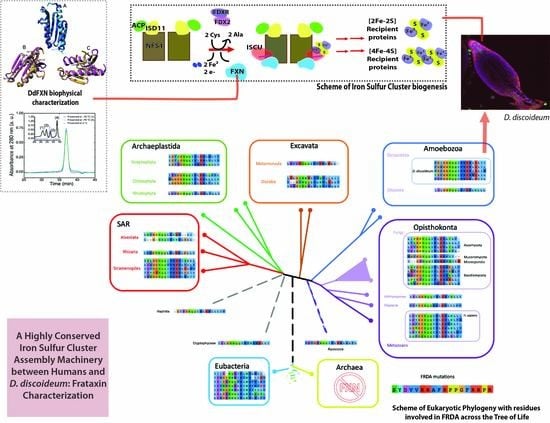
A Highly Conserved Iron-Sulfur Cluster Assembly Machinery between Humans and Amoeba Dictyostelium discoideum: The Characterization of Frataxin
Abstract
1. Introduction
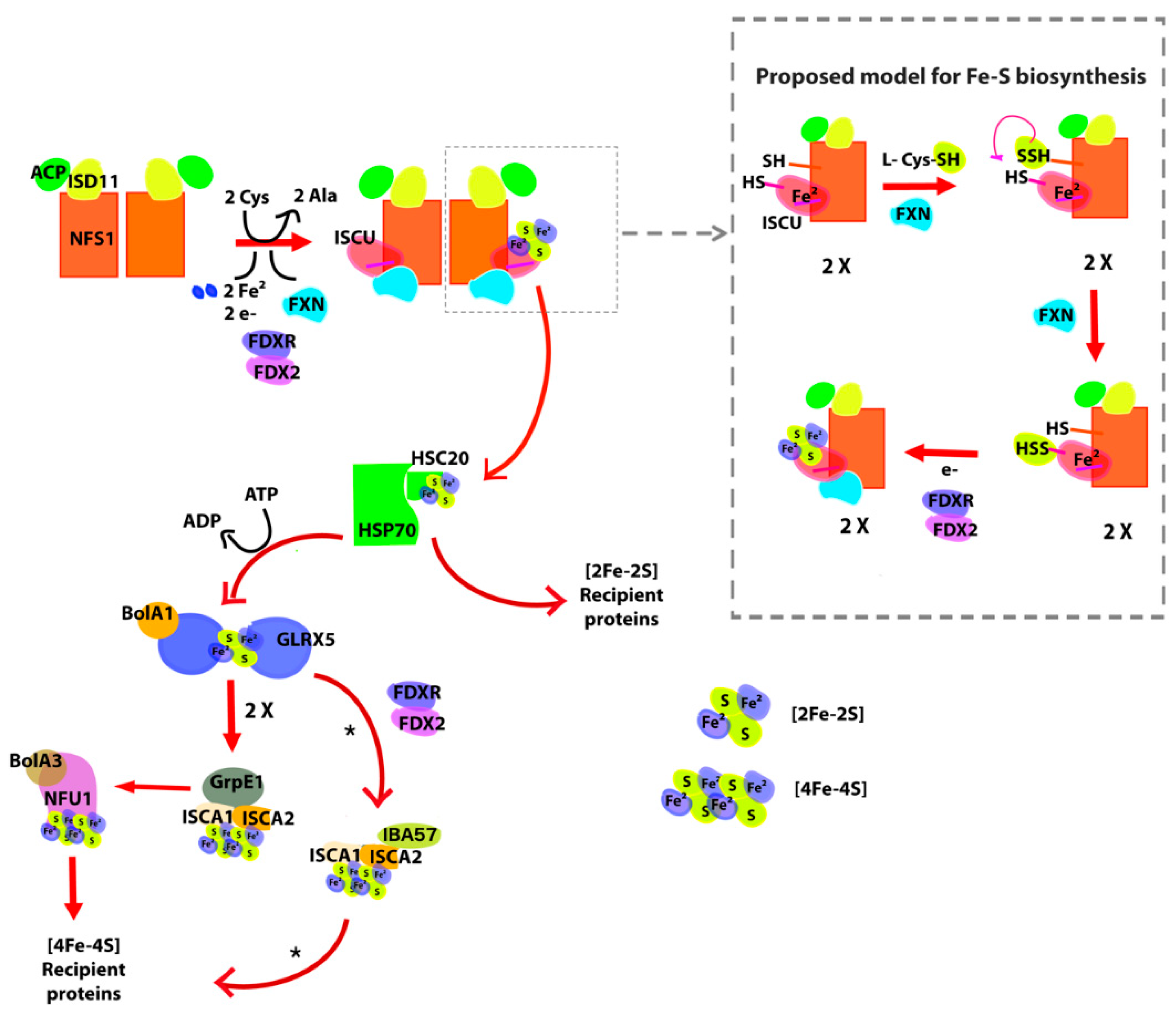
1.1. Iron–Sulfur Cluster Assembly in Mammals
1.2. D. discoideum as A Model to Study Iron-Sulfur Cluster Assembly Physiology
2. Results
2.1. The [Fe-S] Cluster Assembly Supercomplex of D. discoideum
- -
- Arg72 from cysteine desulfurase NFS1. It is involved in a rare disease when it is mutated to Gln (R72Q) [24].
- -
- -
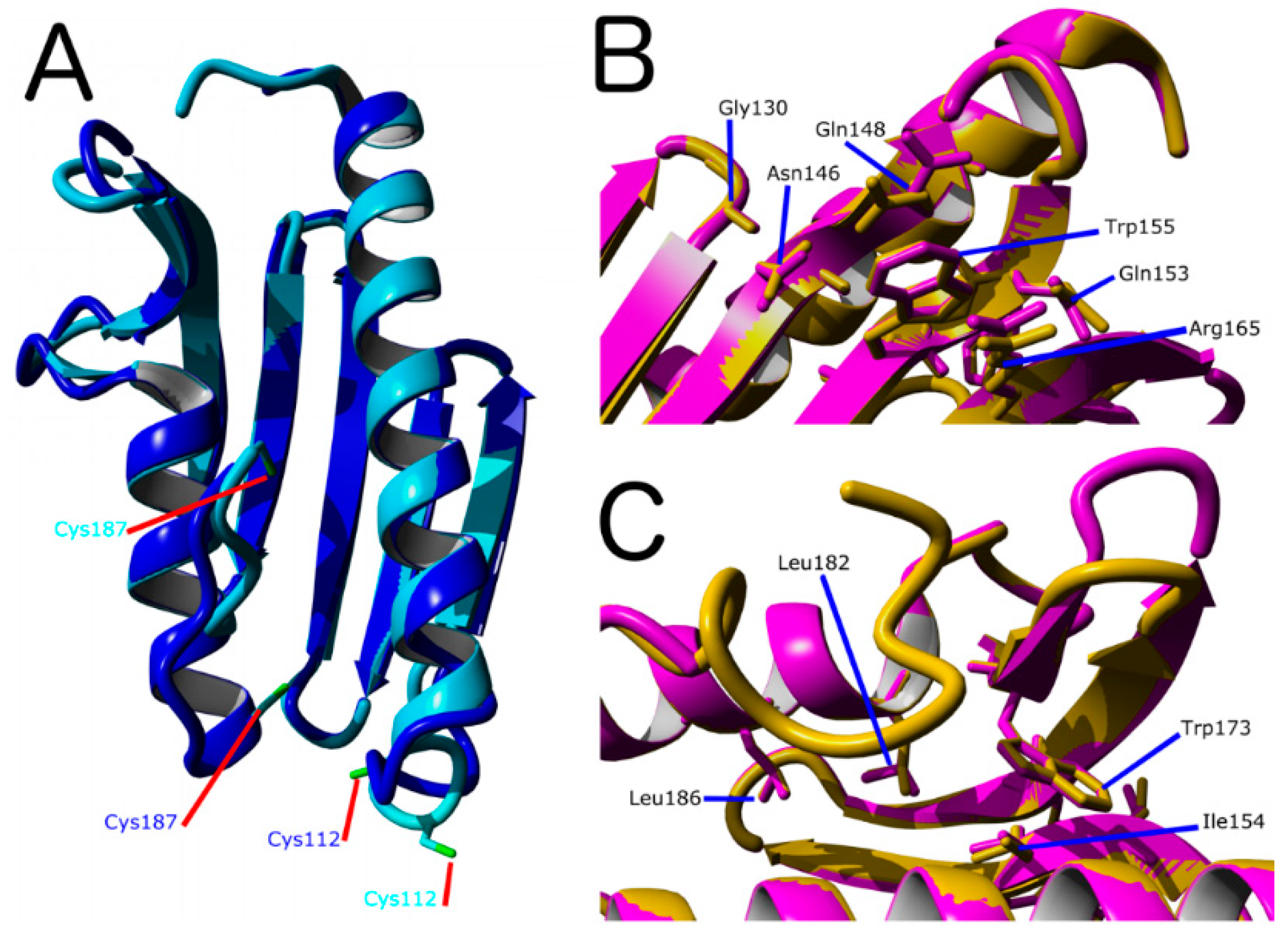
- Gly130, Gly137, Asn146, Gln148, Ile154, Trp155, Arg165, Trp173, Leu182, Leu186 from the kinetic activator FXN (human FXN numbering). Mutation of these residues result in FRDA.
- -
- Arg68 from tutor protein ISD11. Recent studies on human ISD11 showed that a homozygous mutation R68L is implicated in the mitochondrial genetic disorder COXPD19.
2.2. Characterization of D. discoideum Frataxin
- (A).
- Based on PDB ID: 6FCO (Chaetomium thermophilum);
- (B).
- Based on PDB ID: 1EKG (H. sapiens);
- (C).
- Built using PDB ID: 2EFF (E. coli).
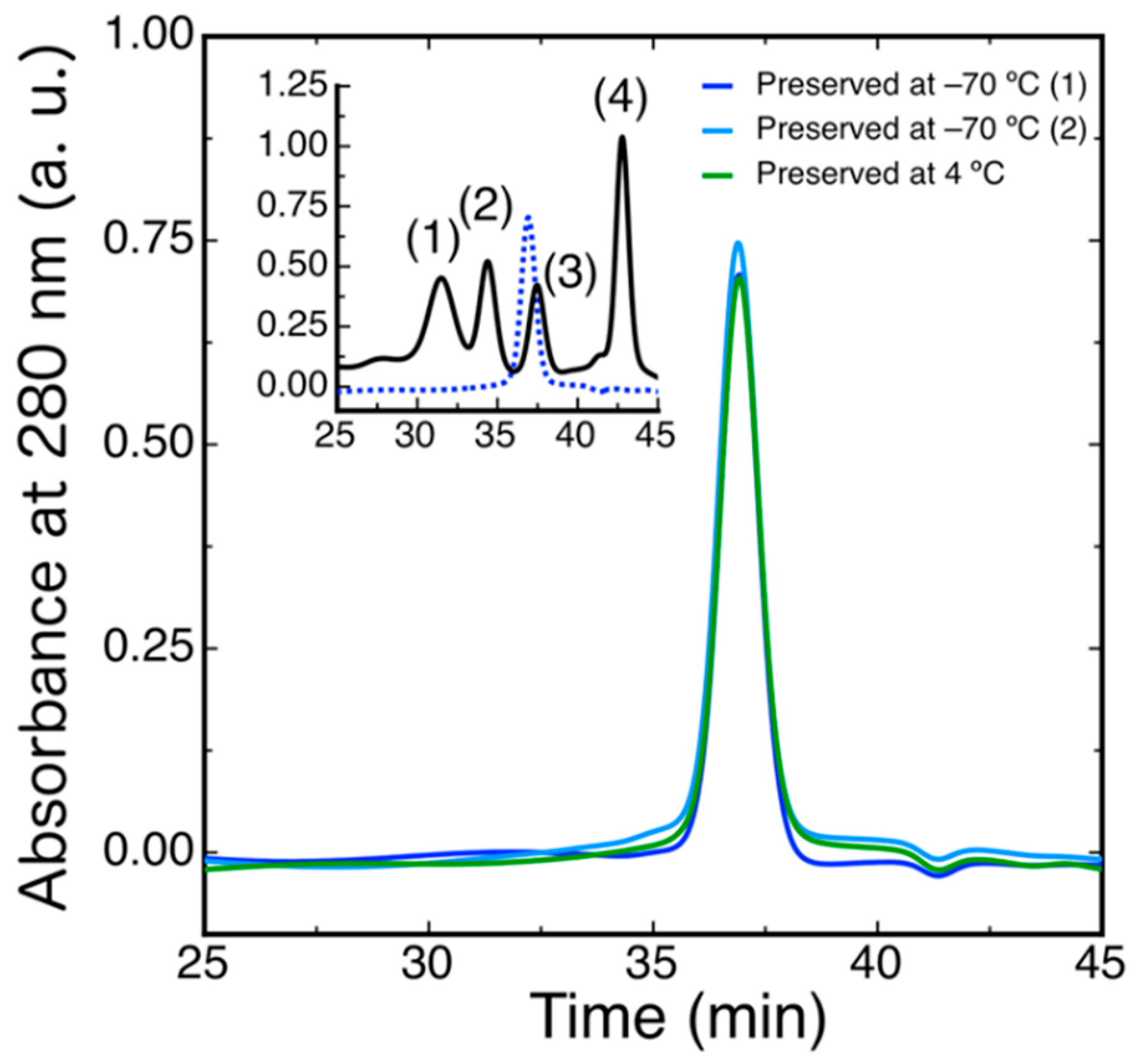
2.3. Protein Expression
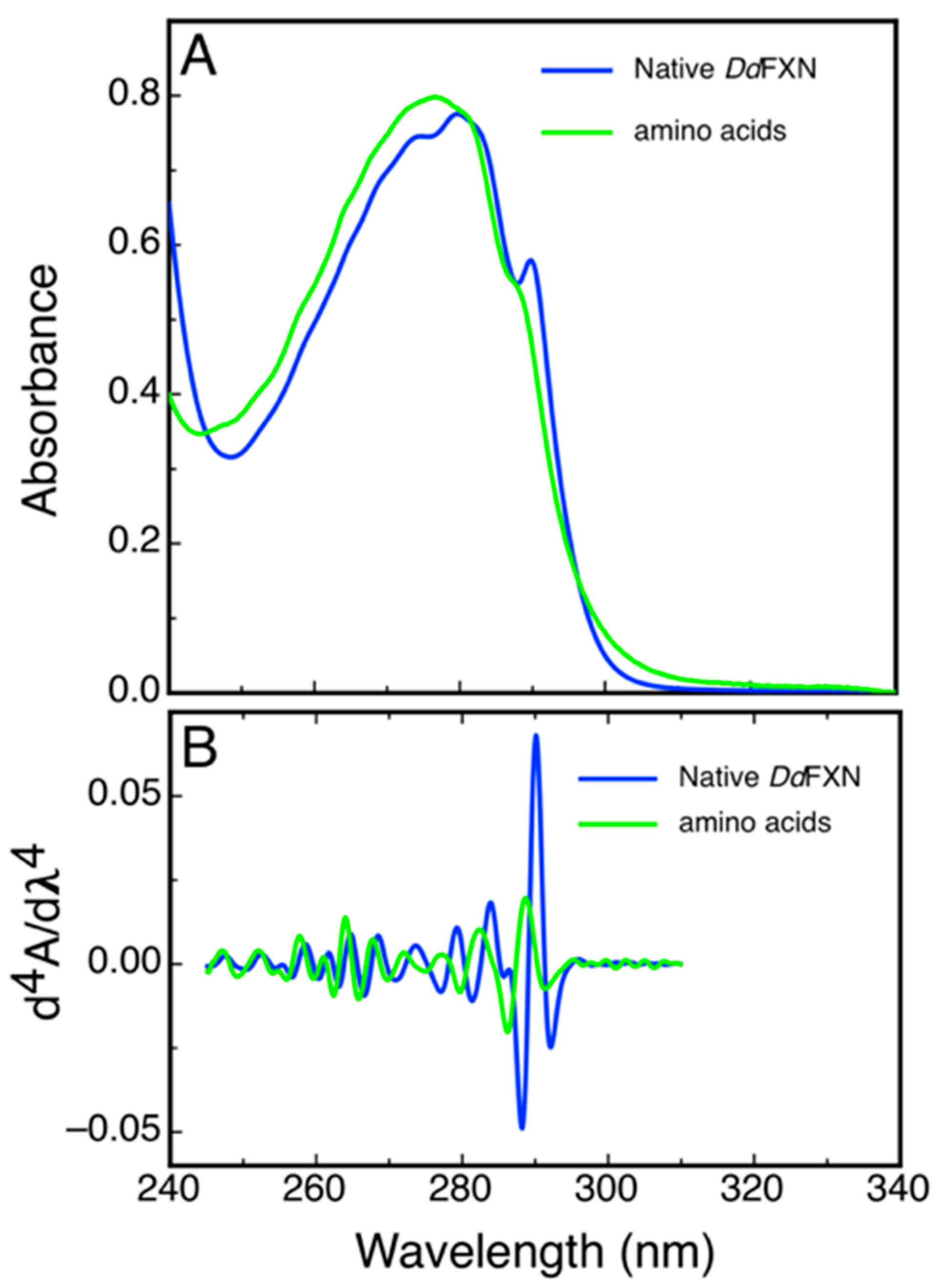
2.4. Spectroscopic Characterization of DdFXN
2.5. Conformational Stability of DdFXN
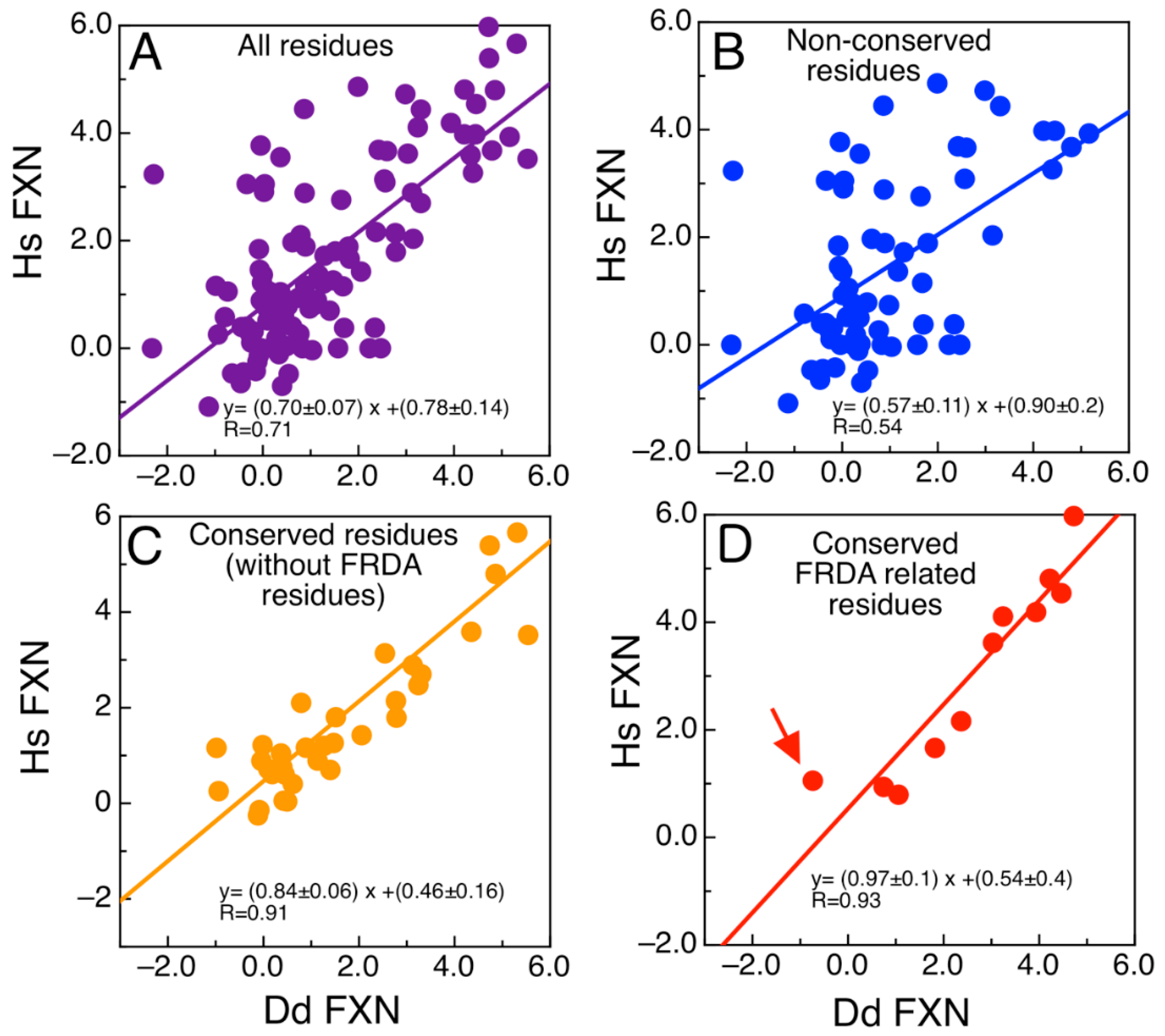
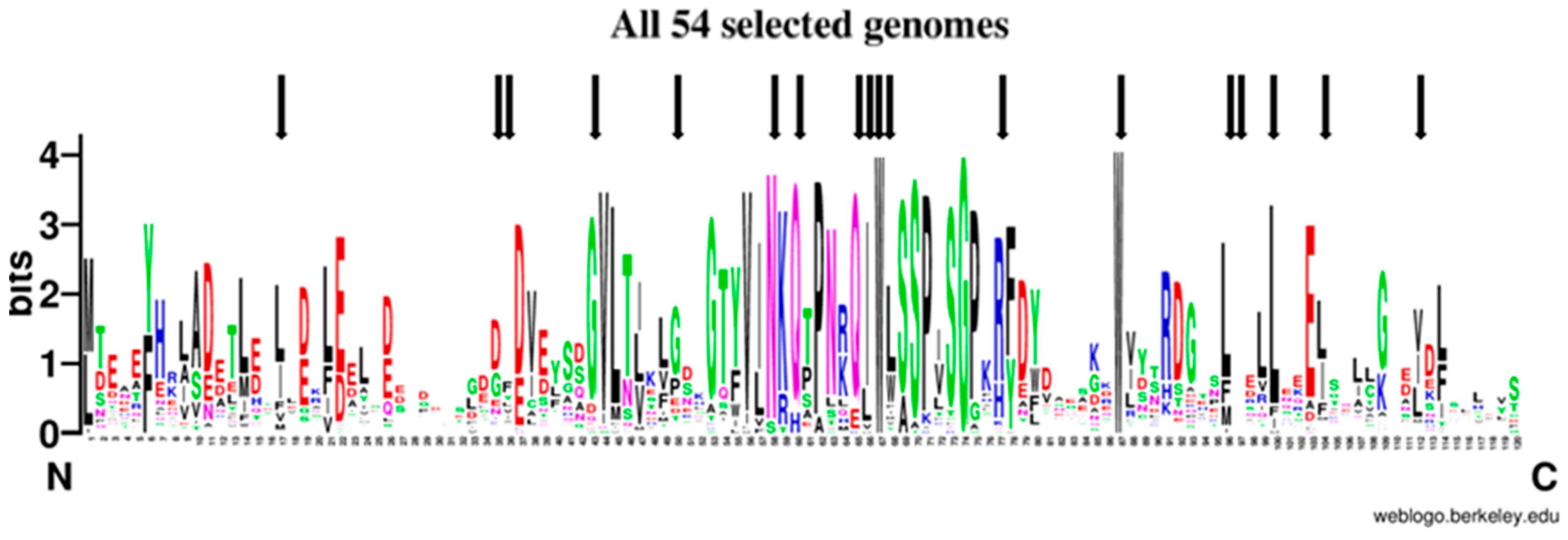
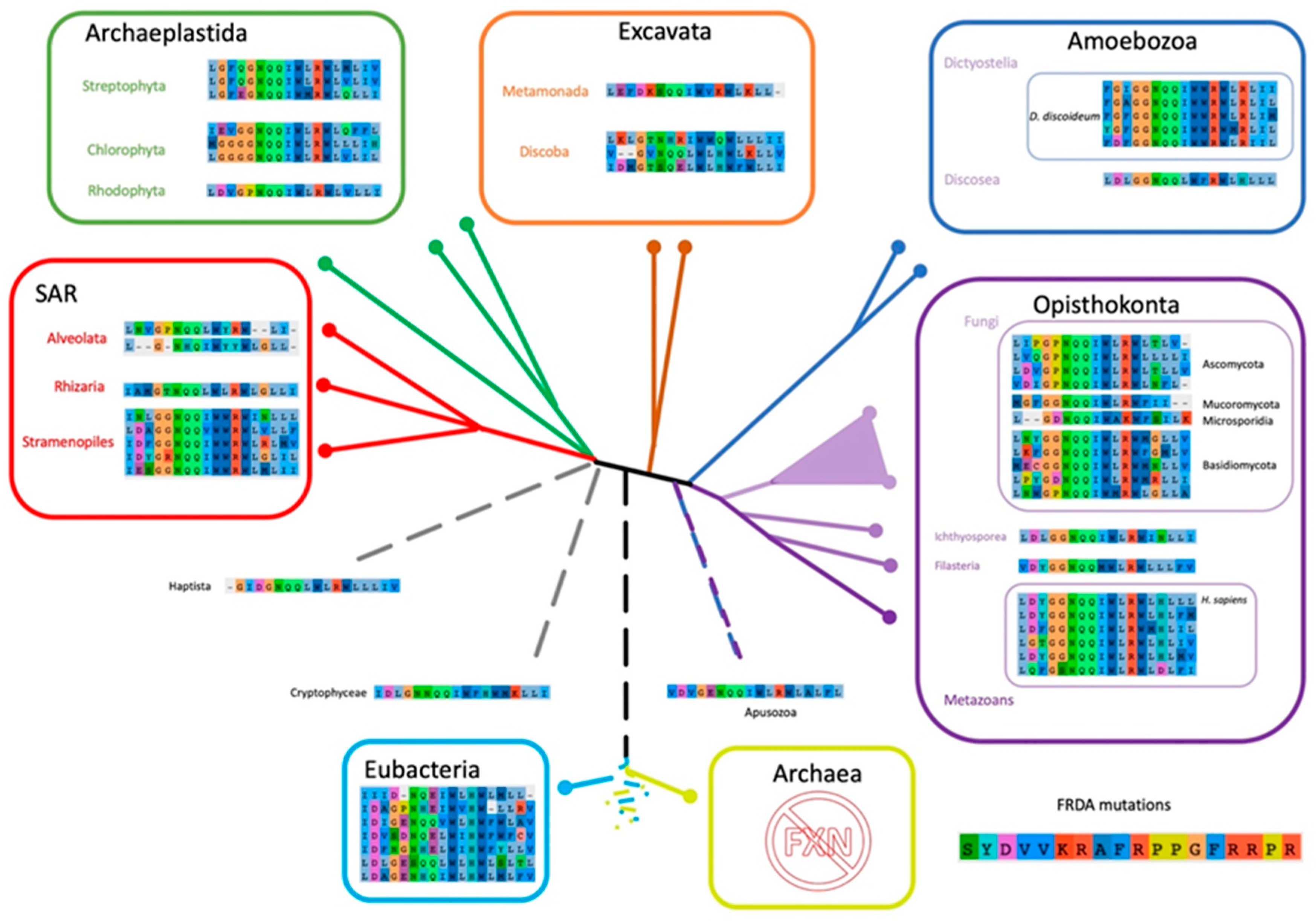
2.6. The Evolutionary Tree of Frataxin and DdFXN: Function and Stability Related Features
3. Discussion
4. Materials and Methods
4.1. Computational Analysis of the Protein Structure
4.2. Molecular Dynamics Simulations
4.3. Protein Expression and Purification
4.4. Thiol Quantification
4.5. Protein Aggregation and the Evaluation of Crystallization Conditions
4.6. Size Exclusion Chromatography
4.7. Characterization by UV 4th Derivative Absorption Spectra Analysis
4.8. Characterization by Circular Dichroism Spectroscopy
4.9. Characterization by Fluorescence Spectroscopy
4.10. Conformational Stability
4.11. Iron-Sulfur Cluster Biogenesis Orthologs in D. discoideum and Frataxin Sequence Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CD | circular dichroism |
| CTR | C-terminal region |
| DdFXN | D. discoideum FXN variant |
| DLS | dynamic light scattering |
| Fe-S | iron–sulfur |
| FRDA | Friedreich’s Ataxia |
| FXN | frataxin |
| HPLC | high-performance liquid chromatography |
| ISCU | Iron–sulfur cluster assembly enzyme |
| ISD11 | LYR motif-containing protein 4 |
| NFS1 | mitochondrial cysteine desulfurase enzyme |
| NATA | N-acetyltryptophanamide |
| NAYA | N-acetyltyrosinamide |
| PAGE | polyacrylamide gel electrophoresis |
| PDB | Protein Data Bank |
| SDS | sodium dodecyl sulfate |
| SEC | size exclusion chromatography |
References
- Lim, S.C.; Friemel, M.; Marum, J.E.; Tucker, E.J.; Bruno, D.L.; Riley, L.G.; Christodoulou, J.; Kirk, E.P.; Boneh, A.; DeGennaro, C.M.; et al. Mutations in LYRM4, encoding iron-sulfur cluster biogenesis factor ISD11, cause deficiency of multiple respiratory chain complexes. Hum. Mol. Genet. 2013, 22, 4460–4473. [Google Scholar] [CrossRef] [PubMed]
- Böttinger, L.; Martensson, C.U.; Song, J.; Zufall, N.; Wiedemann, N.; Becker, T. Respiratory chain supercomplexes associate with the cysteine desulfurase complex of the iron-sulfur cluster assembly machinery. Mol. Biol. Cell 2018, 29, 776–785. [Google Scholar] [CrossRef]
- Adam, A.C.; Bornhövd, C.; Prokisch, H.; Neupert, W.; Hell, K. The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 2006, 25, 174–183. [Google Scholar] [CrossRef]
- Barton, J.K.; Silva, R.M.B.; O’Brien, E. Redox chemistry in the genome: Emergence of the [4fe4s] cofactor in repair and replication. Annu. Rev. Biochem. 2019, 88, 163–190. [Google Scholar] [CrossRef]
- Pandey, A.; Pain, J.; Dziuba, N.; Pandey, A.K.; Dancis, A.; Lindahl, P.A.; Pain, D. Mitochondria Export Sulfur Species Required for Cytosolic tRNA Thiolation. Cell Chem. Biol. 2018, 25, 738–748.e3. [Google Scholar] [CrossRef]
- Pandey, A.K.; Pain, J.; Dancis, A.; Pain, D. Mitochondria export iron-sulfur and sulfur intermediates to the cytoplasm for iron-sulfur cluster assembly and tRNA thiolation in yeast. J. Biol. Chem. 2019, 294, 9489–9502. [Google Scholar] [CrossRef]
- Boniecki, M.T.; Freibert, S.A.; Mühlenhoff, U.; Lill, R.; Cygler, M. Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex. Nat. Commun. 2017, 8, 1287. [Google Scholar] [CrossRef]
- Cai, K.; Frederick, R.O.; Tonelli, M.; Markley, J.L. Mitochondrial Cysteine Desulfurase and ISD11 Coexpressed in Escherichia coli Yield Complex Containing Acyl Carrier Protein. ACS Chem. Biol. 2017, 12, 918–921. [Google Scholar] [CrossRef] [PubMed]
- Cory, S.A.; Van Vranken, J.G.; Brignole, E.J.; Patra, S.; Winge, D.R.; Drennan, C.L.; Rutter, J.; Barondeau, D.P. Structure of human Fe-S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions. Proc. Natl. Acad. Sci. USA 2017, 114, E5325–E5334. [Google Scholar] [CrossRef]
- Fox, N.G.; Yu, X.; Feng, X.; Bailey, H.J.; Martelli, A.; Nabhan, J.F.; Strain-Damerell, C.; Bulawa, C.; Yue, W.W.; Han, S. Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism. Nat. Commun. 2019, 10, 2210. [Google Scholar] [CrossRef] [PubMed]
- Nakai, Y.; Yoshihara, Y.; Hayashi, H.; Kagamiyama, H. cDNA cloning and characterization of mouse nifS-like protein, m-Nfs1: Mitochondrial localization of eukaryotic NifS-like proteins. FEBS Lett. 1998, 433, 143–148. [Google Scholar] [CrossRef]
- Gervason, S.; Larkem, D.; Ben Mansour, A.; Botzanowski, T.; Müller, C.S.; Pecqueur, L.; Le Pavec, G.; Delaunay-Moisan, A.; Brun, O.; Agramunt, J.; et al. Physiologically relevant reconstitution of iron-sulfur cluster biosynthesis uncovers persulfide-processing functions of ferredoxin-2 and frataxin. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; McCabe, J.W.; Russell, D.H.; Barondeau, D.P. Molecular Mechanism of ISC Iron-Sulfur Cluster Biogenesis Revealed by High-Resolution Native Mass Spectrometry. J. Am. Chem. Soc. 2020, 142, 6018–6029. [Google Scholar] [CrossRef] [PubMed]
- Adrover, M.; Howes, B.D.; Iannuzzi, C.; Smulevich, G.; Pastore, A. Anatomy of an iron-sulfur cluster scaffold protein: Understanding the determinants of [2Fe-2S] cluster stability on IscU. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 1448–1456. [Google Scholar] [CrossRef] [PubMed]
- Raulfs, E.C.; O’Carroll, I.P.; Dos Santos, P.C.; Unciuleac, M.C.; Dean, D.R. In vivo iron-sulfur cluster formation. Proc. Natl. Acad. Sci. USA 2008, 105, 8591–8596. [Google Scholar] [CrossRef]
- Tsai, C.-L.; Barondeau, D.P. Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry 2010, 49, 9132–9139. [Google Scholar] [CrossRef]
- Patraa, S.; Barondeaua, D.P. Mechanism of activation of the human cysteine desulfurase complex by frataxin. Proc. Natl. Acad. Sci. USA. 2019, 116, 19421–19430. [Google Scholar] [CrossRef]
- Bridwell-Rabb, J.; Fox, N.G.; Tsai, C.L.; Winn, A.M.; Barondeau, D.P. Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry 2014, 53, 4904–4913. [Google Scholar] [CrossRef]
- Yoon, T.; Cowan, J.A. Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 2003, 125, 6078–6084. [Google Scholar] [CrossRef]
- Cai, K.; Frederick, R.O.; Tonelli, M.; Markley, J.L. Interactions of iron-bound frataxin with ISCU and ferredoxin on the cysteine desulfurase complex leading to Fe-S cluster assembly. J. Inorg. Biochem. 2018, 183, 107–116. [Google Scholar] [CrossRef]
- Parent, A.; Elduque, X.; Cornu, D.; Belot, L.; Le Caer, J.-P.P.; Grandas, A.; Toledano, M.B.; D’Autréaux, B. Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nat. Commun. 2015, 6, 5686. [Google Scholar] [CrossRef]
- Campuzano, V.; Montermini, L.; Moltò, M.D.; Pianese, L.; Cossée, M.; Cavalcanti, F.; Monros, E.; Rodius, F.; Duclos, F.; Monticelli, A.; et al. Friedreich’s ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 1996, 271, 1423–1427. [Google Scholar] [CrossRef] [PubMed]
- Mochel, F.; Knight, M.A.; Tong, W.H.; Hernandez, D.; Ayyad, K.; Taivassalo, T.; Andersen, P.M.; Singleton, A.; Rouault, T.A.; Fischbeck, K.H.; et al. Splice Mutation in the Iron-Sulfur Cluster Scaffold Protein ISCU Causes Myopathy with Exercise Intolerance. Am. J. Hum. Genet. 2008, 82, 652–660. [Google Scholar] [CrossRef]
- Farhan, S.M.K.; Wang, J.; Robinson, J.F.; Lahiry, P.; Siu, V.M.; Prasad, C.; Kronick, J.B.; Ramsay, D.A.; Anthony Rupar, C.; Hegele, R.A. Exome sequencing identifies nfs1 deficiency in a novel fe-s cluster disease, infantile mitochondrial complex ii/iii deficiency. Mol. Genet. Genomic Med. 2014, 2, 73–80. [Google Scholar] [CrossRef]
- Galea, C.A.; Huq, A.; Lockhart, P.J.; Tai, G.; Corben, L.A.; Yiu, E.M.; Gurrin, L.C.; Lynch, D.R.; Gelbard, S.; Durr, A.; et al. Compound heterozygous FXN mutations and clinical outcome in friedreich ataxia. Ann. Neurol. 2016, 79, 485–495. [Google Scholar] [CrossRef]
- Herrera, M.G.; Noguera, M.E.; Sewell, K.E.; Agudelo Suárez, W.A.; Capece, L.; Klinke, S.; Santos, J. Structure of the Human ACP-ISD11 Heterodimer. Biochemistry 2019, 58. [Google Scholar] [CrossRef]
- Georgina Herrera, M.; Florencia Pignataro, M.; Ezequiel Noguera, M.; Magalí Cruz, K.; Santos, J.; Herrera, M.G.; Pignataro, M.F.; Noguera, M.E.; Cruz, K.M.; Santos, J. Rescuing the Rescuer: On the Protein Complex between the Human Mitochondrial Acyl Carrier Protein and ISD11. ACS Chem. Biol. 2018, 13, 1455–1462. [Google Scholar] [CrossRef]
- Melber, A.; Winge, D.R. Steps Toward Understanding Mitochondrial Fe/S Cluster Biogenesis. In Methods in Enzymology; Academic Press Inc.: Cambridge, MA, USA, 2018; Volume 599, pp. 265–292. ISBN 9780128147177. [Google Scholar]
- Sen, S.; Hendricks, A.L.; Cowan, J.A. Cluster exchange reactivity of [2Fe-2S]-bridged heterodimeric BOLA1-GLRX5. FEBS J. 2020. [Google Scholar] [CrossRef]
- Nasta, V.; Suraci, D.; Gourdoupis, S.; Ciofi-Baffoni, S.; Banci, L. A pathway for assembling [4Fe-4S] 2+ clusters in mitochondrial iron–sulfur protein biogenesis. FEBS J. 2020, 287, 2312–2327. [Google Scholar] [CrossRef]
- Cai, K.; Liu, G.; Frederick, R.O.; Xiao, R.; Montelione, G.T.; Markley, J.L. Structural/Functional Properties of Human NFU1, an Intermediate [4Fe-4S] Carrier in Human Mitochondrial Iron-Sulfur Cluster Biogenesis. Structure 2016, 24, 2080–2091. [Google Scholar] [CrossRef]
- Weiler, B.D.; Brück, M.-C.; Kothe, I.; Bill, E.; Lill, R.; Mühlenhoff, U. Mitochondrial protein assembly involves reductive cluster fusion on ISCA1-ISCA2 by electron flow from ferredoxin FDX2. Proc. Natl. Acad. Sci. USA 2020, 117, 20555–20565. [Google Scholar] [CrossRef]
- Kessin, R.H. Dictyostelium; Cambridge University Press: Cambridge, UK, 2001; ISBN 9780511525315. [Google Scholar]
- Williams, J.G. Dictyostelium Finds New Roles to Model. Genetics 2010, 185, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Bozzaro, S. The model organism Dictyostelium discoideum. In Dictyostelium Discoideum Protocols; Humana Press: Totowa, NJ, USA, 2013; pp. 17–37. [Google Scholar]
- Saha, P.P.; Kumar, S.K.P.; Srivastava, S.; Sinha, D.; Pareek, G.; D’Silva, P. The presence of multiple cellular defects associated with a novel G50E iron-sulfur cluster scaffold protein (ISCU) mutation leads to development of mitochondrial myopathy. J. Biol. Chem. 2014, 289, 10359–10377. [Google Scholar] [CrossRef]
- Legati, A.; Reyes, A.; Ceccatelli Berti, C.; Stehling, O.; Marchet, S.; Lamperti, C.; Ferrari, A.; Robinson, A.J.; Mühlenhoff, U.; Lill, R.; et al. A novel de novo dominant mutation in ISCU associated with mitochondrial myopathy. J. Med. Genet. 2017, 54, 815–824. [Google Scholar] [CrossRef]
- Schaf, J.; Damstra-Oddy, J.; Williams, R.S.B. Dictyostelium discoideum as a pharmacological model system to study the mechanisms of medicinal drugs and natural products. Int. J. Dev. Biol. 2019, 63, 541–550. [Google Scholar] [CrossRef]
- Müller-Taubenberger, A.; Kortholt, A.; Eichinger, L. Simple system—substantial share: The use of Dictyostelium in cell biology and molecular medicine. Eur. J. Cell Biol. 2013, 92, 45–53. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Castro, I.H.; Bringas, M.; Doni, D.; Noguera, M.E.; Capece, L.; Aran, M.; Blaustein, M.; Costantini, P.; Santos, J. Relationship between activity and stability: Design and characterization of stable variants of human frataxin. Arch. Biochem. Biophys. 2020, 108491. [Google Scholar] [CrossRef]
- Leidgens, S.; de Smet, S.; Foury, F. Frataxin interacts with Isu1 through a conserved tryptophan in its β-sheet. Hum. Mol. Genet. 2009, 19, 276–286. [Google Scholar] [CrossRef][Green Version]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33. [Google Scholar] [CrossRef]
- Van Durme, J.; Delgado, J.; Stricher, F.; Serrano, L.; Schymkowitz, J.; Rousseau, F. A graphical interface for the FoldX forcefield. Bioinformatics 2011, 27, 1711–1712. [Google Scholar] [CrossRef]
- Bridwell-Rabb, J.; Winn, A.M.; Barondeau, D.P. Structure—Function analysis of friedreich’s ataxia mutants reveals determinants of frataxin binding and activation of the fe—S assembly complex. Biochemistry 2011, 50, 7265–7274. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef]
- Faraj, S.E.; Roman, E.A.; Aran, M.; Gallo, M.; Santos, J. The alteration of the C-terminal region of human frataxin distorts its structural dynamics and function. FEBS J. 2014, 281, 3397–3419. [Google Scholar] [CrossRef] [PubMed]
- Faraj, S.E.; Noguera, M.E.; Delfino, J.M.; Santos, J. Global Implications of Local Unfolding Phenomena, Probed by Cysteine Reactivity in Human Frataxin. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Faraj, S.E.; González-Lebrero, R.M.; Roman, E.A.; Santos, J. Human Frataxin Folds Via an Intermediate State. Role of the C-Terminal Region. Sci. Rep. 2016, 6, 20782. [Google Scholar] [CrossRef]
- Roman, E.A.; Faraj, S.E.; Gallo, M.; Salvay, A.G.; Ferreiro, D.U.; Santos, J. Protein Stability and Dynamics Modulation: The Case of Human Frataxin. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Adinolfi, S.; Nair, M.; Politou, A.; Bayer, E.; Martin, S.; Temussi, P.; Pastore, A. The Factors Governing the Thermal Stability of Frataxin Orthologues: How To Increase a Protein’s Stability. Biochemistry 2004, 43. [Google Scholar] [CrossRef]
- Myers, J.K.; Nick Pace, C.; Martin Scholtz, J. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995, 4, 2138–2148. [Google Scholar] [CrossRef]
- Gibson, T.J.; Koonin, E.V.; Musco, G.; Pastore, A.; Bork, P. Friedreich’s ataxia protein: Phylogenetic evidence for mitochondrial dysfunction. Trends Neurosci. 1996, 19, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Simpson, A.G.B.B.; Lane, C.E.; Lukeš, J.; Bass, D.; Bowser, S.S.; Brown, M.W.; Burki, F.; Dunthorn, M.; Hampl, V.; et al. The Revised Classification of Eukaryotes. J. Eukaryot. Microbiol. 2012, 59, 429–514. [Google Scholar] [CrossRef]
- Andreini, C.; Putignano, V.; Rosato, A.; Banci, L. The human iron-proteome. Metallomics 2018, 10, 1223–1231. [Google Scholar] [CrossRef]
- Abramowitz, N.; Schechter, I.; Berger, A. On the size of the active site in proteases II. Carboxypeptidase-A. Biochem. Biophys. Res. Commun. 1967, 29, 862–867. [Google Scholar] [CrossRef]
- Lane, N. Hot mitochondria? PLoS Biol. 2018, 16. [Google Scholar]
- Chrétien, D.; Bénit, P.; Ha, H.-H.; Keipert, S.; El-Khoury, R.; Chang, Y.-T.; Jastroch, M.; Jacobs, H.T.; Rustin, P.; Rak, M. Mitochondria are physiologically maintained at close to 50 °C. PLoS Biol. 2018, 16, e2003992. [Google Scholar] [CrossRef]
- Arai, S.; Suzuki, M.; Park, S.J.; Yoo, J.S.; Wang, L.; Kang, N.Y.; Ha, H.H.; Chang, Y.T. Mitochondria-targeted fluorescent thermometer monitors intracellular temperature gradient. Chem. Commun. 2015, 51, 8044–8047. [Google Scholar] [CrossRef]
- Faggianelli, N.; Puglisi, R.; Veneziano, L.; Romano, S.; Frontali, M.; Vannocci, T.; Fortuni, S.; Testi, R.; Pastore, A. Analyzing the effects of a G137V mutation in the FXN gene. Front. Mol. Neurosci. 2015, 8, 1–8. [Google Scholar] [CrossRef]
- Bellanda, M.; Maso, L.; Doni, D.; Bortolus, M.; De Rosa, E.; Lunardi, F.; Alfonsi, A.; Noguera, M.E.; Herrera, M.G.; Santos, J.; et al. Exploring iron-binding to human frataxin and to selected Friedreich ataxia mutants by means of NMR and EPR spectroscopies. Biochim. Biophys. Acta Proteins Proteomics 2019, 1867. [Google Scholar] [CrossRef]
- Vazquez, D.S.; Agudelo, W.A.; Yone, A.; Vizioli, N.; Arán, M.; González Flecha, F.L.; González Lebrero, M.C.; Santos, J. A helix-coil transition induced by the metal ion interaction with a grafted iron-binding site of the CyaY protein family. Dalt. Trans. 2015, 44, 2370–2379. [Google Scholar] [CrossRef]
- Correia, A.R.; Wang, T.; Craig, E.A.; Gomes, C.M. Iron-binding activity in yeast frataxin entails a trade off with stability in the α1/β1 acidic ridge region. Biochem. J. 2010, 426, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Fey, P.; Kowal, A.S.; Gaudet, P.; Pilcher, K.E.; Chisholm, R.L. Protocols for growth and development of Dictyostelium discoideum. Nat. Protoc. 2007, 2, 1307–1316. [Google Scholar] [CrossRef]
- Redi, C.A.; Eichinger, L.; Rivero-Crespo, F.; Ludwig, E.; Francisco, R.-C.; Francisco, R.-C.; Walker, J.M.; Ditor, S.E.E.; Methods, M.A. Dictyostelium Discoideum Protocols, 2nd ed.; Eichinger, L., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 983, ISBN 9781627033022. [Google Scholar]
- Peracino, B.; Buracco, S.; Bozzaro, S. The Nramp (Slc11) proteins regulate development, resistance to pathogenic bacteria and iron homeostasis in Dictyostelium discoideum. J. Cell Sci. 2013, 126, 301–311. [Google Scholar] [CrossRef]
- Bozzaro, S.; Buracco, S.; Peracino, B. Iron metabolism and resistance to infection by invasive bacteria in the social amoeba Dictyostelium discoideum. Front. Cell. Infect. Microbiol. 2013, 4. [Google Scholar]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Rasheed, M.; Jamshidiha, M.; Puglisi, R.; Yan, R.; Cota, E.; Pastore, A. Structural and functional characterization of a frataxin from a thermophilic organism. FEBS J. 2019, 286, 495–506. [Google Scholar] [CrossRef]
- Sirano, D.P.; Shigeta, R.; Chi, Y.I.; Ristow, M.; Shoelson, S.E. Crystal structure of human frataxin. J. Biol. Chem. 2000, 275, 30753–30756. [Google Scholar] [CrossRef]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. Kollman (2018) AMBER 2018. University of California, San Francisco. 2018. Available online: https://ambermd.org/doc12/Amber18.pdf (accessed on 17 September 2020).
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Clérico, E.M.; Peisajovich, S.G.; Ceolín, M.; Ghiringhelli, P.D.; Ermácora, M.R. Engineering a compact non-native state of intestinal fatty acid-binding protein. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2000, 1476, 203–218. [Google Scholar] [CrossRef]
- Nozaki, Y. Determination of tryptophan, tyrosine, and phenylalanine by second derivative spectrophotometry. Arch. Biochem. Biophys. 1990, 277, 324–333. [Google Scholar] [CrossRef]
- Noguera, M.E.; Aran, M.; Smal, C.; Vazquez, D.S.; Herrera, M.G.; Roman, E.A.; Alaimo, N.; Gallo, M.; Santos, J. Insights on the conformational dynamics of human frataxin through modifications of loop-1. Arch. Biochem. Biophys. 2017, 636, 123–137. [Google Scholar] [CrossRef]
- Fey, P.; Dodson, R.J.; Basu, S.; Chisholm, R.L. One Stop Shop for Everything Dictyostelium: DictyBase and the Dicty Stock Center in 2012. Dictyostelium Discoideum Protoc. Methods Mol. Biol. 2013, 983, 59–92. [Google Scholar] [PubMed]
- Basu, S.; Fey, P.; Pandit, Y.; Dodson, R.; Kibbe, W.A.; Chisholm, R.L. DictyBase 2013: Integrating multiple Dictyostelid species. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinforma. Appl. 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]



| Protein (Full Sequence) | Conserved Domain | ||||||
|---|---|---|---|---|---|---|---|
| Name Description | ID Dictybase a/NCBI b | ID Human Orthologue c | % Identity/Similarity d | % Coverage/e-Value | ID (Description) e | Interval/e-Value | |
| NFS1—Cysteine desulfurase | ISU COMPLEX | DDB_G0279287/XP_641773 | Q9Y697 | 59.9/77.6 | 87/0 | cl18945 (Aspartate aminotransferase) | 49–448/ 0 |
| ACPM—Acyl carrier protein | DDB_G0291866/XP_629874 | O14561 | 30.1/48 | 55/8.0 × 10−15 | cl09936 (Phosphopantetheine attachment site) | 2–120/ 5.49 × 10−18 | |
| ISCU—Fe-S cluster assembly enzyme | DDB_G0283003/XP_639309 | Q9H1K1 | 50.2/59.4 | 75/1.0 × 10−63 | PRK11325 (scaffold protein; Provisional) | 56–179/ 1.51 × 10−83 | |
| FDX2—Ferredoxin-2 | DDB_G0267486/XP_647073 | Q6P4F2 | 36.5/51.6 | 62/2.0 × 10−40 | PLN02593 (adrenodoxin-like ferredoxin protein) | 44–159/ 3.75 × 10−66 | |
| FXN—Frataxin | DDB_G0293246/XP_629221 | Q16595 | 30/47.2 | 49/4.0 × 10−20 | pfam01491 (Frataxin_Cyay. Frataxin-like domain) | 88–190/ 1.36 × 10−43 | |
| LYRM4 (ISD11) | DDB_G0290725/XP_635573 | Q9HD34 | 43.9/68.1 | 89/1.0 × 10−23 | cd20264 (Complex1_LYR_LYRM4, leucine-tyrosine-arginine motif found in LYR motif-containing protein 4) | 7–73/ 1.42 × 10−25 | |
| HSPA9 (GRP75)—Hsp 70 | CLUSTER RELEASE COMPLEX | DDB_G0293298/XP_629204.1 | P38646/HSPA9 | 58.6/71.6 | 89/0 | PRK00290/cl35085 (dnaK molecular chaperone, Provisional) | 30–658/ 0 |
| GrpE1 (MGE)—PAM complex | DDB_G0283763/XP_638912.1 | Q9HAV7 | 38.2/54.4 | 61/2.00 × 10−35 | pfam01025 (grpE) | 42–209/ 1.99 × 10−52 | |
| GLRX5—Glutaredoxin-related protein | DDB_G0274657/XP_644145 | Q86SX6 | 32.4/58.5 | 59/5.00 × 10−29 | cd03028 (Glutaredoxin family, PKC-interacting cousin of TRX PICOT-like subfamily) | 147–235/ 1.88 × 10−56 | |
| cd02984 (TRX_PICOT, TRX domain, PKC-interacting cousin of TRX) subfamily) | 7–105/ 2.38 × 10−38 | ||||||
| HSC20 (HscB), Fe-S cluster co-chaperone | DDB_G0280889/XP_640965.1 | Q8IWL3 | 21.3/39 | 80/1.00 × 10−25 | PRK05014 (co-chaperone HscB; Provisional) | 136–290/ 1.22 × 10−24 | |
| BolA1 | DDB_G0274169/XP_644296.1 | Q9Y3E2 | 36.8/50.7 | 74/1.00 × 10−31 | pfam01722 (BolA, BolA-like protein; morphoprotein BolA from E. coli) | 15–89/ 1.03 × 10−35 | |
| BolA1 | DDB_G0290319/XP_635799.1 | Q9Y3E2 | 32.9/50.7 | 72/2.00 × 10−24 | pfam01722 (BolA-like protein; morphoprotein BolA from E. coli) | 49–121/ 4.46 × 10−40 | |
| AbcB7 ABC transporter | DDB_G0292554/XP_629496.1 | O75027 | 41.4/59.3 | 84/0 | COG5265 (ATM1, ABC-type transport system involved in Fe-S cluster assembly, permease and ATPase components) | 199–693/0 | |
| ISCA1 | ISA COMPLEX | DDB_G0280173/XP_639260.1 | Q9BUE6 | 32.9/55.5 | 82/1.00 × 10−31 | TIGR00049 (Iron–sulfur cluster assembly accessory protein) | 21–123/1.28 × 10−39 |
| ISCA2 | DDB_G0284809/XP_641284.1 | Q86U28 | 23.3/37.7 | 77/9.00 × 10−26 | TIGR00049 (cluster assembly accessory protein) | 102–205/3.95 × 10−35 | |
| IBA57-Putative transferase CAF17 | DDB_G0285011/XP_639996 | Q5T440 | 24.6/37.5 | 64/3.00 × 10−36 | COG0354 (YgfZ, Folate-binding Fe-S cluster repair protein possible role in tRNA modification) | 13–266/3.99 × 10−42 | |
| NFU1—NIF system Fe-S cluster scaffold protein | LATE STAGE TRANSFER | DDB_G0285593/XP_638146 | Q9UMS0 | 37.4/50.1 | 73/3.00 × 10−64 | pfam08712 (Nfu/NifU N- terminal) | 100–187/6.63 × 10−40 |
| pfam01106 (NifU carboxy-terminal) | 216–282/2.11 × 10−31 | ||||||
| BolA3 | DDB_G0274439/XP_639996.1+ | Q53S33 | 26.6/38.7 | 44/4.00 × 10−13 | COG0271 (BolA Stress-induced morphogen, activity unknown, signal transduction mechanisms) * | 28–99/ 1.78 × 10−21 * | |
| Secondary Structure Type | Human FXN (PDB ID 1EKG) 1 | DdFXN Model (A) 1,3 | DdFXN Model (B) 1,2 | DdFXN Model (C) 1,4 | DdFXN Jpred 4 5 | Bestsel (200–250 nm) 6 | Bestsel (190–250 nm) 7 |
|---|---|---|---|---|---|---|---|
| α-helix | 30.3 | 32.7 | 34.2 | 34.2 | 31.7 | 38.0 | 41.4 |
| β-strand | 30.3 | 35.4 | 34.2 | 35.1 | 24.4 | 22.4 | 15.7 |
| Other | 39.4 | 31.9 | 31.6 | 30.7 | 43.9 | 39.6 | 42.9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olmos, J.; Pignataro, M.F.; Benítez dos Santos, A.B.; Bringas, M.; Klinke, S.; Kamenetzky, L.; Velazquez, F.; Santos, J. A Highly Conserved Iron-Sulfur Cluster Assembly Machinery between Humans and Amoeba Dictyostelium discoideum: The Characterization of Frataxin. Int. J. Mol. Sci. 2020, 21, 6821. https://doi.org/10.3390/ijms21186821
Olmos J, Pignataro MF, Benítez dos Santos AB, Bringas M, Klinke S, Kamenetzky L, Velazquez F, Santos J. A Highly Conserved Iron-Sulfur Cluster Assembly Machinery between Humans and Amoeba Dictyostelium discoideum: The Characterization of Frataxin. International Journal of Molecular Sciences. 2020; 21(18):6821. https://doi.org/10.3390/ijms21186821
Chicago/Turabian StyleOlmos, Justo, María Florencia Pignataro, Ana Belén Benítez dos Santos, Mauro Bringas, Sebastián Klinke, Laura Kamenetzky, Francisco Velazquez, and Javier Santos. 2020. "A Highly Conserved Iron-Sulfur Cluster Assembly Machinery between Humans and Amoeba Dictyostelium discoideum: The Characterization of Frataxin" International Journal of Molecular Sciences 21, no. 18: 6821. https://doi.org/10.3390/ijms21186821
APA StyleOlmos, J., Pignataro, M. F., Benítez dos Santos, A. B., Bringas, M., Klinke, S., Kamenetzky, L., Velazquez, F., & Santos, J. (2020). A Highly Conserved Iron-Sulfur Cluster Assembly Machinery between Humans and Amoeba Dictyostelium discoideum: The Characterization of Frataxin. International Journal of Molecular Sciences, 21(18), 6821. https://doi.org/10.3390/ijms21186821