Silencing of HvGSK1.1—A GSK3/SHAGGY-Like Kinase–Enhances Barley (Hordeum vulgare L.) Growth in Normal and in Salt Stress Conditions
Abstract
:1. Introduction
2. Results
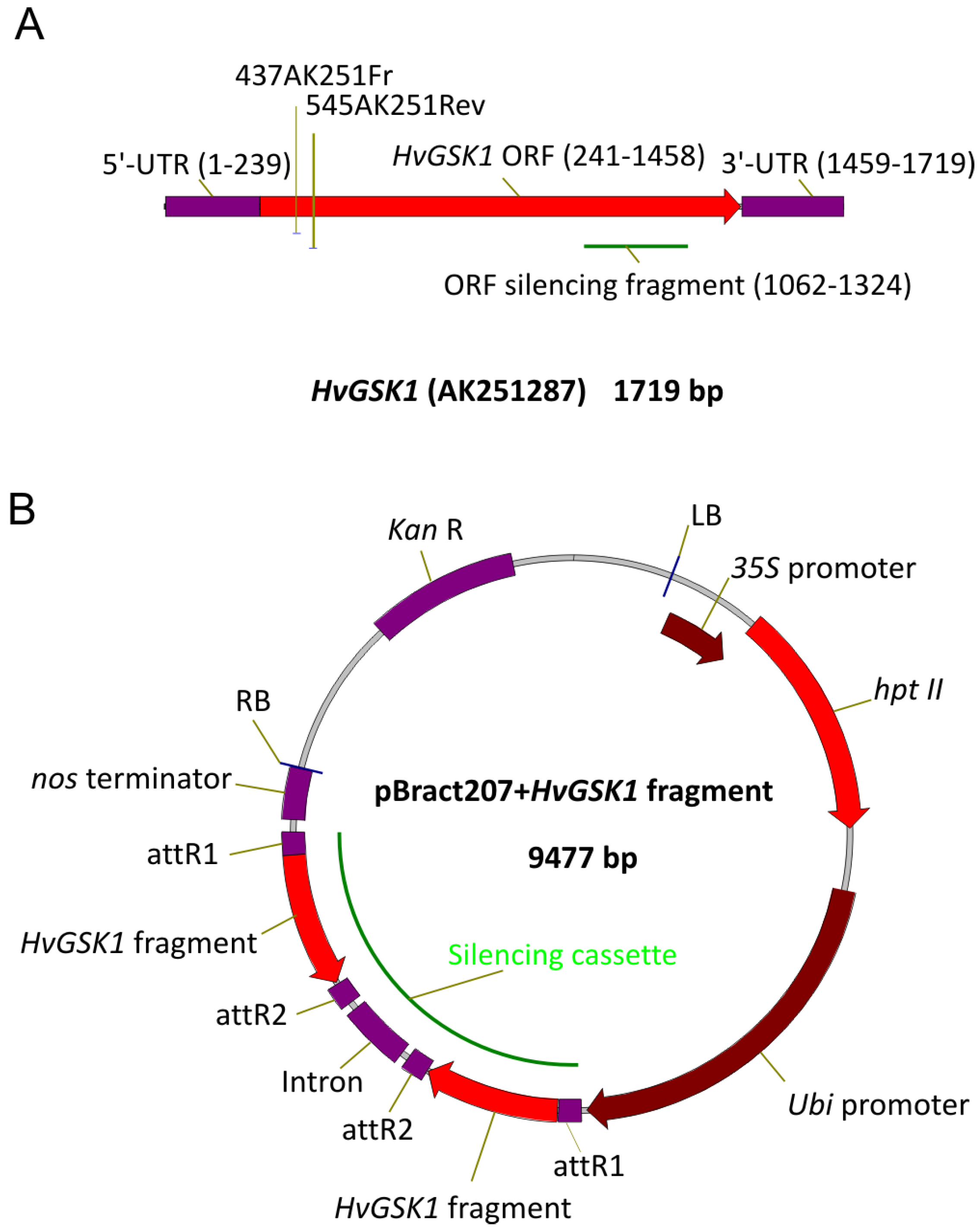
2.1. Silencing Vector Construction and Agro-Based Transformation of Immature Barley Embryos
2.2. Silencing of HvGSK1.1 Modifies Expression of GSK Paralogs in Plants Grown in Normal and Salt Stress Conditions
2.3. Leaf Inclination Biotest
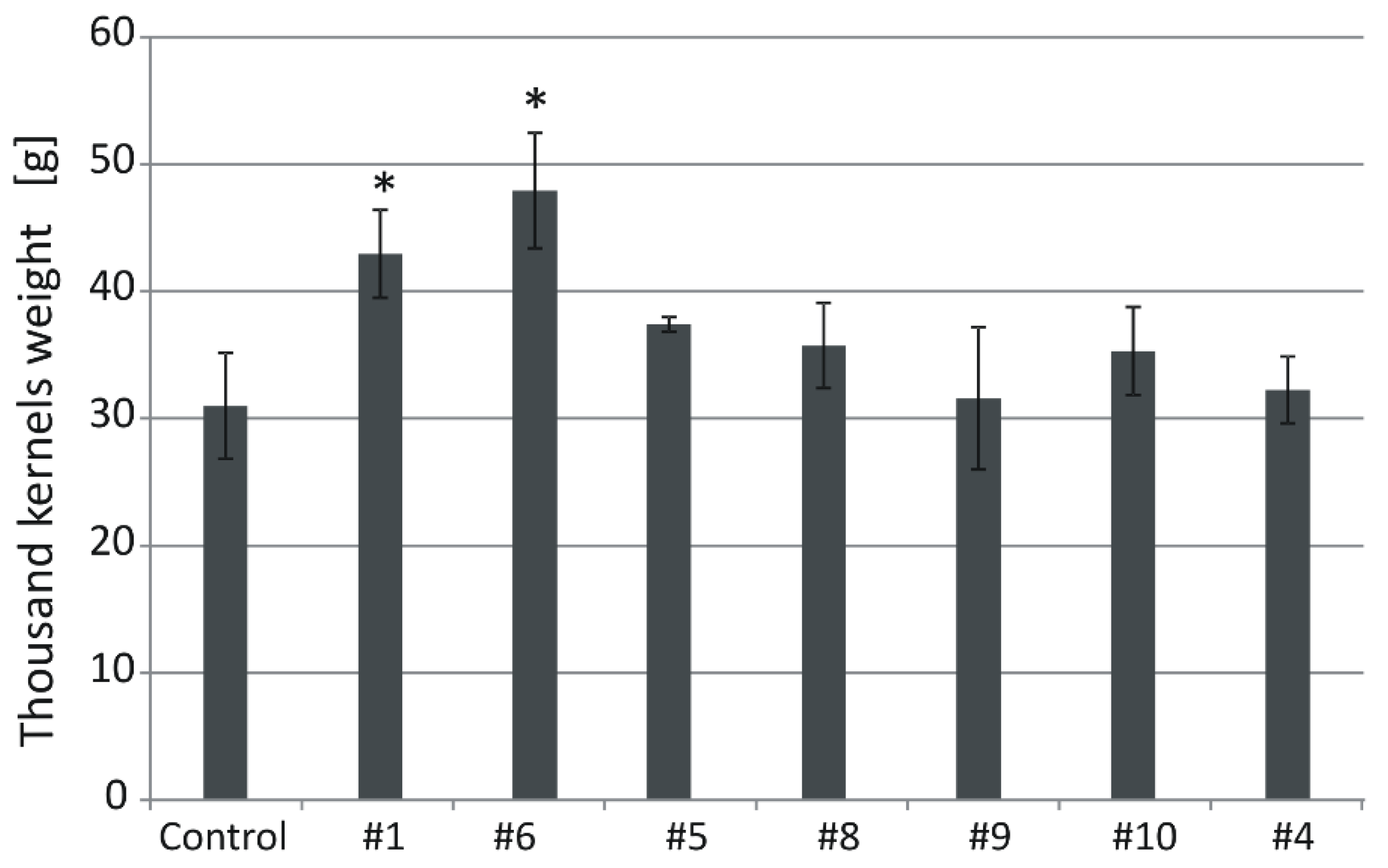
2.4. Silencing of HvGSK1.1 Increases Biomass of Plants Grown in Normal and Salt Stress Conditions and Kernel Weight
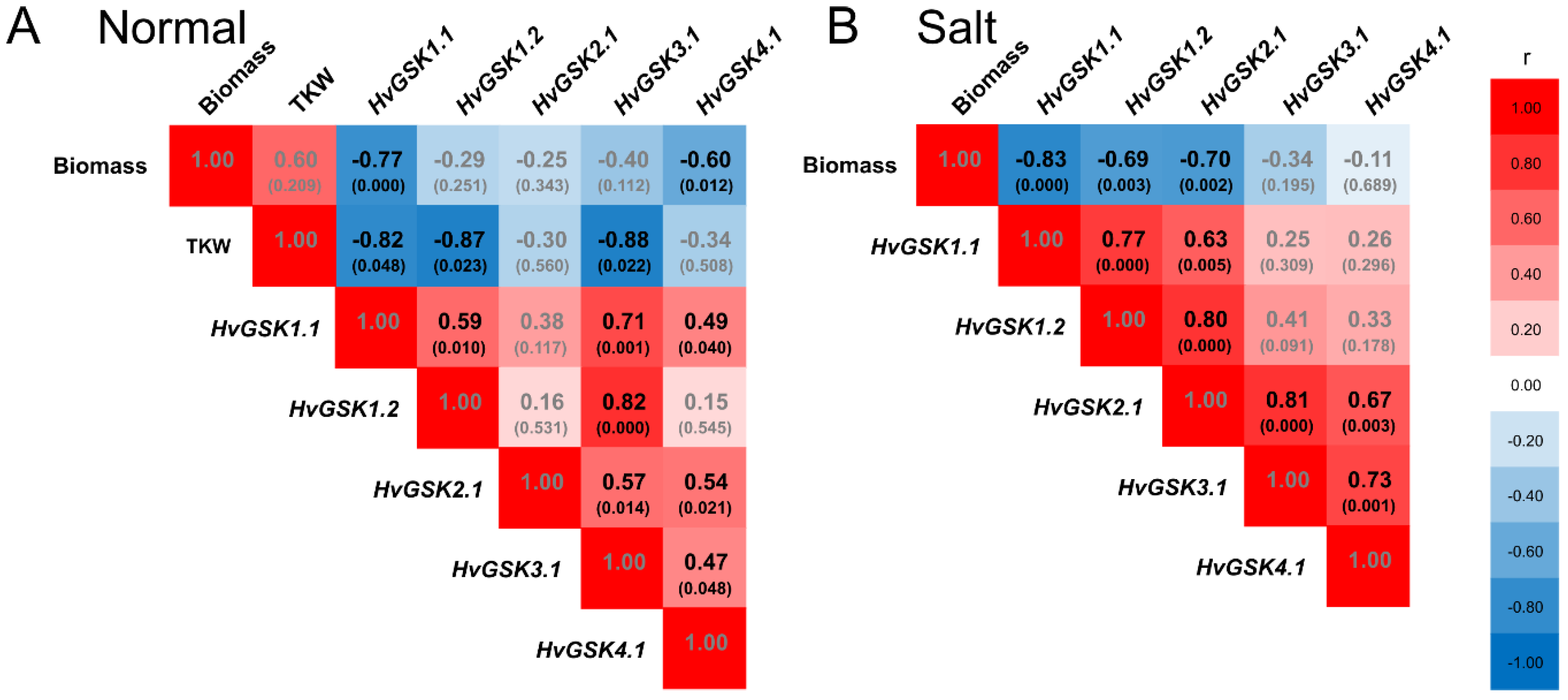
2.5. Silencing of HvGSK1.1 Correlates with Expression of GSK Paralogs, Plant Biomass, and Thousand Kernel Weight
3. Discussion
4. Materials and Methods
4.1. Bioinformatics of HvGSK1.1 and Rice Orthologs
4.2. Plant Material and Growth in Normal and Salt Stress Conditions
4.3. Construction of Silencing Vector and Agrobacterium-Mediated Transformation of Barley
4.4. Nucleic Acid Isolation, Transcript Quantification Leaf Inclination Test
4.5. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Saini, S.; Sharma, I.; Pati, P.K. Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Front. Plant Sci. 2015, 6, 950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.Y.; Sae-Seaw, J.; Wang, Z.Y. Brassinosteroid signalling. Development 2013, 140, 1615–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belkhadir, Y.; Jaillais, Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015, 206, 522–540. [Google Scholar] [CrossRef]
- Kim, T.W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.X.; Sun, Y.; Burlingame, A.L.; Wang, Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009, 11, 1254–1260. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Nakano, T.; Gendron, J.; He, J.X.; Chen, M.; Vafeados, D.; Yang, Y.L.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.H.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.M.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Gampala, S.S.; Kim, T.W.; He, J.X.; Tang, W.Q.; Deng, Z.P.; Bai, M.Y.; Guan, S.H.; Lalonde, S.; Sun, Y.; Gendron, J.M.; et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 2007, 13, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Vert, G.; Chory, J. Downstream nuclear events in brassinosteroid signalling. Nature 2006, 441, 96–100. [Google Scholar] [CrossRef]
- Sun, Y.; Fan, X.Y.; Cao, D.M.; Tang, W.Q.; He, K.; Zhu, J.Y.; He, J.X.; Bai, M.Y.; Zhu, S.W.; Oh, E.; et al. Integration of Brassinosteroid Signal Transduction with the Transcription Network for Plant Growth Regulation in Arabidopsis. Dev. Cell 2010, 19, 765–777. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Zhu, J.Y.; Bai, M.Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 2014, 3, e03031. [Google Scholar] [CrossRef]
- He, J.X.; Gendron, J.M.; Yang, Y.L.; Li, J.M.; Wang, Z.Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10185–10190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.Y.; Li, Y.; Cao, D.M.; Yang, H.; Oh, E.; Bi, Y.; Zhu, S.; Wang, Z.Y. The F-box Protein KIB1 Mediates Brassinosteroid-Induced Inactivation and Degradation of GSK3-like Kinases in Arabidopsis. Mol. Cell 2017, 66, 648–657 e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.Z.; Zhu, J.Y.; Roh, J.; Marchive, C.; Kim, S.K.; Meyer, C.; Sun, Y.; Wang, W.F.; Wang, Z.Y. TOR Signaling Promotes Accumulation of BZR1 to Balance Growth with Carbon Availability in Arabidopsis. Curr. Biol. 2016, 26, 1854–1860. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.J.; Zhang, R.; Meng, Z.G.; Deng, K.X.; Que, Y.M.; Zhuo, F.P.; Feng, L.; Guo, S.D.; Datla, R.; Ren, M.Z. Brassinosteriod Insensitive 2 (BIN2) acts as a downstream effector of the Target of Rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. New Phytol. 2017, 213, 233–249. [Google Scholar] [CrossRef] [Green Version]
- Kim, L.; Kimmel, A.R. GSK3 at the edge: Regulation of developmental specification and cell polarization. Curr. Drug Targets 2006, 7, 1411–1419. [Google Scholar] [CrossRef]
- Jonak, C.; Hirt, H. Glycogen synthase kinase 3/SHAGGY-like kinases in plants: An emerging family with novel functions. Trends Plant Sci. 2002, 7, 457–461. [Google Scholar] [CrossRef]
- Dornelas, M.C.; van Lammeren, A.A.M.; Kreis, M. Arabidopsis thaliana SHAGGY-related protein kinases (AtSK11 and 12) function in perianth and gynoecium development. Plant J. 2000, 21, 419–429. [Google Scholar] [CrossRef]
- Kim, T.W.; Michniewicz, M.; Bergmann, D.C.; Wang, Z.Y. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 2012, 482, 419–422. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.; Rozhon, W.; Bigeard, J.; Pflieger, D.; Husar, S.; Pitzschke, A.; Teige, M.; Jonak, C.; Hirt, H.; Poppenberger, B. Brassinosteroid-regulated GSK3/Shaggy-like Kinases Phosphorylate Mitogen-activated Protein (MAP) Kinase Kinases, Which Control Stomata Development in Arabidopsis thaliana. J. Biol. Chem. 2013, 288, 7519–7527. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.; Ryu, H.; Rho, S.; Hill, K.; Smith, S.; Audenaert, D.; Park, J.; Han, S.; Beeckman, T.; Bennett, M.J.; et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 2014, 16, 66–76. [Google Scholar] [CrossRef]
- Che, R.; Tong, H.; Shi, B.; Liu, Y.; Fang, S.; Liu, D.; Xiao, Y.; Hu, B.; Liu, L.; Wang, H.; et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2015, 2, 15195. [Google Scholar] [CrossRef] [PubMed]
- Eremina, M.; Unterholzner, S.J.; Rathnayake, A.I.; Castellanos, M.; Khan, M.; Kugler, K.G.; May, S.T.; Mayer, K.F.X.; Rozhon, W.; Poppenberger, B. Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad. Sci. USA 2016, 201611477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dal Santo, S.; Stampfl, H.; Krasensky, J.; Kempa, S.; Gibon, Y.; Petutschnig, E.; Rozhon, W.; Heuck, A.; Clausen, T.; Jonak, C. Stress-Induced GSK3 Regulates the Redox Stress Response by Phosphorylating Glucose-6-Phosphate Dehydrogenase in Arabidopsis. Plant Cell 2012, 24, 3380–3392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rybel, B.; Audenaert, D.; Vert, G.; Rozhon, W.; Mayerhofer, J.; Peelman, F.; Coutuer, S.; Denayer, T.; Jansen, L.; Nguyen, L.; et al. Chemical Inhibition of a Subset of Arabidopsis thaliana GSK3-like Kinases Activates Brassinosteroid Signaling. Chem. Biol. 2009, 16, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Saidi, Y.; Hearn, T.J.; Coates, J.C. Function and evolution of ‘green’ GSK3/Shaggy-like kinases. Trends Plant Sci. 2012, 17, 39–46. [Google Scholar] [CrossRef]
- Dornelas, M.C.; Wittich, P.; von Recklinghausen, I.; Lammeren, A.; Kreis, M. Characterization of three novel members of the Arabidopsis SHAGGY-related protein kinase (ASK) multigene family. Plant Mol. Biol. Report. 1999, 39, 137–147. [Google Scholar] [CrossRef]
- Kempa, S.; Rozhon, W.; Samaj, J.; Erban, A.; Baluska, F.; Becker, T.; Haselmayer, J.; Schleiff, E.; Kopka, J.; Hirt, H.; et al. A plastid-localized glycogen synthase kinase 3 modulates stress tolerance and carbohydrate metabolism. Plant J. 2007, 49, 1076–1090. [Google Scholar] [CrossRef] [Green Version]
- Anuradha, S.; Rao, S.S.R. Application of brassinosteroids to rice seeds (Oryza sativa L.) reduced the impact of salt stress on growth, prevented photosynthetic pigment loss and increased nitrate reductase activity. Plant Growth Regul. 2003, 40, 29–32. [Google Scholar] [CrossRef]
- Krishna, P. Brassinosteroid-mediated stress responses. J. Plant Growth Regul. 2003, 22, 289–297. [Google Scholar] [CrossRef]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 2007, 225, 353–364. [Google Scholar] [CrossRef]
- Singh, I.; Shono, M. Physiological and molecular effects of 24-epibrassinolide, a brassinosteroid on thermotolerance of tomato. Plant Growth Regul. 2005, 47, 111–119. [Google Scholar] [CrossRef]
- Dhaubhadel, S.; Chaudhary, S.; Dobinson, K.F.; Krishna, P. Treatment with 24-epibrassinolide, a brassinosteroid, increases the basic thermotolerance of Brassica napus and tomato seedlings. Plant Mol. Biol. 1999, 40, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Pinol, R.; Simon, E. Effect of 24-Epibrassinolide on Chlorophyll Fluorescence and Photosynthetic CO2 Assimilation in Vicia faba Plants Treated with the Photosynthesis-Inhibiting Herbicide Terbutryn. J. Plant Growth Regul. 2009, 28, 97–105. [Google Scholar] [CrossRef]
- Sharma, I.; Bhardwaj, R.; Pati, P.K. Stress modulation response of 24-epibrassinolide against imidacloprid in an elite indica rice variety Pusa Basmati-1. Pestic. Biochem. Physiol. 2013, 105, 144–153. [Google Scholar] [CrossRef]
- Zhu, T.; Deng, X.G.; Tan, W.R.; Zhou, X.; Luo, S.S.; Han, X.Y.; Zhang, D.W.; Lin, H.H. Nitric oxide is involved in brassinosteroid-induced alternative respiratory pathway in Nicotiana benthamiana seedlings’ response to salt stress. Physiol. Plant. 2016, 156, 150–163. [Google Scholar] [CrossRef]
- Sharma, I.; Ching, E.; Saini, S.; Bhardwaj, R.; Pati, P.K. Exogenous application of brassinosteroid offers tolerance to salinity by altering stress responses in rice variety Pusa Basmati-1. Plant Physiol. Biochem. Ppb/Soc. Fr. Physiol. Veg. 2013, 69, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, M.; Fariduddin, Q.; Khan, T.A.; Hayat, S. Epibrassinolide reverses the stress generated by combination of excess aluminum and salt in two wheat cultivars through altered proline metabolism and antioxidants. S. Afr. J. Bot. 2017, 112, 391–398. [Google Scholar] [CrossRef]
- Cui, F.; Liu, L.J.; Zhao, Q.Z.; Zhang, Z.H.; Li, Q.L.; Lin, B.Y.; Wu, Y.R.; Tang, S.Y.; Xie, Q. Arabidopsis Ubiquitin Conjugase UBC32 Is an ERAD Component That Functions in Brassinosteroid-Mediated Salt Stress Tolerance. Plant Cell 2012, 24, 233–244. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.J.; Lu, S.J.; Wang, M.J.; He, H.H.; Sun, L.; Wang, H.R.; Liu, X.H.; Jiang, L.; Sun, J.L.; Xin, X.Y.; et al. A Novel QTL qTGW3 Encodes the GSK3/SHAGGY-Like Kinase OsGSK5/OsSK41 that Interacts with OsARF4 to Negatively Regulate Grain Size and Weight in Rice. Mol. Plant 2018, 11, 736–749. [Google Scholar] [CrossRef] [Green Version]
- Xia, D.; Zhou, H.; Liu, R.J.; Dan, W.H.; Li, P.B.; Wu, B.; Chen, J.X.; Wang, L.Q.; Gao, G.J.; Zhang, Q.L.; et al. GL3.3, a Novel QTL Encoding a GSK3/SHAGGY-like Kinase, Epistatically Interacts with GS3 to Produce Extra-long Grains in Rice. Mol. Plant 2018, 11, 754–756. [Google Scholar] [CrossRef] [Green Version]
- Thitisaksakul, M.; Arias, M.C.; Dong, S.; Beckles, D.M. Overexpression of GSK3-like Kinase 5 (OsGSK5) in rice (Oryza sativa) enhances salinity tolerance in part via preferential carbon allocation to root starch. Funct. Plant Biol. 2017, 44, 705. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.Y.; Wang, T.; Wang, L.L.; Li, X.M.; Jia, Y.C.; Liu, C.; Huang, X.H.; Xie, W.B.; Wang, X.L. Natural selection of a GSK3 determines rice mesocotyl domestication by coordinating strigolactone and brassinosteroid signaling. Nat Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Sun, S.Y.; Chen, D.H.; Li, X.M.; Qiao, S.L.; Shi, C.N.; Li, C.X.; Shen, H.Y.; Wang, X.L. Brassinosteroid Signaling Regulates Leaf Erectness in Oryza sativa via the Control of a Specific U-Type Cyclin and Cell Proliferation. Dev. Cell 2015, 34, 220–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.L.; Yang, Z.E.; Zhang, B.; Yu, D.Q.; Liu, J.; Gong, Q.; Qanmber, G.; Li, Y.; Lu, L.L.; Lin, Y.J.; et al. Genome-wide characterization and phylogenetic analysis of GSK gene family in three species of cotton: Evidence for a role of some GSKs in fiber development and responses to stress. BMC Plant Biol. 2018, 18, 330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groszyk, J.; Yanushevska, Y.; Zielezinski, A.; Nadolska-Orczyk, A.; Karlowski, W.M.; Orczyk, W. Annotation and profiling of barley GLYCOGEN SYNTHASE3/Shaggy-like genes indicated shift in organ-preferential expression. PLoS ONE 2018, 13, e0199364. [Google Scholar] [CrossRef] [PubMed]
- Koh, S.; Lee, S.C.; Kim, M.K.; Koh, J.H.; Lee, S.; An, G.; Choe, S.; Kim, S.R. T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol. Biol. 2007, 65, 453–466. [Google Scholar] [CrossRef]
- Wada, K.; Marumo, S.; Abe, H.; Morishita, T.; Nakamura, K.; Uchiyama, M.; Mori, K. A rice lamina inclination test—A micro quantitative bioassay for brassinosteroids. Agric. Biol. Chem. 1984, 48, 719–726. [Google Scholar]
- Gan, L.J.; Wu, H.; Wu, D.P.; Zhang, Z.F.; Guo, Z.F.; Yang, N.; Xia, K.; Zhou, X.; Oh, K.; Matsuoka, M.; et al. Methyl jasmonate inhibits lamina joint inclination by repressing brassinosteroid biosynthesis and signaling in rice. Plant Sci. 2015, 241, 238–245. [Google Scholar] [CrossRef]
- Bartlett, J.G.; Alves, S.C.; Smedley, M.; Snape, J.W.; Harwood, W.A. High-throughput Agrobacterium-mediated barley transformation. Plant Methods 2008, 4, 22. [Google Scholar] [CrossRef] [Green Version]
- Zalewski, W.; Orczyk, W.; Gasparis, S.; Nadolska-Orczyk, A. HvCKX2 gene silencing by biolistic or Agrobacterium-mediated transformation in barley leads to different phenotypes. BMC Plant Biol. 2012, 12, 206. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Jablonski, B.; Ogonowska, H.; Szala, K.; Bajguz, A.; Orczyk, W.; Nadolska-Orczyk, A. Silencing of TaCKX1 Mediates Expression of Other TaCKX Genes to Increase Yield Parameters in Wheat. Int. J. Mol. Sci. 2020, 21, 4809. [Google Scholar] [CrossRef] [PubMed]
- Rozhon, W.; Wang, W.; Berthiller, F.; Mayerhofer, J.; Chen, T.; Petutschnig, E.; Sieberer, T.; Poppenberger, B.; Jonak, C. Bikinin-like inhibitors targeting GSK3/Shaggy-like kinases: Characterisation of novel compounds and elucidation of their catabolism in planta. BMC Plant Biol. 2014, 14, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamuro, C.; Ihara, Y.; Wu, X.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 2000, 12, 1591–1605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajguz, A.; Hayat, S. Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol. Biochem. Ppb/Soc. Fr. Physiol. Veg. 2009, 47, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Vriet, C.; Russinova, E.; Reuzeau, C. Boosting crop yields with plant steroids. Plant Cell 2012, 24, 842–857. [Google Scholar] [CrossRef] [Green Version]
- Perez-Perez, J.M.; Ponce, M.R.; Micol, J.L. The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev. Biol. 2002, 242, 161–173. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.Y.; Jiang, W.B.; Hu, Y.W.; Wu, P.; Zhu, J.Y.; Liang, W.Q.; Wang, Z.Y.; Lin, W.H. BR signal influences Arabidopsis ovule and seed number through regulating related genes expression by BZR1. Mol. Plant 2013, 6, 456–469. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.B.; Huang, H.Y.; Hu, Y.W.; Zhu, S.W.; Wang, Z.Y.; Lin, W.H. Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 2013, 162, 1965–1977. [Google Scholar] [CrossRef]
- Wu, C.Y.; Trieu, A.; Radhakrishnan, P.; Kwok, S.F.; Harris, S.; Zhang, K.; Wang, J.; Wan, J.; Zhai, H.; Takatsuto, S.; et al. Brassinosteroids regulate grain filling in rice. Plant Cell 2008, 20, 2130–2145. [Google Scholar] [CrossRef] [Green Version]
- Wan, T.T.; Li, Q.Z.; Lou, S.T.; Yang, Y.; Peng, L.F.; Lin, Z.Z.; Hu, Q.; Ma, L.Y. GSK3/shaggy-like kinase 1 ubiquitously regulates cell growth from Arabidopsis to Moso bamboo (Phyllostachys edulis). Plant Sci. 2019, 283, 290–300. [Google Scholar]
- Gearing, L.J.; Cumming, H.E.; Chapman, R.; Finkel, A.M.; Woodhouse, I.B.; Luu, K.; Gould, J.A.; Forster, S.C.; Hertzog, P.J. CiiiDER: A tool for predicting and analysing transcription factor binding sites. PLoS ONE 2019, 14, e0215495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 1950, 347, 1–32. [Google Scholar]
- Himmelbach, A.; Zierold, U.; Hensel, G.; Riechen, J.; Douchkov, D.; Schweizer, P.; Kumlehn, J. A set of modular binary vectors for transformation of cereals. Plant Physiol. 2007, 145, 1192–1200. [Google Scholar] [CrossRef] [Green Version]
- Hellens, R.P.; Edwards, E.A.; Leyland, N.R.; Bean, S.; Mullineaux, P.M. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000, 42, 819–832. [Google Scholar] [CrossRef]
- Zalewski, W.; Galuszka, P.; Gasparis, S.; Orczyk, W.; Nadolska-Orczyk, A. Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity. J. Exp. Bot. 2010, 61, 1839–1851. [Google Scholar] [CrossRef] [Green Version]
- Murray, M.G.; Thompson, W.F. Rapid Isolation of High Molecular-Weight Plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Goto, K.; Kanazawa, A.; Kusaba, M.; Masuta, C. A Simple and Rapid Method to Detect Plant siRNAs Using Nonradioactive Probes. Plant Mol. Biol. Report. 2003, 21, 51–58. [Google Scholar] [CrossRef]






| Vector | Cultured Immature Embryos | Regenerated Plants | Plants with Confirmed t-DNA | Plants with Single T-DNA Integration Locus | Plants with Multiple T-DNA Integration Loci |
|---|---|---|---|---|---|
| T-DNA of silencing vector | 1975 | 14 | 11 | 10 | 1 |
| Control of in vitro culture and plant regeneration | 107 | Good plant regeneration on hygromycin-free medium, 20–25 shoots from one immature embryo. | |||
| Control of in vitro culture and hygromycin selection | 107 | No shoots were regenerated on hygromycin-containing medium. | |||
| Gene Name Ensemble Plants ID NCBI ID | Primers | Amplicon Length and Reaction Conditions | |
|---|---|---|---|
| Symbol | Sequence | ||
| HvGSK1.1 HORVU3Hr1G034440.2 AK251287 | 1FRGSK1 | GGTGGTGGTAGAGTAGGAGTA | 1719 bp, 95 °C 5 min, 35 cycles (95 °C 30 s, 60 °C 30 s, 72 °C 30 s) 72 °C 5 min. |
| 4RevGSK1 | TTACAGATCAGCTATGGCAAT | ||
| ORFgskFR | TGGTGAAAGTGGTGTGGACC | 262 bp, 95 °C 5 min, 35 cycles (95 °C 30 s, 56 °C 30 s, 72 °C 30 s) 72 °C 5 min. | |
| ORFgskRe | GGTCCCGAAGCTCATCAAAG | ||
| 437AK251_Fr | AGGGAACAGAGACTGGTCACAT | 108 bp, 95 °C 15 min, 45 cycles (95 °C 25 s, 60 °C 25 s, 72 °C 25 s) 72 °C 1 min. | |
| 545AK251_Rev | AATGAACCTTGACCAACAATCC | ||
| HvGSK1.2 HORVU5Hr1G117030.1 AK368391 | AK368_FR | TCTGGGCACACCTACAAGGG | 139 bp, 95 °C 15 min, 45 cycles (95 °C 25 s, 60 °C 25 s, 72 °C 25 s) 72 °C 1 min. |
| AK368_Rev | TGGAGACCAGGTCCACTGCT | ||
| HvGSK2.1 HORVU3Hr1G026020.1 AK364823 | 403AL364_FR | AGTGCTTGGAGACTGGAGAGAC | 122 bp, 95 °C 15 min, 45 cycles (95 °C 25 s, 60 °C 25 s, 72 °C 25 s) 72 °C 1 min. |
| 524AK364_Re | GTGCTTCAGAGAGACGACATTG | ||
| HvGSK3.1 HORVU1Hr1G048580.7AK362547 | 1270AK362_Fr | AAAGTGGCGTTGATCAGTTGG | 123 bp, 95 °C 15 min, 45 cycles (95 °C 25 s, 60 °C 25 s, 72 °C 25 s) 72 °C 1 min. |
| 1393AK362_Rev | CAGGGATGAGCTTTTATCTGAGG | ||
| HvGSK4.1 HORVU5Hr1G119790.18AK358344 | AK358_FR | GCGAGAAGGCAGAACCTGTT | 133 bp, 95 °C 15 min, 45 cycles (95 °C 25 s, 60 °C 25 s, 72 °C 25 s) 72 °C 1 min. |
| AK358_Rev | TGTCACCCACCCACACAAAG | ||
| ARF AJ508228 | Ref2_FR | GCTCTCCAACAACATTGCCAAC | 162 bp, 95 °C 15 min, 45 cycles (95 °C 25 s, 60 °C 25 s, 72 °C 25 s) 72 °C 1 min. |
| Ref2_Rev | GCTTCTGCCTGTCACATACGC | ||
| ACTB (actin) AY145451 | qAct1 | AGCAACTGGGATGACATGGAG | 172 bp, 95 °C 15 min, 45 cycles (95 °C 25 s, 60 °C 25 s, 72 °C 25 s) 72 °C 1 min. |
| qAct2a | CGTACATGGCAGGAACATTG | ||
| hptII | pBr_Hyg_F2 | GACGGCAATTTCGATGATG | 205 bp, 95 °C 15 min, 45 cycles (95 °C 25 s, 60 °C 25 s, 72 °C 25 s) 72 °C 1 min. |
| pBr_Hyg_R2 | CCGGTCGGCATCTACTCTAT | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kloc, Y.; Dmochowska-Boguta, M.; Zielezinski, A.; Nadolska-Orczyk, A.; Karlowski, W.M.; Orczyk, W. Silencing of HvGSK1.1—A GSK3/SHAGGY-Like Kinase–Enhances Barley (Hordeum vulgare L.) Growth in Normal and in Salt Stress Conditions. Int. J. Mol. Sci. 2020, 21, 6616. https://doi.org/10.3390/ijms21186616
Kloc Y, Dmochowska-Boguta M, Zielezinski A, Nadolska-Orczyk A, Karlowski WM, Orczyk W. Silencing of HvGSK1.1—A GSK3/SHAGGY-Like Kinase–Enhances Barley (Hordeum vulgare L.) Growth in Normal and in Salt Stress Conditions. International Journal of Molecular Sciences. 2020; 21(18):6616. https://doi.org/10.3390/ijms21186616
Chicago/Turabian StyleKloc, Yuliya, Marta Dmochowska-Boguta, Andrzej Zielezinski, Anna Nadolska-Orczyk, Wojciech M. Karlowski, and Waclaw Orczyk. 2020. "Silencing of HvGSK1.1—A GSK3/SHAGGY-Like Kinase–Enhances Barley (Hordeum vulgare L.) Growth in Normal and in Salt Stress Conditions" International Journal of Molecular Sciences 21, no. 18: 6616. https://doi.org/10.3390/ijms21186616





