Protein Databases Related to Liquid–Liquid Phase Separation
Abstract
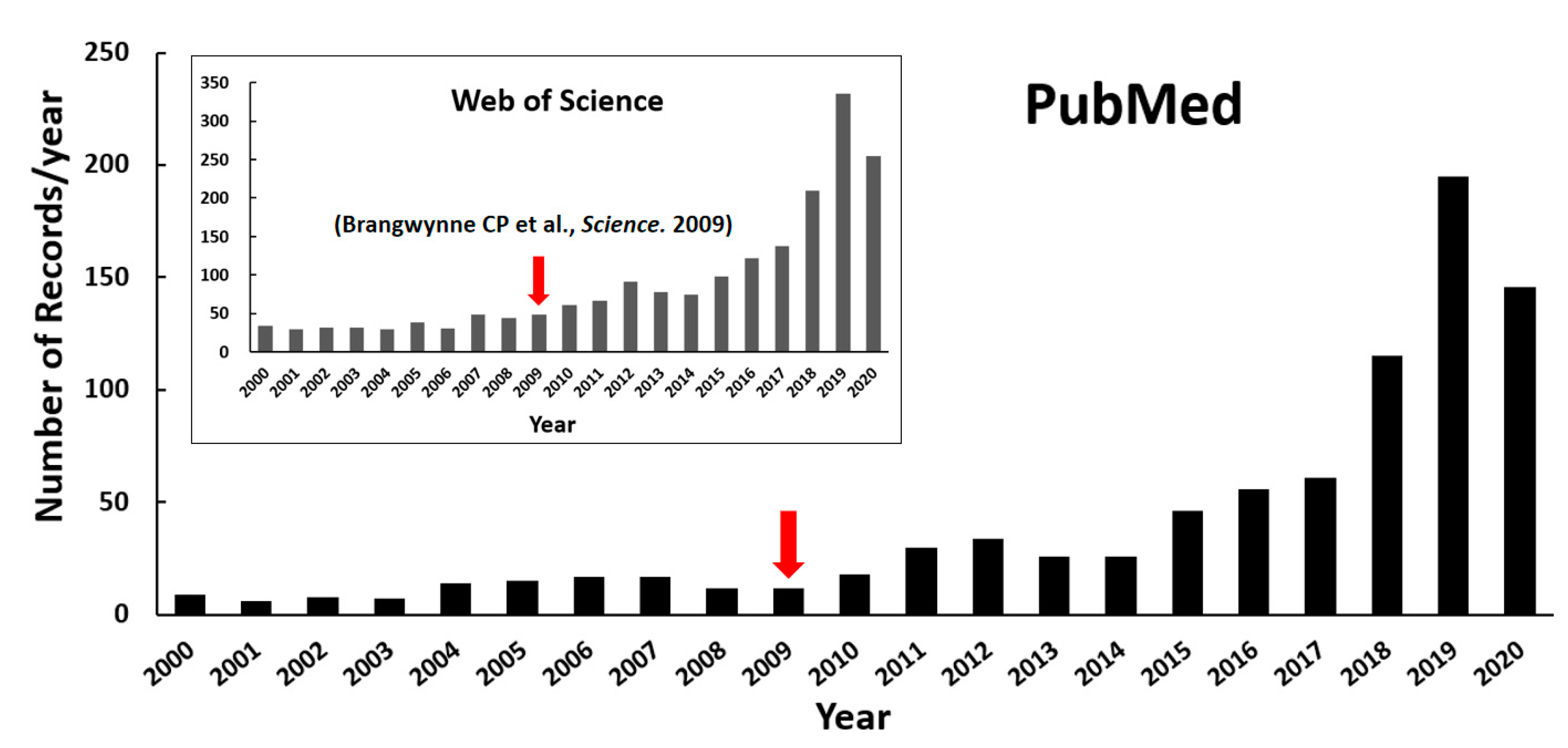
:1. Introduction
2. Databases Related to LLPS
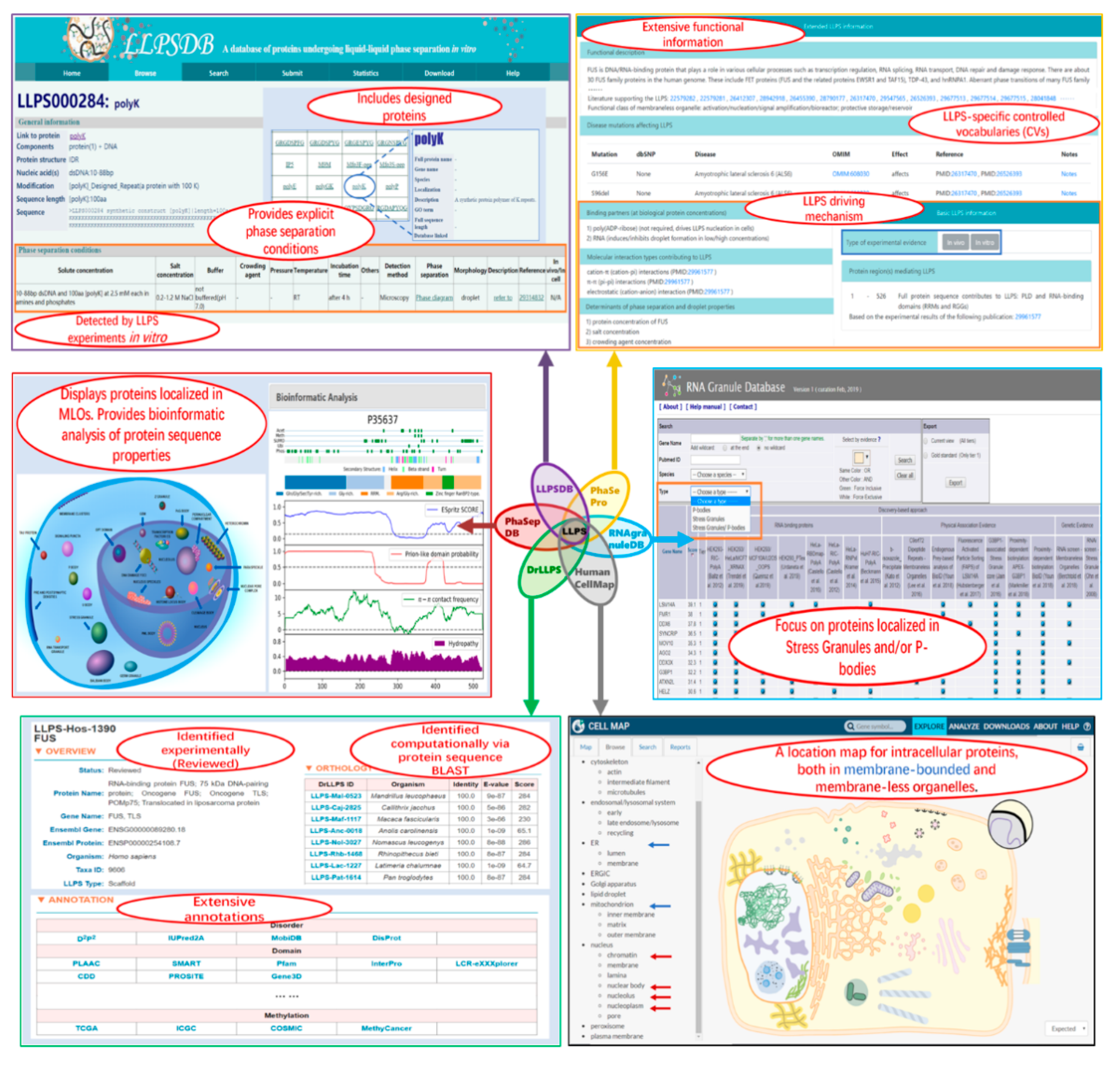
2.1. LLPSDB
2.2. PhaSePro
2.3. PhaSepDB
2.4. DrLLPS
2.5. RNAgranuleDB and HUMAN CELL MAP
3. Comparison of the Databases
4. Summary
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| LLPS | Liquid−liquid phase separation |
| MLOs | membraneless organelles |
| SG | stress granule |
| PB | P-body |
| PTMs | post-translational modifications |
| ALS | amyotrophic lateral sclerosis |
| FTD | frontotemporal dementia |
| AD | Alzheimer’s disease |
| IDRs | intrinsically disordered regions |
| IDPs | intrinsically disordered proteins |
| LCRs | low-complexity regions |
| CVs | controlled vocabularies |
References
- Uversky, V.N. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv. Colloid Interface Sci. 2017, 239, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Mitrea, D.M.; Kriwacki, R.W. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 2016, 14, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Julicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. What lava lamps and vinaigrette can teach us about cell biology. Nature 2018, 555, 300–302. [Google Scholar] [CrossRef] [Green Version]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gui, X.R.; Luo, F.; Li, Y.C.; Zhou, H.; Qin, Z.H.; Liu, Z.Y.; Gu, J.G.; Xie, M.Y.; Zhao, K.; Dai, B.; et al. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat. Commun. 2019, 10, 2006–2017. [Google Scholar] [CrossRef] [Green Version]
- Patel, A.; Lee, H.O.; Jawerth, L.; Maharana, S.; Jahnel, M.; Hein, M.Y.; Stoynov, S.; Mahamid, J.; Saha, S.; Franzmann, T.M.; et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 2015, 162, 1066–1077. [Google Scholar] [CrossRef] [Green Version]
- Mann, J.R.; Gleixner, A.M.; Mauna, J.C.; Gomes, E.; DeChellis-Marks, M.R.; Needham, P.G.; Copley, K.E.; Hurtle, B.; Portz, B.; Pyles, N.J.; et al. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 2019, 102, 321–338.e8. [Google Scholar] [CrossRef] [Green Version]
- Kostylev, M.A.; Tuttle, M.D.; Lee, S.; Klein, L.E.; Takahashi, H.; Cox, T.O.; Gunther, E.C.; Zilm, K.W.; Strittmatter, S.M. Liquid and Hydrogel Phases of PrP(C) Linked to conformation shifts and triggered by Alzheimer’s amyloid-beta oligomers. Mol. Cell 2018, 72, 426–443.e12. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.S.; Lai, L. Physiochemical mechanisms of biomolecular liquid-liquid phase separation. Acta Phys.-Chim. Sin. 2020, 36, 1907050–1907053. [Google Scholar] [CrossRef]
- Dignon, G.L.; Best, R.B.; Mittal, J. Biomolecular Phase separation: From molecular driving forces to macroscopic properties. Annu. Rev. Phys. Chem. 2020, 71, 53–75. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.M.; Holehouse, A.S.; Pappu, R.V. Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 2020, 49, 107–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, B.S.; Reed, E.H.; Parthasarathy, R.; Jahnke, C.N.; Caldwell, R.M.; Bermudez, J.G.; Ramage, H.; Good, M.C.; Hammer, D.A. Controllable protein phase separation and modular recruitment to form responsive membraneless organelles. Nat. Commun. 2018, 9, 2985. [Google Scholar] [CrossRef] [PubMed]
- Elbaum-Garfinkle, S.; Kim, Y.; Szczepaniak, K.; Chen, C.C.; Eckmann, C.R.; Myong, S.; Brangwynne, C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 7189–7194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monahan, Z.; Ryan, V.H.; Janke, A.M.; Burke, K.A.; Rhoads, S.N.; Zerze, G.H.; O’Meally, R.; Dignon, G.L.; Conicella, A.E.; Zheng, W.; et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017, 36, 2951–2967. [Google Scholar] [CrossRef]
- McGurk, L.; Gomes, E.; Guo, L.; Mojsilovic-Petrovic, J.; Tran, V.; Kalb, R.G.; Shorter, J.; Bonini, N.M. Poly(ADP-ribose) prevents pathological phase separation of TDP-43 by promoting liquid demixing and stress granule localization. Mol. Cell 2018, 71, 703–717.e9. [Google Scholar] [CrossRef] [Green Version]
- Fung, H.Y.J.; Birol, M.; Rhoades, E. IDPs in macromolecular complexes: The roles of multivalent interactions in diverse assemblies. Curr. Opin. Struct. Biol. 2018, 49, 36–43. [Google Scholar] [CrossRef]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.H.; Forman-Kay, J.D.; Chan, H.S. Theories for sequence-dependent phase behaviors of biomolecular condensates. Biochemistry 2018, 57, 2499–2508. [Google Scholar] [CrossRef]
- Martin, E.W.; Mittag, T. Relationship of sequence and phase separation in protein low-complexity regions. Biochemistry 2018, 57, 2478–2487. [Google Scholar] [CrossRef]
- Murthy, A.C.; Dignon, G.L.; Kan, Y.; Zerze, G.H.; Parekh, S.H.; Mittal, J.; Fawzi, N.L. Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol. 2019, 26, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Harmon, T.S.; Holehouse, A.S.; Pappu, R.V. Differential solvation of intrinsically disordered linkers drives the formation of spatially organized droplets in ternary systems of linear multivalent proteins. New J. Phys. 2018, 20, 045002. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Song, Z.; Zhong, S.; Zuo, L.; Qi, Z.; Qu, L.J.; Lai, L. Mechanism of DNA-induced phase separation for transcriptional repressor VRN1. Angew. Chem. Int. Ed. Engl. 2019, 58, 4858–4862. [Google Scholar] [CrossRef]
- Du, M.; Chen, Z.J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 2018, 361, 704–709. [Google Scholar] [CrossRef] [Green Version]
- Drino, A.; Schaefer, M.R. RNAs, Phase separation, and membrane-Less organelles: Are post-transcriptional modifications modulating organelle dynamics? Bioessays 2018, 40, e1800085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darling, A.L.; Zaslavsky, B.Y.; Uversky, V.N. Intrinsic disorder-based emergence in cellular biology: Physiological and pathological liquid-liquid phase transitions in cells. Polymers 2019, 11, 990–1012. [Google Scholar] [CrossRef] [Green Version]
- Posey, A.E.; Holehouse, A.S.; Pappu, R.V. Phase separation of intrinsically disordered proteins. Methods Enzymol. 2018, 611, 1–30. [Google Scholar]
- Ruff, K.M.; Roberts, S.; Chilkoti, A.; Pappu, R.V. Advances in understanding stimulus-responsive phase behavior of intrinsically disordered protein polymers. J. Mol. Biol. 2018, 430, 4619–4635. [Google Scholar] [CrossRef]
- Wang, J.; Choi, J.M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 2018, 174, 688–699 e16. [Google Scholar] [CrossRef] [Green Version]
- Schuster, B.S.; Dignon, G.L.; Tang, W.S.; Kelley, F.M.; Ranganath, A.K.; Jahnke, C.N.; Simpkins, A.G.; Regy, R.M.; Hammer, D.A.; Good, M.C.; et al. Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc. Natl. Acad. Sci. USA 2020, 117, 11421–11431. [Google Scholar] [CrossRef]
- Uversky, V.N.; Kuznetsova, I.M.; Turoverov, K.K.; Zaslavsky, B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 2015, 589, 15–22. [Google Scholar] [CrossRef] [Green Version]
- Banani, S.F.; Rice, A.M.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional control of phase-separated cellular bodies. Cell 2016, 166, 651–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, A.; Mazarakos, K.; Zhou, H.X. Three archetypical classes of macromolecular regulators of protein liquid-liquid phase separation. Proc. Natl. Acad. Sci. USA 2019, 116, 19474–19483. [Google Scholar] [CrossRef] [Green Version]
- Murakami, T.; Qamar, S.; Lin, J.Q.; Schierle, G.S.; Rees, E.; Miyashita, A.; Costa, A.R.; Dodd, R.B.; Chan, F.T.; Michel, C.H.; et al. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 2015, 88, 678–690. [Google Scholar] [CrossRef] [Green Version]
- Dao, T.P.; Martyniak, B.; Canning, A.J.; Lei, Y.; Colicino, E.G.; Cosgrove, M.S.; Hehnly, H.; Castaneda, C.A. ALS-linked mutations affect UBQLN2 oligomerization and phase separation in a position- and amino acid-dependent manner. Structure 2019, 27, 937–951.e5. [Google Scholar] [CrossRef]
- Ryan, V.H.; Dignon, G.L.; Zerze, G.H.; Chabata, C.V.; Silva, R.; Conicella, A.E.; Amaya, J.; Burke, K.A.; Mittal, J.; Fawzi, N.L. Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell 2018, 69, 465–479.e7. [Google Scholar] [CrossRef]
- Cinar, H.; Fetahaj, Z.; Cinar, S.; Vernon, R.M.; Chan, H.S.; Winter, R.H.A. Temperature, hydrostatic pressure, and osmolyte effects on liquid-liquid phase separation in protein condensates: Physical chemistry and biological implications. Chemistry 2019, 25, 13049–13069. [Google Scholar] [CrossRef] [PubMed]
- Ning, W.; Guo, Y.; Lin, S.; Mei, B.; Wu, Y.; Jiang, P.; Tan, X.; Zhang, W.; Chen, G.; Peng, D.; et al. DrLLPS: A data resource of liquid-liquid phase separation in eukaryotes. Nucleic Acids Res. 2020, 48, D288–D295. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef]
- Lee, K.H.; Zhang, P.; Kim, H.J.; Mitrea, D.M.; Sarkar, M.; Freibaum, B.D.; Cika, J.; Coughlin, M.; Messing, J.; Molliex, A.; et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 2016, 167, 774–788.e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Youn, J.Y.; Dunham, W.H.; Hong, S.J.; Knight, J.D.R.; Bashkurov, M.; Chen, G.I.; Bagci, H.; Rathod, B.; MacLeod, G.; Eng, S.W.M.; et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 2018, 69, 517–532.e11. [Google Scholar] [CrossRef]
- Jain, S.; Wheeler, J.R.; Walters, R.W.; Agrawal, A.; Barsic, A.; Parker, R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 2016, 164, 487–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Peng, X.; Li, Y.; Tang, W.; Zhu, J.; Huang, J.; Qi, Y.; Zhang, Z. LLPSDB: A database of proteins undergoing liquid-liquid phase separation in vitro. Nucleic Acids Res. 2020, 48, D320–D327. [Google Scholar] [CrossRef]
- Meszaros, B.; Erdos, G.; Szabo, B.; Schad, E.; Tantos, A.; Abukhairan, R.; Horvath, T.; Murvai, N.; Kovacs, O.P.; Kovacs, M.; et al. PhaSePro: The database of proteins driving liquid-liquid phase separation. Nucleic Acids Res. 2020, 48, D360–D367. [Google Scholar] [PubMed]
- You, K.; Huang, Q.; Yu, C.; Shen, B.; Sevilla, C.; Shi, M.; Hermjakob, H.; Chen, Y.; Li, T. PhaSepDB: A database of liquid-liquid phase separation related proteins. Nucleic Acids Res. 2020, 48, D354–D359. [Google Scholar] [CrossRef]
- Youn, J.Y.; Dyakov, B.J.A.; Zhang, J.; Knight, J.D.R.; Vernon, R.M.; Forman-Kay, J.D.; Gingras, A.C. Properties of stress granule and P-body proteomes. Mol. Cell 2019, 76, 286–294. [Google Scholar] [CrossRef]
- Go, C.D.; Knight, J.D.R.; Rajasekharan, A.; Rathod, B.; Hesketh, G.G.; Abe, K.T.; Youn, J.-Y.; Samavarchi-Tehrani, P.; Zhang, H. A proximity biotinylation map of a human cell. bioRxiv 2019, 796391. [Google Scholar] [CrossRef] [Green Version]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Alpi, E.; Bely, B.; Bingley, M.; Britto, R.; Bursteinas, B.; Busiello, G.; et al. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar]
- Piovesan, D.; Tabaro, F.; Paladin, L.; Necci, M.; Micetic, I.; Camilloni, C.; Davey, N.; Dosztanyi, Z.; Meszaros, B.; Monzon, A.M.; et al. MobiDB 3.0: More annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res. 2018, 46, D471–D476. [Google Scholar] [CrossRef]
- Piovesan, D.; Tabaro, F.; Micetic, I.; Necci, M.; Quaglia, F.; Oldfield, C.J.; Aspromonte, M.C.; Davey, N.E.; Davidovic, R.; Dosztanyi, Z.; et al. DisProt 7.0: A major update of the database of disordered proteins. Nucleic Acids Res. 2017, 45, D219–D227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amberger, J.S.; Bocchini, C.A.; Scott, A.F.; Hamosh, A. OMIM.org: Leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019, 47, D1038–D1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuchi, S.; Amemiya, T.; Sakamoto, S.; Nobe, Y.; Hosoda, K.; Kado, Y.; Murakami, S.D.; Koike, R.; Hiroaki, H.; Ota, M. IDEAL in 2014 illustrates interaction networks composed of intrinsically disordered proteins and their binding partners. Nucleic Acids Res. 2014, 42, D320–D325. [Google Scholar] [CrossRef] [PubMed]
- Miskei, M.; Antal, C.; Fuxreiter, M. FuzDB: Database of fuzzy complexes, a tool to develop stochastic structure-function relationships for protein complexes and higher-order assemblies. Nucleic Acids Res. 2017, 45, D228–D235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varadi, M.; De Baets, G.; Vranken, W.F.; Tompa, P.; Pancsa, R. AmyPro: A database of proteins with validated amyloidogenic regions. Nucleic Acids Res. 2018, 46, D387–D392. [Google Scholar] [CrossRef] [Green Version]
- Meszaros, B.; Erdos, G.; Dosztanyi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Kornhauser, J.M.; Latham, V.; Murray, B.; Nandhikonda, V.; Nord, A.; Skrzypek, E.; Wheeler, T.; Zhang, B.; Gnad, F. 15 years of PhosphoSitePlus((R)): Integrating post-translationally modified sites, disease variants and isoforms. Nucleic Acids Res. 2019, 47, D433–D441. [Google Scholar] [CrossRef] [Green Version]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Tosatto, S.C. ESpritz: Accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, A.K.; Nutter-Upham, A.; Lindquist, S.; King, O.D. PLAAC: A web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 2014, 30, 2501–2502. [Google Scholar] [CrossRef] [Green Version]
- Vernon, R.M.; Chong, P.A.; Tsang, B.; Kim, T.H.; Bah, A.; Farber, P.; Lin, H.; Forman-Kay, J.D. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 2018, 7, e31486. [Google Scholar] [CrossRef] [PubMed]
- Holehouse, A.S.; Das, R.K.; Ahad, J.N.; Richardson, M.O.; Pappu, R.V. CIDER: Resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophys. J. 2017, 112, 16–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunningham, F.; Achuthan, P.; Akanni, W.; Allen, J.; Amode, M.R.; Armean, I.M.; Bennett, R.; Bhai, J.; Billis, K.; Boddu, S.; et al. Ensembl 2019. Nucleic Acids Res. 2019, 47, D745–D751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeya, M.; Yamasaki, F.; Uzuhashi, S.; Aoki, T.; Sawada, H.; Nagai, T.; Tomioka, K.; Tomooka, N.; Sato, T.; Kawase, M. NIASGBdb: NIAS Genebank databases for genetic resources and plant disease information. Nucleic Acids Res. 2011, 39, D1108–D1113. [Google Scholar] [CrossRef] [Green Version]
- Pruitt, K.D.; Brown, G.R.; Hiatt, S.M.; Thibaud-Nissen, F.; Astashyn, A.; Ermolaeva, O.; Farrell, C.M.; Hart, J.; Landrum, M.J.; McGarvey, K.M.; et al. RefSeq: An update on mammalian reference sequences. Nucleic Acids Res. 2014, 42, D756–D763. [Google Scholar] [CrossRef]
- Vernon, R.M.; Forman-Kay, J.D. First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol. 2019, 58, 88–96. [Google Scholar] [CrossRef]
- Sun, T.; Li, Q.; Xu, Y.; Zhang, Z.; Lai, L.; Pei, J. Prediction of liquid-liquid phase separation proteins using machine learning. bioRxiv 2019, 842336. [Google Scholar] [CrossRef]
- Orlando, G.; Raimondi, D.; Tabaro, F.; Codice, F.; Moreau, Y.; Vranken, W.F. Computational identification of prion-like RNA-binding proteins that form liquid phase-separated condensates. Bioinformatics 2019, 35, 4617–4623. [Google Scholar] [CrossRef]
| Databases | Organization | Data Contents | Data Sources | Outstanding Features | Availability | Ref. |
|---|---|---|---|---|---|---|
| LLPSDB | Entries are defined by specific protein and/or nucleic acid constructs. Classified by
| 273 proteins 1175 entries | Validated by LLPS experiments in vitro |
| http://bio-comp.ucas.ac.cn/llpsdb or http://bio-comp.org.cn/llpsdb | [44] |
| PhaSePro | Entries are defined by specific proteins. | 121 proteins (109 from eukaryotes, 5 from bacteria, and 7 from viruses) | Validated by LLPS experiments in vitro and/or in vivo |
| https://phasepro.elte.hu | [45] |
| PhaSepDB | Entries are defined by specific proteins. Classified by
| 2914 proteins (352 are detected by LLPS experiments; 378 are reviewed from UniProt according to protein localization in MLOs; 2572 are identified to be localized in MLOs based on high throughput experiments) |
|
| http://db.phasep.pro/ | [46] |
| DrLLPS | Entries are defined by specific genes. Classified by
| 437,887 proteins in 164 eukaryotes (9285 are identified experimentally, 428,602 are identified computationally via protein sequence blast) |
|
| http://llps.biocuckoo.cn/ | [39] |
| RNAgranuleDB | Entries are defined by specific proteins. Three hierarchical levels:
| 4385 proteins (368 proteins were assigned to Tier 1 with highest confidence SG-PB proteins, 475 to Tier 2, 428 to Tier 3, and 3114 to Tier 4) | Localized in stress granule and P body, validated by experiments |
| http://rnagranuledb.lunenfeld.ca | [47] |
| HUMAN CELL MAP | Entries are defined by specific genes. Classified by organelle type (membrane-bound or membraneless) | 4145 proteins | Localized in membrane-bound or membraneless organelles through identification based on experiments combined with analysis. |
| https://cell-map.org/ or https://humancellmap.org/ | [48] |
| LLPSDB | PhaSePro | PhaSepDB | DrLLPS | RNAgranuleDB | HUMAN CELL MAP | |
|---|---|---|---|---|---|---|
| LLPSDB | 273 | |||||
| PhaSePro | 65 | 121 | ||||
| PhaSepDB | 94 | 82 | 2957 | |||
| DrLLPS | 115 | 83 | 1520 | 9281 | ||
| RNAgranuleDB | 75 | 56 | 491 | 2440 | 4386 | |
| HUMAN CELL MAP | 45 | 35 | 1056 | 1825 | 2519 | 4424 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Wang, X.; Dou, Z.; Yang, W.; Huang, B.; Lou, J.; Zhang, Z. Protein Databases Related to Liquid–Liquid Phase Separation. Int. J. Mol. Sci. 2020, 21, 6796. https://doi.org/10.3390/ijms21186796
Li Q, Wang X, Dou Z, Yang W, Huang B, Lou J, Zhang Z. Protein Databases Related to Liquid–Liquid Phase Separation. International Journal of Molecular Sciences. 2020; 21(18):6796. https://doi.org/10.3390/ijms21186796
Chicago/Turabian StyleLi, Qian, Xi Wang, Zhihui Dou, Weishan Yang, Beifang Huang, Jizhong Lou, and Zhuqing Zhang. 2020. "Protein Databases Related to Liquid–Liquid Phase Separation" International Journal of Molecular Sciences 21, no. 18: 6796. https://doi.org/10.3390/ijms21186796
APA StyleLi, Q., Wang, X., Dou, Z., Yang, W., Huang, B., Lou, J., & Zhang, Z. (2020). Protein Databases Related to Liquid–Liquid Phase Separation. International Journal of Molecular Sciences, 21(18), 6796. https://doi.org/10.3390/ijms21186796






