L.; Wu, W.L.; Jang, C.W.; Yen, Y.C.; Wang, S.H.; Tsai, F.Y.; Shen, Y.Y.; Chen, Y.W. Interferon-stimulated gene 15 modulates cell migration by interacting with Rac1 and contributes to lymph node metastasis of oral squamous cell carcinoma cells. Oncogene 2019, 38, 4480–4495. [Google Scholar] [CrossRef]
- Aguilar-Cazares, D.; Chavez-Dominguez, R.; Carlos-Reyes, A.; Lopez-Camarillo, C.; Hernadez de la Cruz, O.N.; Lopez-Gonzalez, J.S. Contribution of Angiogenesis to Inflammation and Cancer. Front. Oncol. 2019, 9, 1399. [Google Scholar] [CrossRef] [Green Version]
- Smola, S. Immune deviation and cervical carcinogenesis. Papillomavirus Res. 2019, 7, 164–167. [Google Scholar] [CrossRef]
- Pan, L.; Yan, G.; Chen, W.; Sun, L.; Wang, J.; Yang, J. Distribution of circulating tumor cell phenotype in early cervical cancer. Cancer Manag. Res. 2019, 11, 5531–5536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.; Qi, S.; Wang, P.; Li, W.; Liu, C.; Li, F. Diagnosis and Prognostic Significance of c-Met in Cervical Cancer: A Meta-Analysis. Dis. Markers 2016, 2016, 6594016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boromand, N.; Hasanzadeh, M.; ShahidSales, S.; Farazestanian, M.; Gharib, M.; Fiuji, H.; Behboodi, N.; Ghobadi, N.; Hassanian, S.M.; Ferns, G.A.; et al. Clinical and prognostic value of the C-Met/HGF signaling pathway in cervical cancer. J. Cell. Physiol. 2018, 233, 4490–4496. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.C.; Pan, M.R.; Hung, W.C. Two Birds, One Stone: Double Hits on Tumor Growth and Lymphangiogenesis by Targeting Vascular Endothelial Growth Factor Receptor 3. Cells 2019, 8, 270. [Google Scholar] [CrossRef] [Green Version]
- Titmarsh, H.F.; O’Connor, R.; Dhaliwal, K.; Akram, A.R. The Emerging Role of the c-MET-HGF Axis in Non-small Lung Cancer Tumor Immunology and Immunotherapy. Front. Oncol. 2020, 10, 54. [Google Scholar] [CrossRef] [Green Version]
- Papaccio, F.; Della Corte, C.M.; Viscardi, G.; Di Liello, R.; Esposito, G.; Sparano, F.; Ciardiello, F.; Morgillo, F. HGF/MET and the Immune System: Relevance for Cancer Immunotherapy. Int. J. Mol. Sci. 2018, 19, 3595. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.F.; Wang, S.S.; Tang, Y.J.; Chen, Y.; Zheng, M.; Tang, Y.L.; Liang, X.H. The Double-Edged Sword-How Human Papillomaviruses Interact with Immunity in Head and Neck Cancer. Front. Immunol. 2019, 10, 653. [Google Scholar] [CrossRef]
- Campbell, J.D.; Yau, C.; Bowlby, R.; Liu, Y.; Brennan, K.; Fan, H.; Taylor, A.M.; Wang, C.; Walter, V.; Akbani, R.; et al. Genomic, Pathway Network, and Immunologic Features Distinguishing Squamous Carcinomas. Cell. Rep. 2018, 23, 194–212.e6. [Google Scholar] [CrossRef] [Green Version]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef] [Green Version]
- Piersma, S.J.; Jordanova, E.S.; van Poelgeest, M.I.; Kwappenberg, K.M.; van der Hulst, J.M.; Drijfhout, J.W.; Melief, C.J.; Kenter, G.G.; Fleuren, G.J.; Offringa, R.; et al. High number of intraepithelial CD8+ tumor-infiltrating lymphocytes is associated with the absence of lymph node metastases in patients with large early-stage cervical cancer. Cancer Res. 2007, 67, 354–361. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.B.; Lu, Y.; Huang, J.L.; Long, Y.; Yao, D.S. Prognostic genes in the tumor microenvironment in cervical squamous cell carcinoma. Aging (Albany NY) 2019, 11, 10154–10166. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.C.; Ismail, M.P.; Duski, D.R.; Othman, N.H.; Bhavaraju, V.M.; Ankathil, R. Identification of Optimal Reference Genes for Normalization of RT-qPCR Data in Cancerous and Non-Cancerous Tissues of Human Uterine Cervix. Cancer Investig. 2017, 35, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Leitão, M.C.; Coimbra, E.C.; de Lima, R.C.; Guimarães, M.L.; Heráclio, S.A.; Silva Neto, J.C.; de Freitas, A.C. Quantifying mRNA and microRNA with qPCR in cervical carcinogenesis: A validation of reference genes to ensure accurate data. PLoS ONE 2014, 9, e111021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rihani, A.; Van Maerken, T.; Pattyn, F.; Van Peer, G.; Beckers, A.; De Brouwer, S.; Kumps, C.; Mets, E.; Van der Meulen, J.; Rondou, P.; et al. Effective Alu repeat based RT-Qpcr normalization in cancer cell perturbation experiments. PLoS ONE 2013, 8, e71776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roszik, J.; Ring, K.L.; Wani, K.M.; Lazar, A.J.; Yemelyanova, A.V.; Soliman, P.T.; Frumovitz, M.; Jazaeri, A.A. Gene Expression Analysis Identifies Novel Targets for Cervical Cancer Therapy. Front. Immunol. 2018, 9, 2102. [Google Scholar] [CrossRef] [Green Version]
- Ndiaye, P.D.; Dufies, M.; Giuliano, S.; Douguet, L.; Grépin, R.; Durivault, J.; Lenormand, P.; Glisse, N.; Mintcheva, J.; Vouret-Craviari, V.; et al. VEGFC acts as a double-edged sword in renal cell carcinoma aggressiveness. Theranostics 2019, 9, 661–675. [Google Scholar] [CrossRef]
- Peng, J.; Xu, H.; Chen, Y.; Wang, W.; Zhu, L.; Shao, Y.; Wang, J. Screening for therapeutic targets of tumor angiogenesis signatures in 31 cancer types and potential insights. Biochem. Biophys. Res. Commun. 2019, 508, 465–471. [Google Scholar] [CrossRef]















| Sample ID 1 | Degree/Stage |
|---|---|
| Norm | morphologically normal cervical epithelium |
| CIN_1 | cervical intraepithelial neoplasia grade 3 (CIN3) |
| CIN_2 | CIN3 (carcinoma in situ, CIS) |
| CIN_3 | CIN2/3 (high-grade squamous intraepithelial lesion) |
| CIN_4 | CIN3 (CIS) |
| CR_1 | invasive carcinoma at IA1 stage |
| CR_2 | invasive carcinoma at IB1 stage |
| CR_3 | invasive carcinoma at IA1 stage |
| CR_4 | invasive carcinoma at IA1 stage |
| CR_5 | invasive carcinoma at IB1 stage |
| CR_6 | invasive carcinoma at IB2/IIA1 |
| CR_7 | invasive carcinoma at IIB stage |
| Gene in List | Total Genes | Functional Category | Enrichment FDR | Genes |
|---|---|---|---|---|
| 5 | 36 | DNA replication | 0.0011 | LIG1, POLA1, MCM3, MCM5, RFC2/4 |
| 6 | 166 | Influenza A | 0.024 | CXCL10, OAS, IRF9, CIITA, IL18, PML |
| 7 | 181 | Axon guidance | 0.026 | EPHA, EPHB, SLIT2, SLIT3, UNC5, PTCH1, BMPR1B |
| 5 | 57 | Pyrimidine metabolism | 0.029 | ENPP3, NT5E, CDA, UPP1, TYMP |
| 4 | 69 | Renin secretion | 0.046 | EDN, EDNRA, CLCA, ADCYAP1R1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurmyshkina, O.; Kovchur, P.; Schegoleva, L.; Volkova, T. Markers of Angiogenesis, Lymphangiogenesis, and Epithelial–Mesenchymal Transition (Plasticity) in CIN and Early Invasive Carcinoma of the Cervix: Exploring Putative Molecular Mechanisms Involved in Early Tumor Invasion. Int. J. Mol. Sci. 2020, 21, 6515. https://doi.org/10.3390/ijms21186515
Kurmyshkina O, Kovchur P, Schegoleva L, Volkova T. Markers of Angiogenesis, Lymphangiogenesis, and Epithelial–Mesenchymal Transition (Plasticity) in CIN and Early Invasive Carcinoma of the Cervix: Exploring Putative Molecular Mechanisms Involved in Early Tumor Invasion. International Journal of Molecular Sciences. 2020; 21(18):6515. https://doi.org/10.3390/ijms21186515
Chicago/Turabian StyleKurmyshkina, Olga, Pavel Kovchur, Ludmila Schegoleva, and Tatyana Volkova. 2020. "Markers of Angiogenesis, Lymphangiogenesis, and Epithelial–Mesenchymal Transition (Plasticity) in CIN and Early Invasive Carcinoma of the Cervix: Exploring Putative Molecular Mechanisms Involved in Early Tumor Invasion" International Journal of Molecular Sciences 21, no. 18: 6515. https://doi.org/10.3390/ijms21186515




