Downregulation of CDC20 Increases Radiosensitivity through Mcl-1/p-Chk1-Mediated DNA Damage and Apoptosis in Tumor Cells
Abstract
:1. Introduction
2. Results
2.1. CDC20 Is Overexpressed in Human Cancer Cells and Upregulated after Radiation
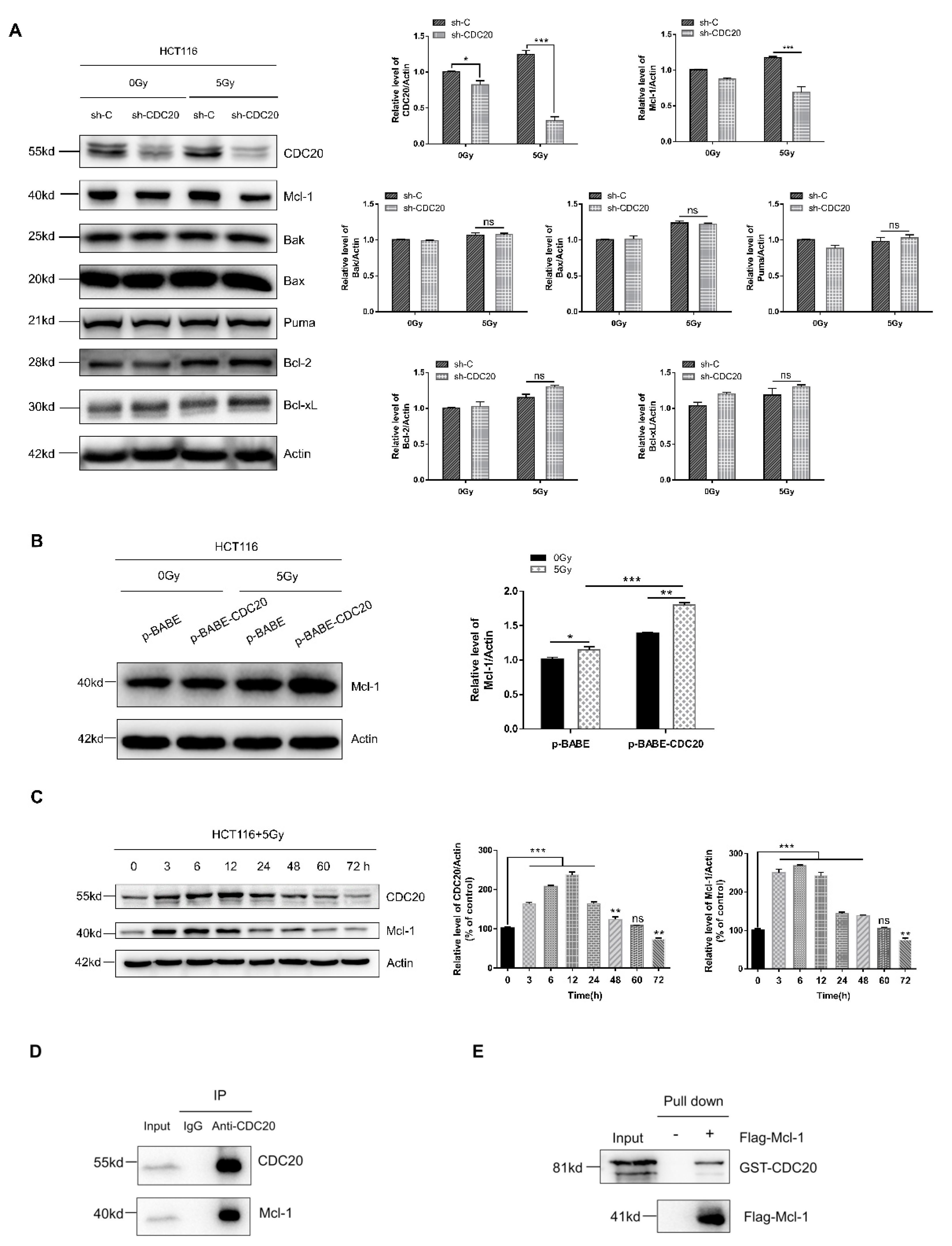
2.2. Knockdown of CDC20 Sensitizes CRC Cells to Radiation through Stimulating DNA Damage and Intrinsic Apoptotic Pathway
2.3. Antiapoptotic Protein Mcl-1 Is a CDC20-Interacting Protein
2.4. Mcl-1 Increases Chk1 Phosphorylation to Regulate CDC20-Mediated DNA Damage and Apoptosis in Radiation
2.5. Reducing CDC20 Expression Enhanced the Radiosensitivity of CRC Xenografts by Inducing Apoptosis in Tumors
3. Discussion
4. Materials and Methods
4.1. Cell Culure and Irradiaion
4.2. Reagents
4.3. Cell Viability Assay
4.4. Colony Formation Assay
4.5. Caspase Assays
4.6. Transfection of Small Interfering RNA (siRNA) and shRNA Sequence
4.7. Lentivirus Production and Cell Culture
4.8. Quantitative Real-Time PCR
4.9. Western Blot Analysis
4.10. Immunoprecipitation Assay
4.11. Protein Expression and Purification
4.12. Xenograft Studies of Nude Mice
4.13. Immunohistochemistry Analysis
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yan, K. Recent Developments in Radiotherapy. N. Engl. J. Med. 2017, 377, 2200. [Google Scholar] [CrossRef]
- Xiao, C.; Wang, Y.P.; Zheng, M.; Chen, J.; Song, G.H.; Zhou, Z.J.; Zhou, C.Z.; Sun, X.; Zhong, L.; Ding, E.X.; et al. RBBP6 increases radioresistance and serves as a therapeutic target for preoperative radiotherapy in colorectal cancer. Cancer Sci. 2018, 109, 1075–1087. [Google Scholar] [CrossRef]
- Manchado, E.; Guillamot, M.; Malumbres, M. Killing cells by targeting mitosis. Cell Death Differ. 2012, 19, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Peters, J. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006, 7, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Tan, M.; Yang, J.; Inuzuka, H.; Dai, X.; Wu, T.; Liu, J.; Shaik, S.; Chen, G.; Deng, J.; et al. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Dev. Cell 2014, 29, 377–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagting, A.; Den Elzen, N.; Vodermaier, H.; Waizenegger, I.; Peters, J.; Pines, J. Human securin proteolysis is controlled by the spindle checkpoint and reveals when the APC/C switches from activation by Cdc20 to Cdh1. J. Cell Biol. 2002, 157, 1125–1137. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, J.; Wan, L.; Zhou, X.; Wang, Z.; Wei, W. Targeting Cdc20 as a novel cancer therapeutic strategy. Pharmacol. Ther. 2015, 151, 141–151. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Chen, B.; Zhu, Z.; Zhang, L.; Zeng, J.; Xu, G.; Liu, G.; Xiong, D.; Luo, Q.; Huang, Z. CDC20 overexpression leads to poor prognosis in solid tumors: A system review and meta-analysis. Medicine 2018, 97, e13832. [Google Scholar] [CrossRef]
- Kidokoro, T.; Tanikawa, C.; Furukawa, Y.; Katagiri, T.; Nakamura, Y.; Matsuda, K. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene 2008, 27, 1562–1571. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.J.; Hu, K.S.; Wang, D.S.; Zeng, Z.L.; Zhang, D.S.; Chen, D.L.; Bai, L.; Xu, R.H. CDC20 overexpression predicts a poor prognosis for patients with colorectal cancer. J. Transl. Med. 2013, 11. [Google Scholar] [CrossRef] [Green Version]
- Manchado, E.; Guillamot, M.; de Carcer, G.; Eguren, M.; Trickey, M.; Garcia-Higuera, I.; Moreno, S.; Yamano, H.; Canamero, M.; Malumbres, M. Targeting mitotic exit leads to tumor regression in vivo: Modulation by Cdk1, Mastl, and the PP2A/B55alpha,delta phosphatase. Cancer Cell 2010, 18, 641–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Yu, Y.; Fu, D.; Li, Z.; Niu, X.; Liao, M.; Lu, S. Functional roles of PC-PLC and Cdc20 in the cell cycle, proliferation, and apoptosis. Cell Biochem. Funct. 2010, 28, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, C.M.; Rossanese, O.W.; Olejniczak, E.T.; Fesik, S.W. Myeloid cell leukemia-1 is an important apoptotic survival factor in triple-negative breast cancer. Cell Death Differ. 2015, 22, 2098–2106. [Google Scholar] [CrossRef] [Green Version]
- Sloss, O.; Topham, C.; Diez, M.; Taylor, S. Mcl-1 dynamics influence mitotic slippage and death in mitosis. Oncotarget 2016, 7, 5176–5192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, W.; Yang, C.Y.; Bai, L. MCL-1 inhibition in cancer treatment. Onco Targets Ther. 2018, 11, 7301–7314. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Guttikonda, S.; Roberts, L.; Uziel, T.; Semizarov, D.; Elmore, S.W.; Leverson, J.D.; Lam, L.T. Mcl-1 is critical for survival in a subgroup of non-small-cell lung cancer cell lines. Oncogene 2011, 30, 1963–1968. [Google Scholar] [CrossRef] [Green Version]
- Wieczorek, S.A.; Breitenbuecher, F.; Soni, A.; Paul-Konietzko, K.; Ziegler, S.; Sak, A.; Iliakis, G.; Schuler, M. Deregulated BCL-2 family proteins impact on repair of DNA double-strand breaks and are targets to overcome radioresistance in lung cancer. J. Cancer Res. Clin. Oncol. 2017, 143, 1733–1744. [Google Scholar] [CrossRef] [PubMed]
- Li, X.H.; Ha, C.T.; Xiao, M. MicroRNA-30 inhibits antiapoptotic factor Mcl-1 in mouse and human hematopoietic cells after radiation exposure. Apoptosis 2016, 21, 708–720. [Google Scholar] [CrossRef] [Green Version]
- Harley, M.E.; Allan, L.A.; Sanderson, H.S.; Clarke, P.R. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J. 2010, 29, 2407–2420. [Google Scholar] [CrossRef] [Green Version]
- Sancar, A.; Lindsey-Boltz, L.A.; Ünsal-Kaçmaz, K.; Linn, S. Molecular Mechanisms of Mammalian DNA Repair and the DNA Damage Checkpoints. Annu. Rev. Biochem. 2004, 73, 39–85. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Hunter, T. Roles of Chk1 in cell biology and cancer therapy. Int. J. Cancer 2014, 134, 1013–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirbu, B.M.; Cortez, D. DNA damage response: Three levels of DNA repair regulation. Cold Spring Harb. Perspect. Biol. 2013, 5, a012724. [Google Scholar] [CrossRef] [PubMed]
- Jamil, S.; Mojtabavi, S.; Hojabrpour, P.; Cheah, S.; Duronio, V. An essential role for MCL-1 in ATR-mediated CHK1 phosphorylation. Mol. Biol. Cell 2008, 19, 3212–3220. [Google Scholar] [CrossRef] [PubMed]
- Jamil, S.; Stoica, C.; Hackett, T.L.; Duronio, V. MCL-1 localizes to sites of DNA damage and regulates DNA damage response. Cell Cycle 2010, 9, 2843–2855. [Google Scholar] [CrossRef] [Green Version]
- Sackton, K.; Dimova, N.; Zeng, X.; Tian, W.; Zhang, M.; Sackton, T.; Meaders, J.; Pfaff, K.; Sigoillot, F.; Yu, H.; et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature 2014, 514, 646–649. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Zhang, B.; Wang, Y.; Shang, G. Cdc20 inhibitor apcin inhibits the growth and invasion of osteosarcoma cells. Oncol. Rep. 2018, 40, 841–848. [Google Scholar] [CrossRef] [Green Version]
- Birkinshaw, R.W.; Czabotar, P.E. The BCL-2 family of proteins and mitochondrial outer membrane permeabilisation. Semin. Cell Dev. Biol. 2017, 72, 152–162. [Google Scholar] [CrossRef]
- Pearce, M.C.; Satterthwait, A.C.; Zhang, X.K.; Kolluri, S. Cancer therapeutics based on BCL-2 functional conversion. Apoptosis Int. J. Prog. Cell Death 2019, 24, 1–2. [Google Scholar] [CrossRef]
- Zabludoff, S.; Deng, C.; Grondine, M.; Sheehy, A.; Ashwell, S.; Caleb, B.; Green, S.; Haye, H.; Horn, C.; Janetka, J.; et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol. Cancer Ther. 2008, 7, 2955–2966. [Google Scholar] [CrossRef] [Green Version]
- Morgan, M.A.; Parsels, L.A.; Zhao, L.; Parsels, J.D.; Davis, M.A.; Hassan, M.C.; Arumugarajah, S.; Hylander-Gans, L.; Morosini, D.; Simeone, D.M.; et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 2010, 70, 4972–4981. [Google Scholar] [CrossRef] [Green Version]
- Bartucci, M.; Svensson, S.; Romania, P.; Dattilo, R.; Patrizii, M.; Signore, M.; Navarra, S.; Lotti, F.; Biffoni, M.; Pilozzi, E.; et al. Therapeutic targeting of Chk1 in NSCLC stem cells during chemotherapy. Cell Death Differ. 2012, 19, 768–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Zou, H.; Yu, W.; Huang, Y.; Liu, B.; Li, T.; Liang, C.; Tao, H. Checkpoint kinase inhibitor AZD7762 enhance cisplatin-induced apoptosis in osteosarcoma cells. Cancer Cell Int. 2019, 19, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; Lin, Y.; Cui, P.; Li, H.; Zhang, L.; Sun, Z.; Huang, S.; Li, S.; Huang, S.; Zhao, Q.; et al. Cdc20/p55 mediates the resistance to docetaxel in castration-resistant prostate cancer in a Bim-dependent manner. Cancer Chemother. Pharmacol. 2018, 81, 999–1006. [Google Scholar] [CrossRef]
- Huang, H.C.; Shi, J.; Orth, J.D.; Mitchison, T.J. Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell 2009, 16, 347–358. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Wen, P.; Fu, Y.; Gao, Y.; Qi, X.; Chen, B.; Tao, Y.; Wu, L.; Xu, A.; Lu, H.; et al. Radiation induces apoptosis primarily through the intrinsic pathway in mammalian cells. Cell Signal 2019, 62, 109337. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Kvansakul, M.; Hinds, M. The Bcl-2 family: Structures, interactions and targets for drug discovery. Apoptosis Int. J. Program. Cell Death 2015, 20, 136–150. [Google Scholar] [CrossRef]
- Yamaguchi, R.; Lartigue, L.; Perkins, G. Targeting Mcl-1 and other Bcl-2 family member proteins in cancer therapy. Pharmacol. Ther. 2019, 195, 13–20. [Google Scholar] [CrossRef]
- Kolodner, R.D.; Putnam, C.D.; Myung, K. Maintenance of genome stability in Saccharomyces cerevisiae. Science 2002, 297, 552–557. [Google Scholar] [CrossRef] [Green Version]
- Patil, M.; Pabla, N.; Dong, Z. Checkpoint kinase 1 in DNA damage response and cell cycle regulation. Cell Mol. Life Sci. 2013, 70, 4009–4021. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, C.S.; Hansen, L.T.; Dziegielewski, J.; Syljuasen, R.G.; Lundin, C.; Bartek, J.; Helleday, T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 2005, 7, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Wang, J.; Wang, X.; Chen, S.; Zhao, Y.; Gu, F.; Xu, A.; Wu, L. Mutagenicity of PFOA in mammalian cells: Role of mitochondria-dependent reactive oxygen species. Environ. Sci. Technol. 2011, 45, 1638–1644. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhao, K.; Yu, S.; Xu, A.; Han, W.; Mei, Y. RNF12 catalyzes BRF1 ubiquitination and regulates RNA polymerase III-dependent transcription. J. Biol. Chem. 2019, 294, 130–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, Y.; Wen, P.; Chen, B.; Hu, G.; Wu, L.; Xu, A.; Zhao, G. Downregulation of CDC20 Increases Radiosensitivity through Mcl-1/p-Chk1-Mediated DNA Damage and Apoptosis in Tumor Cells. Int. J. Mol. Sci. 2020, 21, 6692. https://doi.org/10.3390/ijms21186692
Gao Y, Wen P, Chen B, Hu G, Wu L, Xu A, Zhao G. Downregulation of CDC20 Increases Radiosensitivity through Mcl-1/p-Chk1-Mediated DNA Damage and Apoptosis in Tumor Cells. International Journal of Molecular Sciences. 2020; 21(18):6692. https://doi.org/10.3390/ijms21186692
Chicago/Turabian StyleGao, Yang, Pengbo Wen, Bin Chen, Guanshuo Hu, Lijun Wu, An Xu, and Guoping Zhao. 2020. "Downregulation of CDC20 Increases Radiosensitivity through Mcl-1/p-Chk1-Mediated DNA Damage and Apoptosis in Tumor Cells" International Journal of Molecular Sciences 21, no. 18: 6692. https://doi.org/10.3390/ijms21186692




