Non-Coding RNAs as Cancer Hallmarks in Chronic Lymphocytic Leukemia
Abstract
1. Introduction: Current Status of Chronic Lymphocytic Leukemia Research
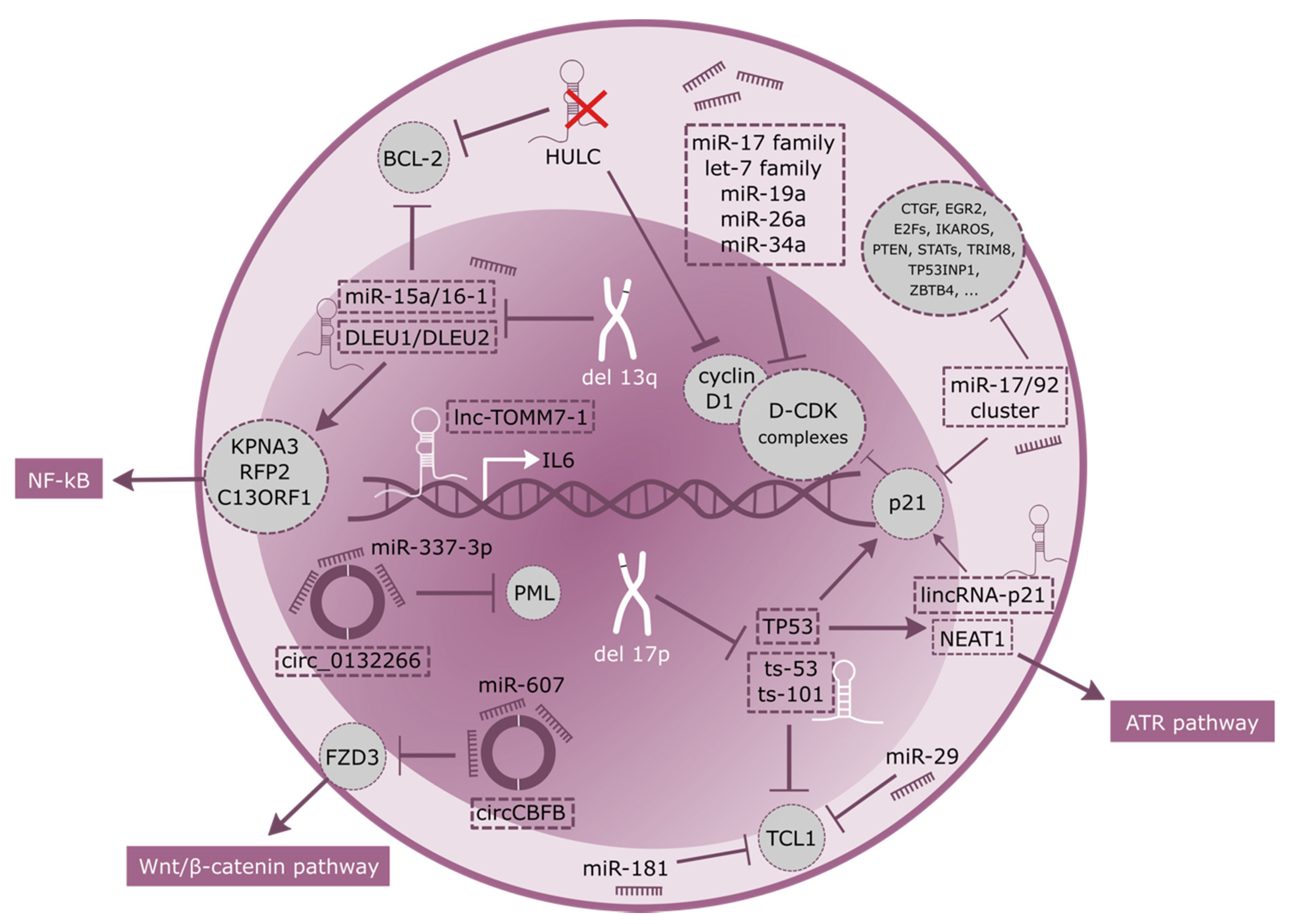
2. Sustaining Proliferative Signaling and Resisting Cell Death
3. Genomic Instability and Mutations
4. Enabling Replicative Immortality
5. Angiogenesis
6. Tumor Microenvironment
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Kipps, T.J.; Stevenson, F.K.; Wu, C.J.; Croce, C.M.; Packham, G.; Wierda, W.G.; O’Brien, S.; Gribben, J.; Rai, K. Chronic lymphocytic leukaemia. Nat. Rev. Dis. Primers 2017, 3, 16096. [Google Scholar] [CrossRef]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A microrna signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M. The emerging role of lncrnas in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Dahl, M.; Kristensen, L.S.; Grønbæk, K. Long Non-Coding RNAs Guide the Fine-Tuning of Gene Regulation in B-Cell Development and Malignancy. Int. J. Mol. Sci. 2018, 19, 2475. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Messmer, B.T.; Messmer, D.; Allen, S.L.; Kolitz, J.E.; Kudalkar, P.; Cesar, D.; Murphy, E.J.; Koduru, P.; Ferrarini, M.; Zupo, S.; et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia b cells. J. Clin. Invest. 2005, 115, 755–764. [Google Scholar] [CrossRef]
- Herndon, T.M.; Chen, S.S.; Saba, N.S.; Valdez, J.; Emson, C.; Gatmaitan, M.; Tian, X.; Hughes, T.E.; Sun, C.; Arthur, D.C.; et al. Direct in vivo evidence for increased proliferation of cll cells in lymph nodes compared to bone marrow and peripheral blood. Leukemia 2017, 31, 1340–1347. [Google Scholar] [CrossRef]
- Bomben, R.; Gobessi, S.; Dal Bo, M.; Volinia, S.; Marconi, D.; Tissino, E.; Benedetti, D.; Zucchetto, A.; Rossi, D.; Gaidano, G.; et al. The mir-17 approximately 92 family regulates the response to toll-like receptor 9 triggering of cll cells with unmutated ighv genes. Leukemia 2012, 26, 1584–1593. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, S.K.; Fassan, M.; Volinia, S.; Lovat, F.; Balatti, V.; Pekarsky, Y.; Croce, C.M. B-cell malignancies in microrna emu-mir-17~92 transgenic mice. Proc. Natl. Acad. Sci. USA 2013, 110, 18208–18213. [Google Scholar] [CrossRef] [PubMed]
- Moyo, T.K.; Wilson, C.S.; Moore, D.J.; Eischen, C.M. Myc enhances b-cell receptor signaling in precancerous b cells and confers resistance to btk inhibition. Oncogene 2017, 36, 4653–4661. [Google Scholar] [CrossRef] [PubMed]
- Mogilyansky, E.; Rigoutsos, I. The mir-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef]
- Weber, A.; Marquardt, J.; Elzi, D.; Forster, N.; Starke, S.; Glaum, A.; Yamada, D.; Defossez, P.A.; Delrow, J.; Eisenman, R.N.; et al. Zbtb4 represses transcription of p21cip1 and controls the cellular response to p53 activation. EMBO J. 2008, 27, 1563–1574. [Google Scholar] [CrossRef]
- Majumder, S.; Bhowal, A.; Basu, S.; Mukherjee, P.; Chatterji, U.; Sengupta, S. Deregulated e2f5/p38/smad3 circuitry reinforces the pro-tumorigenic switch of tgfbeta signaling in prostate cancer. J. Cell Physiol 2016, 231, 2482–2492. [Google Scholar] [CrossRef]
- Bueno, M.J.; Malumbres, M. Micrornas and the cell cycle. Biochim. Biophys. Acta 2011, 1812, 592–601. [Google Scholar] [CrossRef]
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Saito, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a novel non-coding rna, miat, that confers risk of myocardial infarction. J. Hum. Genet. 2006, 51, 1087–1099. [Google Scholar] [CrossRef]
- Ohnishi, Y.; Tanaka, T.; Yamada, R.; Suematsu, K.; Minami, M.; Fujii, K.; Hoki, N.; Kodama, K.; Nagata, S.; Hayashi, T.; et al. Identification of 187 single nucleotide polymorphisms (snps) among 41 candidate genes for ischemic heart disease in the japanese population. Hum. Genet. 2000, 106, 288–292. [Google Scholar] [CrossRef]
- Sun, C.; Huang, L.; Li, Z.; Leng, K.; Xu, Y.; Jiang, X.; Cui, Y. Long non-coding rna miat in development and disease: A new player in an old game. J. Biomed. Sci. 2018, 25, 23. [Google Scholar] [CrossRef]
- Sattari, A.; Siddiqui, H.; Moshiri, F.; Ngankeu, A.; Nakamura, T.; Kipps, T.J.; Croce, C.M. Upregulation of long noncoding rna miat in aggressive form of chronic lymphocytic leukemias. Oncotarget 2016, 7, 54174–54182. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Balatti, V.; Croce, C.M. Bcl2 and mir-15/16: From gene discovery to treatment. Cell Death Differ. 2018, 25, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- rna genes mir15 and mir16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. Mir-15 and mir-16 induce apoptosis by targeting bcl2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Cimmino, A.; Fabbri, M.; Ferracin, M.; Wojcik, S.E.; Shimizu, M.; Taccioli, C.; Zanesi, N.; Garzon, R.; Aqeilan, R.I.; et al. Mir-15a and mir-16-1 cluster functions in human leukemia. Proc. Natl. Acad. Sci. USA 2008, 105, 5166–5171. [Google Scholar] [CrossRef] [PubMed]
- Dal Bo, M.; Rossi, F.M.; Rossi, D.; Deambrogi, C.; Bertoni, F.; Del Giudice, I.; Palumbo, G.; Nanni, M.; Rinaldi, A.; Kwee, I.; et al. 13q14 deletion size and number of deleted cells both influence prognosis in chronic lymphocytic leukemia. Genes Chromosomes Cancer 2011, 50, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.; Li, H.; Li, Z.; Xiong, W.; Liu, H.; Liu, W.; Lv, R.; Yu, Z.; Zou, D.; Xu, Y.; et al. The prognostic significance of 13q deletions of different sizes in patients with b-cell chronic lymphoproliferative disorders: A retrospective study. Int. J. Hematol. 2017, 106, 418–425. [Google Scholar] [CrossRef]
- Bullrich, F.; Fujii, H.; Calin, G.; Mabuchi, H.; Negrini, M.; Pekarsky, Y.; Rassenti, L.; Alder, H.; Reed, J.C.; Keating, M.J.; et al. Characterization of the 13q14 tumor suppressor locus in cll: Identification of alt1, an alternative splice variant of the leu2 gene. Cancer Res. 2001, 61, 6640–6648. [Google Scholar]
- Garding, A.; Bhattacharya, N.; Claus, R.; Ruppel, M.; Tschuch, C.; Filarsky, K.; Idler, I.; Zucknick, M.; Caudron-Herger, M.; Oakes, C.; et al. Epigenetic upregulation of lncrnas at 13q14.3 in leukemia is linked to the in cis downregulation of a gene cluster that targets nf-kb. PLoS Genet. 2013, 9, e1003373. [Google Scholar] [CrossRef]
- Buccheri, V.; Barreto, W.G.; Fogliatto, L.M.; Capra, M.; Marchiani, M.; Rocha, V. Prognostic and therapeutic stratification in cll: Focus on 17p deletion and p53 mutation. Ann. Hematol. 2018, 97, 2269–2278. [Google Scholar] [CrossRef]
- Rossi, D.; Cerri, M.; Deambrogi, C.; Sozzi, E.; Cresta, S.; Rasi, S.; De Paoli, L.; Spina, V.; Gattei, V.; Capello, D.; et al. The Prognostic Value of TP53 Mutations in Chronic Lymphocytic Leukemia Is Independent of Del17p13: Implications for Overall Survival and Chemorefractoriness. Clin. Cancer Res. 2009, 15, 995–1004. [Google Scholar] [CrossRef]
- Blume, C.J.; Hotz-Wagenblatt, A.; Hullein, J.; Sellner, L.; Jethwa, A.; Stolz, T.; Slabicki, M.; Lee, K.; Sharathchandra, A.; Benner, A.; et al. P53-dependent non-coding rna networks in chronic lymphocytic leukemia. Leukemia 2015, 29, 2015–2023. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Wu, H.; Zhu, X.; Guo, X.; Hutchins, A.P.; Luo, Z.; Song, H.; Chen, Y.; Lai, K.; Yin, M.; et al. The p53-induced lincrna-p21 derails somatic cell reprogramming by sustaining h3k9me3 and cpg methylation at pluripotency gene promoters. Cell Res. 2015, 25, 80–92. [Google Scholar] [CrossRef]
- Dimitrova, N.; Zamudio, J.R.; Jong, R.M.; Soukup, D.; Resnick, R.; Sarma, K.; Ward, A.J.; Raj, A.; Lee, J.T.; Sharp, P.A.; et al. Lincrna-p21 activates p21 in cis to promote polycomb target gene expression and to enforce the g1/s checkpoint. Mol. Cell 2014, 54, 777–790. [Google Scholar] [CrossRef]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.; Radaelli, E.; Vermi, W.; et al. P53 induces formation of neat1 lncrna-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016, 22, 861–868. [Google Scholar] [CrossRef]
- Choudhry, H.; Albukhari, A.; Morotti, M.; Haider, S.; Moralli, D.; Smythies, J.; Schodel, J.; Green, C.M.; Camps, C.; Buffa, F.; et al. Tumor hypoxia induces nuclear paraspeckle formation through hif-2alpha dependent transcriptional activation of neat1 leading to cancer cell survival. Oncogene 2015, 34, 4546. [Google Scholar] [CrossRef]
- Kwok, M.; Davies, N.; Agathanggelou, A.; Smith, E.; Petermann, E.; Yates, E.; Brown, J.; Lau, A.; Stankovic, T. Synthetic lethality in chronic lymphocytic leukaemia with DNA damage response defects by targeting the atr pathway. Lancet 2015, 385, S58. [Google Scholar] [CrossRef]
- Kim, H.K. Transfer rna-derived small non-coding rna: Dual regulator of protein synthesis. Mol. Cells 2019, 42, 687–692. [Google Scholar] [PubMed]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of trna derived rna fragments reveals that they are evolutionarily conserved and associate with ago proteins to recognize specific rna targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Balatti, V.; Palamarchuk, A.; Rizzotto, L.; Veneziano, D.; Nigita, G.; Rassenti, L.Z.; Pass, H.I.; Kipps, T.J.; Liu, C.G.; et al. Dysregulation of a family of short noncoding rnas, tsrnas, in human cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 5071–5076. [Google Scholar] [CrossRef] [PubMed]
- Paduano, F.; Gaudio, E.; Mensah, A.A.; Pinton, S.; Bertoni, F.; Trapasso, F. T-cell leukemia/lymphoma 1 (tcl1): An oncogene regulating multiple signaling pathways. Front. Oncol. 2018, 8, 317. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, X.; Wu, H.; Ni, P.; Gu, Z.; Qiao, Y.; Chen, N.; Sun, F.; Fan, Q. Creb up-regulates long non-coding rna, hulc expression through interaction with microrna-372 in liver cancer. Nucleic Acids Res. 2010, 38, 5366–5383. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Wu, J.; Feng, J. Long noncoding rna hulc predicts poor clinical outcome and represents pro-oncogenic activity in diffuse large b-cell lymphoma. Biomed. Pharmacother. 2016, 79, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Vigorito, E.; Perks, K.L.; Abreu-Goodger, C.; Bunting, S.; Xiang, Z.; Kohlhaas, S.; Das, P.P.; Miska, E.A.; Rodriguez, A.; Bradley, A.; et al. Microrna-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007, 27, 847–859. [Google Scholar] [CrossRef]
- Elton, T.S.; Selemon, H.; Elton, S.M.; Parinandi, N.L. Regulation of the mir155 host gene in physiological and pathological processes. Gene 2013, 532, 1–12. [Google Scholar] [CrossRef]
- Ronchetti, D.; Manzoni, M.; Agnelli, L.; Vinci, C.; Fabris, S.; Cutrona, G.; Matis, S.; Colombo, M.; Galletti, S.; Taiana, E.; et al. Lncrna profiling in early-stage chronic lymphocytic leukemia identifies transcriptional fingerprints with relevance in clinical outcome. Blood Cancer J. 2016, 6, e468. [Google Scholar] [CrossRef]
- Xia, L.; Wu, L.; Bao, J.; Li, Q.; Chen, X.; Xia, H.; Xia, R. Circular rna circ-cbfb promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating mir-607/fzd3/wnt/beta-catenin pathway. Biochemical Biophys. Res. Commun. 2018, 503, 385–390. [Google Scholar] [CrossRef]
- Wu, W.; Wu, Z.; Xia, Y.; Qin, S.; Li, Y.; Wu, J.; Liang, J.; Wang, L.; Zhu, H.; Fan, L.; et al. Downregulation of circ_0132266 in chronic lymphocytic leukemia promoted cell viability through mir-337-3p/pml axis. Aging 2019, 11, 3561–3573. [Google Scholar] [CrossRef]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA damage as a source of genomic instability in cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef]
- Knittel, G.; Liedgens, P.; Reinhardt, H.C. Targeting atm-deficient cll through interference with DNA repair pathways. Frontiers Genet. 2015, 6, 207. [Google Scholar] [CrossRef]
- Bagnara, D.; Kaufman, M.S.; Calissano, C.; Marsilio, S.; Patten, P.E.; Simone, R.; Chum, P.; Yan, X.J.; Allen, S.L.; Kolitz, J.E.; et al. A novel adoptive transfer model of chronic lymphocytic leukemia suggests a key role for t lymphocytes in the disease. Blood 2011, 117, 5463–5472. [Google Scholar] [CrossRef]
- Wang, L.Q.; Wong, K.Y.; Li, Z.H.; Chim, C.S. Epigenetic silencing of tumor suppressor long non-coding rna bm742401 in chronic lymphocytic leukemia. Oncotarget 2016, 7, 82400–82410. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.R.; Ruppert, A.S.; Fobare, S.; Chen, T.L.; Liu, C.; Lehman, A.; Blachly, J.S.; Zhang, X.; Lucas, D.M.; Grever, M.R.; et al. The long noncoding rna, trerna, decreases DNA damage and is associated with poor response to chemotherapy in chronic lymphocytic leukemia. Oncotarget 2017, 8, 25942–25954. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Kang, Q.; Zhu, X.; Chen, Q.; Wang, X.; Chen, Y.; Ouyang, J.; Zhang, L.; Tan, H.; Chen, R.; et al. A long noncoding rna critically regulates bcr-abl-mediated cellular transformation by acting as a competitive endogenous rna. Oncogene 2015, 34, 1768–1779. [Google Scholar] [CrossRef]
- Xia, T.; Chen, S.; Jiang, Z.; Shao, Y.; Jiang, X.; Li, P.; Xiao, B.; Guo, J. Long noncoding rna fer1l4 suppresses cancer cell growth by acting as a competing endogenous rna and regulating pten expression. Sci. Rep. 2015, 5, 13445. [Google Scholar] [CrossRef] [PubMed]
- Zenz, T.; Krober, A.; Scherer, K.; Habe, S.; Buhler, A.; Benner, A.; Denzel, T.; Winkler, D.; Edelmann, J.; Schwanen, C.; et al. Monoallelic tp53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: Results from a detailed genetic characterization with long-term follow-up. Blood 2008, 112, 3322–3329. [Google Scholar] [CrossRef]
- El-Mabhouh, A.A.; Ayres, M.L.; Shpall, E.J.; Baladandayuthapani, V.; Keating, M.J.; Wierda, W.G.; Gandhi, V. Evaluation of bendamustine in combination with fludarabine in primary chronic lymphocytic leukemia cells. Blood 2014, 123, 3780–3789. [Google Scholar] [CrossRef]
- Cerna, K.; Oppelt, J.; Chochola, V.; Musilova, K.; Seda, V.; Pavlasova, G.; Radova, L.; Arigoni, M.; Calogero, R.A.; Benes, V.; et al. Microrna mir-34a downregulates foxp1 during DNA damage response to limit bcr signalling in chronic lymphocytic leukaemia b cells. Leukemia 2019, 33, 403–414. [Google Scholar] [CrossRef]
- Martinez, P.; Blasco, M.A. Telomere-driven diseases and telomere-targeting therapies. J. Cell Biol. 2017, 216, 875–887. [Google Scholar] [CrossRef]
- Cusanelli, E.; Chartrand, P. Telomeric repeat-containing rna terra: A noncoding rna connecting telomere biology to genome integrity. Frontiers Genet. 2015, 6, 143. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing rna and rna surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent rna polymerase ii. Nat. Cell Biol. 2008, 10, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Artandi, S.E.; DePinho, R.A. Telomeres and telomerase in cancer. Carcinogenesis 2010, 31, 9–18. [Google Scholar] [CrossRef]
- Landau, D.A.; Tausch, E.; Taylor-Weiner, A.N.; Stewart, C.; Reiter, J.G.; Bahlo, J.; Kluth, S.; Bozic, I.; Lawrence, M.; Bottcher, S.; et al. Mutations driving cll and their evolution in progression and relapse. Nature 2015, 526, 525–530. [Google Scholar] [CrossRef]
- Schmidt, T.; Carmeliet, P. Angiogenesis: A target in solid tumors, also in leukemia? Hematol. Am. Soc. Hematol Educ Program 2011, 2011, 1–8. [Google Scholar] [CrossRef]
- Aguirre Palma, L.M.; Gehrke, I.; Kreuzer, K.A. Angiogenic factors in chronic lymphocytic leukaemia (cll): Where do we stand? Crit. Rev. Oncol. Hematol. 2015, 93, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Shanafelt, T.D.; Cimmino, A.; Taccioli, C.; Volinia, S.; Liu, C.G.; Calin, G.A.; Croce, C.M.; Chan, D.A.; Giaccia, A.J.; et al. Aberrant regulation of pvhl levels by microrna promotes the hif/vegf axis in cll b cells. Blood 2009, 113, 5568–5574. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, S.G.; Kontos, C.K.; Diamantopoulos, M.A.; Bouchla, A.; Glezou, E.; Bazani, E.; Pappa, V.; Scorilas, A. Microrna-155-5p overexpression in peripheral blood mononuclear cells of chronic lymphocytic leukemia patients is a novel, independent molecular biomarker of poor prognosis. Dis. Markers 2017, 2017, 2046545. [Google Scholar] [CrossRef] [PubMed]
- Marton, S.; Garcia, M.R.; Robello, C.; Persson, H.; Trajtenberg, F.; Pritsch, O.; Rovira, C.; Naya, H.; Dighiero, G.; Cayota, A. Small rnas analysis in cll reveals a deregulation of mirna expression and novel mirna candidates of putative relevance in cll pathogenesis. Leukemia 2008, 22, 330–338. [Google Scholar] [CrossRef]
- Willimott, S.; Wagner, S.D. Stromal cells and cd40 ligand (cd154) alter the mirnome and induce mirna clusters including, mir-125b/mir-99a/let-7c and mir-17-92 in chronic lymphocytic leukaemia. Leukemia 2012, 26, 1113–1116. [Google Scholar] [CrossRef]
- Dews, M.; Fox, J.L.; Hultine, S.; Sundaram, P.; Wang, W.; Liu, Y.Y.; Furth, E.; Enders, G.H.; El-Deiry, W.; Schelter, J.M.; et al. The myc-mir-17~92 axis blunts tgf{beta} signaling and production of multiple tgf{beta}-dependent antiangiogenic factors. Cancer Res. 2010, 70, 8233–8246. [Google Scholar] [CrossRef]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Paggetti, J.; Haderk, F.; Seiffert, M.; Janji, B.; Distler, U.; Ammerlaan, W.; Kim, Y.J.; Adam, J.; Lichter, P.; Solary, E.; et al. Exosomes released by chronic lymphocytic leukemia cells induce the transition of stromal cells into cancer-associated fibroblasts. Blood 2015, 126, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Herman, S.E.; Sun, X.; McAuley, E.M.; Hsieh, M.M.; Pittaluga, S.; Raffeld, M.; Liu, D.; Keyvanfar, K.; Chapman, C.M.; Chen, J.; et al. Modeling tumor-host interactions of chronic lymphocytic leukemia in xenografted mice to study tumor biology and evaluate targeted therapy. Leukemia 2013, 27, 2311–2321. [Google Scholar] [CrossRef]
- Burger, J.A.; Chiorazzi, N. B cell receptor signaling in chronic lymphocytic leukemia. Trends Immunol. 2013, 34, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A. Nurture versus nature: The microenvironment in chronic lymphocytic leukemia. Hematol. Am. Soc. Hematol. Educ. Program 2011, 2011, 96–103. [Google Scholar] [CrossRef]
- Choi, M.Y.; Kashyap, M.K.; Kumar, D. The chronic lymphocytic leukemia microenvironment: Beyond the b-cell receptor. Best Pract. Res. Clin Haematol. 2016, 29, 40–53. [Google Scholar] [CrossRef]
- Purroy, N.; Abrisqueta, P.; Carabia, J.; Carpio, C.; Palacio, C.; Bosch, F.; Crespo, M. Co-culture of primary cll cells with bone marrow mesenchymal cells, cd40 ligand and cpg odn promotes proliferation of chemoresistant cll cells phenotypically comparable to those proliferating in vivo. Oncotarget 2015, 6, 7632–7643. [Google Scholar] [CrossRef]
- Carabia, J.; Carpio, C.; Abrisqueta, P.; Jimenez, I.; Purroy, N.; Calpe, E.; Palacio, C.; Bosch, F.; Crespo, M. Microenvironment regulates the expression of mir-21 and tumor suppressor genes pten, pias3 and pdcd4 through zap-70 in chronic lymphocytic leukemia. Sci. Rep. 2017, 7, 12262. [Google Scholar] [CrossRef]
- Rossi, S.; Shimizu, M.; Barbarotto, E.; Nicoloso, M.S.; Dimitri, F.; Sampath, D.; Fabbri, M.; Lerner, S.; Barron, L.L.; Rassenti, L.Z.; et al. Microrna fingerprinting of cll patients with chromosome 17p deletion identify a mir-21 score that stratifies early survival. Blood 2010, 116, 945–952. [Google Scholar] [CrossRef]
- Medina, P.P.; Nolde, M.; Slack, F.J. Oncomir addiction in an in vivo model of microrna-21-induced pre-b-cell lymphoma. Nature 2010, 467, 86–90. [Google Scholar] [CrossRef]
- Cui, B.; Chen, L.; Zhang, S.; Mraz, M.; Fecteau, J.F.; Yu, J.; Ghia, E.M.; Zhang, L.; Bao, L.; Rassenti, L.Z.; et al. Microrna-155 influences b-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood 2014, 124, 546–554. [Google Scholar] [CrossRef]
- Fonte, E.; Apollonio, B.; Scarfo, L.; Ranghetti, P.; Fazi, C.; Ghia, P.; Caligaris-Cappio, F.; Muzio, M. In vitro sensitivity of cll cells to fludarabine may be modulated by the stimulation of toll-like receptors. Clin. Cancer Res. 2013, 19, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Davis, B.N. Control of microrna biogenesis by tgfbeta signaling pathway-a novel role of smads in the nucleus. Cytokine Growth Factor Rev. 2009, 20, 517–521. [Google Scholar] [CrossRef]
- Carrella, S.; Barbato, S.; D’Agostino, Y.; Salierno, F.G.; Manfredi, A.; Banfi, S.; Conte, I. Tgf-beta controls mir-181/erk regulatory network during retinal axon specification and growth. PLoS ONE 2015, 10, e0144129. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Matveeva, A.; Kovalevska, L.; Kholodnyuk, I.; Ivanivskaya, T.; Kashuba, E. The tgf-beta - smad pathway is inactivated in cronic lymphocytic leukemia cells. Exp. Oncol. 2017, 39, 286–290. [Google Scholar] [CrossRef]
- Zhu, D.X.; Zhu, W.; Fang, C.; Fan, L.; Zou, Z.J.; Wang, Y.H.; Liu, P.; Hong, M.; Miao, K.R.; Liu, P.; et al. Mir-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis 2012, 33, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Simon-Gabriel, C.P.; Foerster, K.; Saleem, S.; Bleckmann, D.; Benkisser-Petersen, M.; Thornton, N.; Umezawa, K.; Decker, S.; Burger, M.; Veelken, H.; et al. Microenvironmental stromal cells abrogate nf-kappab inhibitor-induced apoptosis in chronic lymphocytic leukemia. Haematologica 2018, 103, 136–147. [Google Scholar] [CrossRef]
- Gagez, A.L.; Duroux-Richard, I.; Lepretre, S.; Orsini-Piocelle, F.; Letestu, R.; De Guibert, S.; Tuaillon, E.; Leblond, V.; Khalifa, O.; Gouilleux-Gruart, V.; et al. Mir-125b and mir-532-3p predict the efficiency of rituximab-mediated lymphodepletion in chronic lymphocytic leukemia patients. A french innovative leukemia organization study. Haematologica 2017, 102, 746–754. [Google Scholar] [CrossRef]
- Zhou, L.; Bai, H.; Wang, C.; Wei, D.; Qin, Y.; Xu, X. Microrna125b promotes leukemia cell resistance to daunorubicin by inhibiting apoptosis. Mol. Med. Rep. 2014, 9, 1909–1916. [Google Scholar] [CrossRef]
- Herishanu, Y.; Katz, B.Z.; Lipsky, A.; Wiestner, A. Biology of chronic lymphocytic leukemia in different microenvironments: Clinical and therapeutic implications. Hematol. Oncol. Clin. North Am. 2013, 27, 173–206. [Google Scholar] [CrossRef] [PubMed]
- Sato-Kuwabara, Y.; Melo, S.A.; Soares, F.A.; Calin, G.A. The fusion of two worlds: Non-coding rnas and extracellular vesicles--diagnostic and therapeutic implications (review). Int. J. Oncol. 2015, 46, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Mao, R.; Yang, J. Nf-kappab and stat3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 2013, 4, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, M.; Emerson, B.M. P50-associated cox-2 extragenic rna (pacer) activates cox-2 gene expression by occluding repressive nf-kappab complexes. Elife 2014, 3, e01776. [Google Scholar] [CrossRef]
- Hu, G.; Gong, A.Y.; Wang, Y.; Ma, S.; Chen, X.; Chen, J.; Su, C.J.; Shibata, A.; Strauss-Soukup, J.K.; Drescher, K.M.; et al. Lincrna-cox2 promotes late inflammatory gene transcription in macrophages through modulating swi/snf-mediated chromatin remodeling. J. Immunol. 2016, 196, 2799–2808. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. Micrornas bind to toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef]
- Curtale, G.; Mirolo, M.; Renzi, T.A.; Rossato, M.; Bazzoni, F.; Locati, M. Negative regulation of toll-like receptor 4 signaling by il-10-dependent microrna-146b. Proc. Natl. Acad. Sci. USA 2013, 110, 11499–11504. [Google Scholar] [CrossRef]
- Lopez-Collazo, E.; del Fresno, C. Pathophysiology of endotoxin tolerance: Mechanisms and clinical consequences. Crit Care 2013, 17, 242. [Google Scholar] [CrossRef]
- Jurado-Camino, T.; Cordoba, R.; Esteban-Burgos, L.; Hernandez-Jimenez, E.; Toledano, V.; Hernandez-Rivas, J.A.; Ruiz-Sainz, E.; Cobo, T.; Siliceo, M.; Perez de Diego, R.; et al. Chronic lymphocytic leukemia: A paradigm of innate immune cross-tolerance. J. Immunol. 2015, 194, 719–727. [Google Scholar] [CrossRef]
- Jurj, A.; Pop, L.; Petrushev, B.; Pasca, S.; Dima, D.; Frinc, I.; Deak, D.; Desmirean, M.; Trifa, A.; Fetica, B.; et al. Exosome-carried microrna-based signature as a cellular trigger for the evolution of chronic lymphocytic leukemia into richter syndrome. Crit Rev. Clin Lab Sci. 2018, 55, 501–515. [Google Scholar] [CrossRef]
- Deng, M.; Yuan, H.; Liu, S.; Hu, Z.; Xiao, H. Exosome-transmitted linc00461 promotes multiple myeloma cell proliferation and suppresses apoptosis by modulating microrna/bcl-2 expression. Cytotherapy 2019, 21, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Farahani, M.; Rubbi, C.; Liu, L.; Slupsky, J.R.; Kalakonda, N. Cll exosomes modulate the transcriptome and behaviour of recipient stromal cells and are selectively enriched in mir-202-3p. PLoS ONE 2015, 10, e0141429. [Google Scholar] [CrossRef] [PubMed]
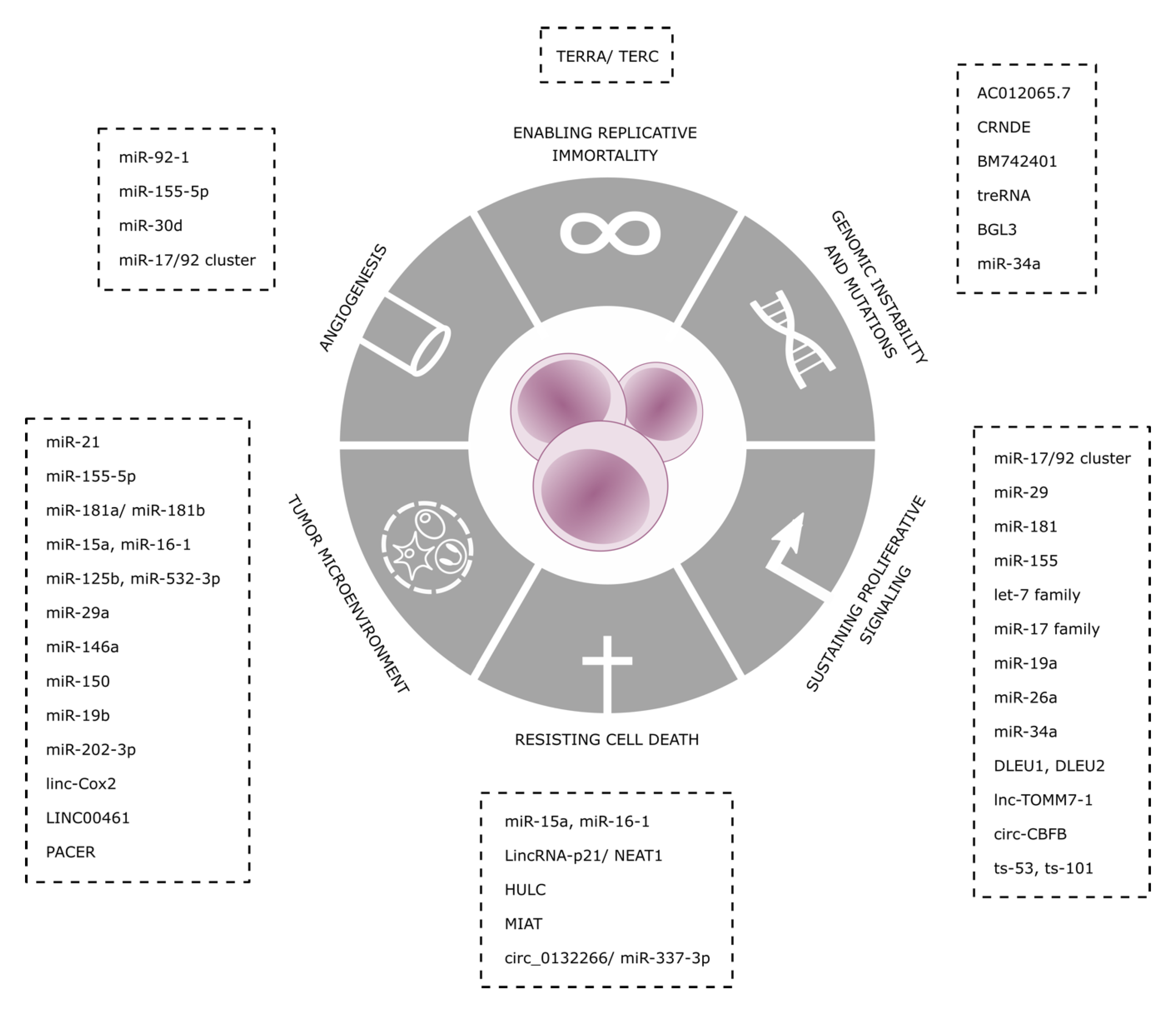
| Non Coding RNA | Mechanism | Levels IN CLL | Ref | |
|---|---|---|---|---|
| Angiogenesis | miR-92-1 | Represses pVHL and stabilizes HIF-1α, activating VEGF expression | overexpressed | [67] |
| miR-155-5p | Targets HIF-1α | High in aggressive disease | [68] | |
| miR-30d | Inhibits MYPT1, increases phosphorylation levels of c-JUN and activates VEGFA signaling cascade | Reduction | [69] | |
| miR-17/92 cluster | Targets SMAD4 and inhibits TGF-β responses | overexpression | [71,72] | |
| Enabling Replicative Immortality | TERRA/TERC | Inhibition of TERT | Overexpressed in CLL | [60,61,62] |
| Genomic Instability | AC012065.7 | positive expression correlation with GDF7 | Promoter hypo-methylated | [80] |
| CRNDE | Interacts with PRC2 and CoREST to modulate transcriptional repression | Promoter hyper -methylation | [80] | |
| BM742401 | Its expression leads to inhibition of cellular proliferation and enhanced apoptosis through caspase-9-dependent intrinsic but not caspase-8-dependent extrinsic apoptosis pathways | Methylated in CLL | [52] | |
| treRNA | decreases DNA damage and sensitivity to chemotherapy, | highly expressed, correlates with shorter overall survival (OS) | [53] | |
| BGL3 | regulates the oncogenic expression of BCR-ABL fusion gene through c-Myc mediated signaling | Decrease in CLL | [54] | |
| miR-34a | Downregulation of FOXP1, limiting BCR signaling | Upregulated during DNA damage response | [58] | |
| Resisting Cell Death | miR-15a, miR-16-1 | Target and deregulate BCL2 | Deleted in 13q- CLL | [24,25] |
| lincRNA-p21 | Induced by p53Induces p21 through hnRNP-K binding | Increased in p53WT samples | [32,33,34] | |
| HULC | endogenous sponge downregulating miRNAs, including miR-372 and miR-200a-3p | Upregulated in CLL | [42] | |
| MIAT | Constitution of a regulatory loop with OCT4 | Increased in patients with poor OS | [19,20] | |
| Circ_0132266 | endogenous sponge of hsa-miR-337-3p resulting in a downstream change of target-gene promyelocytic leukemia protein (PML) | decreased in the PBMCs of CLL patients | [48] | |
| Sustaining Proliferative Signaling | miR-17/92 cluster | mechanism is poorly understood, but up-regulation of miRNAs belonging to the miR-17-92 cluster is preceded by induction of MYC | Overexpressed in CLL | [11,12,13,14] |
| miR-29, miR-181 | Targeting TCL1 | Downregulated in CLL | [40] | |
| miR-155 | transcriptionally activated by MYB, leads to downregulation of several tumor suppressor genes. | Increased in CLL | [45] | |
| DLEU1, DLEU2 | NF-kB activation. Host of miR-15a/16-1 cluster targeting BCL2 | Homozygosis loss | [26,27,29] | |
| lnc-TOMM7-1 | It maps to chromosome 7p antisense to the interleukin-6 (IL6) gene and participate to its transcriptional regulation | downregulation | [46] | |
| circ_CBFB | acts as a sponge for miR-607, and contributes to the regulation of the Wnt/β-catenin pathway | highly upregulated | [47] | |
| ts-53, ts-101 | tsRNAs that play regulatory roles associating with Argonaute proteins | down-regulated in all CLL types | [40] | |
| Tumor Microenvironment | miR-21 | Transcriptionally activated by ZAP70, enhancer of BCR signaling | overexpression | [77,78,79] |
| miR-155-5p | Decreases SHIP1, resulting in higher responsiveness to BCR ligatio | upregulated | [81] | |
| miR-181a/miR-181b | regulated by TGF-β, they target BCL− 2, MCL-1 and XIAP | Downregulated | [84,85] | |
| miR-125-b | negatively regulates MS4A1 | levels in circulation are inversely correlated with rituximab-induced lymphodepletion | [87,88] | |
| miR-19b | Upregulates Ki67 and downregulates TP53 | Upregulated in plasma-derived exosomes in CLL | [100] | |
| miR-202-3p | It targets Sufu, a negative regulator of Hedgehog signaling | selectively enriched in CLL-derived EVs | [102] | |
| Linc-Cox2 | Acts as scaffold molecule recruiting SWI/SNF complex, activating the late primary inflammatory NF-κB-dependent genes | highly induced in macrophages upon TLR ligation | [93] | |
| LINC00461 | Directly correlates with decrease ERK1/2 and AKT activities and expression levels of miR-9, MEF2C and TMEM161B | highly expressed in MSCs-derived exosomes | [101] | |
| PACER | regulates COX-2 expression and acts as a decoy lncRNA for NF-kB signaling | n.d. | [92] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabris, L.; Juracek, J.; Calin, G. Non-Coding RNAs as Cancer Hallmarks in Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2020, 21, 6720. https://doi.org/10.3390/ijms21186720
Fabris L, Juracek J, Calin G. Non-Coding RNAs as Cancer Hallmarks in Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences. 2020; 21(18):6720. https://doi.org/10.3390/ijms21186720
Chicago/Turabian StyleFabris, Linda, Jaroslav Juracek, and George Calin. 2020. "Non-Coding RNAs as Cancer Hallmarks in Chronic Lymphocytic Leukemia" International Journal of Molecular Sciences 21, no. 18: 6720. https://doi.org/10.3390/ijms21186720
APA StyleFabris, L., Juracek, J., & Calin, G. (2020). Non-Coding RNAs as Cancer Hallmarks in Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences, 21(18), 6720. https://doi.org/10.3390/ijms21186720







