Systematic Analysis of Cold Stress Response and Diurnal Rhythm Using Transcriptome Data in Rice Reveals the Molecular Networks Related to Various Biological Processes
Abstract
:1. Introduction
2. Results
2.1. Genome-wide Identification of Cold Stress Response Genes Exhibiting Diurnal Rhythm Expression Patterns Using Meta-Expression Datasets
2.2. Literature Analysis to Identify Characterized Gene Functions Associated with CD Genes
2.3. GO and KEGG Enrichment Analyses of the CD Genes Reveal that Photosynthesis and Light Harvesting Are Closely Related to the Cold Stress Response and Circadian Clock
2.4. Construction of Protein-protein Interactions of CD Genes Reveals the Molecular Network Interplay with Various Biological Processes
2.5. Validation of the Cold Stress Response and Circadian Clock Network with a Case Study Using the Stay-green (sgr) Mutant
3. Discussion
4. Materials and Methods
4.1. Collection of Microarray Data and Meta-expression Analysis
4.2. Literature Search for Functionally Characterized Genes
4.3. GO and KEGG Enrichment Analyses
4.4. MapMan Analysis
4.5. Protein–Protein Interaction Network Construction
4.6. Plant Material and Stress Treatment
4.7. RNA Extraction and Quantitative RT-PCR (qRT-PCR) Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ray, D.K.; Ramankutty, N.; Mueller, N.D.; West, P.C.; Foley, J.A. Recent Patterns of Crop Yield Growth and Stagnation. Nature Commun. 2012, 3, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [Green Version]
- Chinnusamy, V.; Zhu, J.; Zhu, J. Cold Stress Regulation of Gene Expression in Plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Cruz, R.P.d.; Sperotto, R.A.; Cargnelutti, D.; Adamski, J.M.; de FreitasTerra, T.; Fett, J.P. Avoiding Damage and Achieving Cold Tolerance in Rice Plants. Food Energy Secur. 2013, 2, 96–119. [Google Scholar] [CrossRef]
- Kuroki, M.; Saito, K.; Matsuba, S.; Yokogami, N.; Shimizu, H.; Ando, I.; Sato, Y. A Quantitative Trait Locus for Cold Tolerance at the Booting Stage on Rice Chromosome 8. Theor. Appl. Genet. 2007, 115, 593–600. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, Q.; Wang, S.; Hong, Y.; Wang, Z. Rice and Cold Stress: Methods for its Evaluation and Summary of Cold Tolerance-Related Quantitative Trait Loci. Rice 2014, 7, 24. [Google Scholar] [CrossRef] [Green Version]
- Kinmonth-Schultz, H.A.; Golembeski, G.S.; Imaizumi, T. Circadian Clock-Regulated Physiological Outputs: Dynamic Responses in Nature. Semin. Cell Dev. Biol. 2013, 24, 407–413. [Google Scholar] [CrossRef] [Green Version]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between Abiotic and Biotic Stress Responses: A Current View from the Points of Convergence in the Stress Signaling Networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Urwin, P.E. The Interaction of Plant Biotic and Abiotic Stresses: From Genes to the Field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef] [Green Version]
- McClung, C.R. Plant Circadian Rhythms. Plant Cell 2006, 18, 792–803. [Google Scholar] [CrossRef] [Green Version]
- Bell-Pedersen, D.; Cassone, V.M.; Earnest, D.J.; Golden, S.S.; Hardin, P.E.; Thomas, T.L.; Zoran, M.J. Circadian Rhythms from Multiple Oscillators: Lessons from Diverse Organisms. Nat. Rev. Genet. 2005, 6, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Mas, P.; Yanovsky, M.J. Time for Circadian Rhythms: Plants Get Synchronized. Curr. Opin. Plant Biol. 2009, 12, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Harmer, S.L. The Circadian System in Higher Plants. Annu. Rev. Plant Biol. 2009, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kévei, E.; Tóth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A. Plant Circadian Clocks Increase Photosynthesis, Growth, Survival, and Competitive Advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turck, F.; Fornara, F.; Coupland, G. Regulation and Identity of Florigen: FLOWERING LOCUS T Moves Center Stage. Annu.Rev. Plant Biol. 2008, 59, 573–594. [Google Scholar] [CrossRef] [Green Version]
- Grundy, J.; Stoker, C.; Carré, I.A. Circadian Regulation of Abiotic Stress Tolerance in Plants. Front. Plant Sci. 2015, 6, 648. [Google Scholar] [CrossRef]
- Bieniawska, Z.; Espinoza, C.; Schlereth, A.; Sulpice, R.; Hincha, D.K.; Hannah, M.A. Disruption of the Arabidopsis Circadian Clock is Responsible for Extensive Variation in the Cold-Responsive Transcriptome. Plant Physiol. 2008, 147, 263–279. [Google Scholar] [CrossRef] [Green Version]
- Nakamichi, N.; Kusano, M.; Fukushima, A.; Kita, M.; Ito, S.; Yamashino, T.; Saito, K.; Sakakibara, H.; Mizuno, T. Transcript Profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR Arrhythmic Triple Mutant Reveals a Role for the Circadian Clock in Cold Stress Response. Plant Cell Physiol. 2009, 50, 447–462. [Google Scholar] [CrossRef] [Green Version]
- Kumar, K.; Rao, K.P.; Biswas, D.K.; Sinha, A.K. Rice WNK1 is Regulated by Abiotic Stress and Involved in Internal Circadian Rhythm. Plant Signal. Behav. 2011, 6, 316–320. [Google Scholar] [CrossRef] [Green Version]
- Ahamed, A.; Murai-Hatano, M.; Ishikawa-Sakurai, J.; Hayashi, H.; Kawamura, Y.; Uemura, M. Cold Stress-Induced Acclimation in Rice is Mediated by Root-Specific Aquaporins. Plant Cell Physiol. 2012, 53, 1445–1456. [Google Scholar] [CrossRef]
- Duan, M.; Huang, P.; Yuan, X.; Chen, H.; Huang, J.; Zhang, H. CMYB1 Encoding a MYB Transcriptional Activator is Involved in Abiotic Stress and Circadian Rhythm in Rice. Sci. World J. 2014, 2014, 178038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro Version 3.0: Expanding the Informatics Resource for Rice Transcriptome. Nucleic Acids Res. 2013, 41, D1206–D1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, E.; Yonemaru, J.; Yamamoto, T.; Yano, M. OGRO: The Overview of Functionally Characterized Genes in Rice Online Database. Rice 2012, 5, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Wu, Y.; Wang, X. bZIP Transcription Factor OsbZIP52/RISBZ5: A Potential Negative Regulator of Cold and Drought Stress Response in Rice. Planta 2012, 235, 1157–1169. [Google Scholar] [CrossRef]
- Chou, W.; Huang, L.; Fang, J.; Yeh, C.; Hong, C.; Wu, S.; Lu, C. Divergence of the Expression and Subcellular Localization of CCR4-Associated Factor 1 (CAF1) Deadenylase Proteins in Oryza Sativa. Plant Mol. Biol. 2014, 85, 443–458. [Google Scholar] [CrossRef]
- Xiong, L.; Yang, Y. Disease Resistance and Abiotic Stress Tolerance in Rice are Inversely Modulated by an Abscisic Acid–inducible Mitogen-Activated Protein Kinase. Plant Cell 2003, 15, 745–759. [Google Scholar] [CrossRef] [Green Version]
- Ito, Y.; Katsura, K.; Maruyama, K.; Taji, T.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of Rice DREB1/CBF-Type Transcription Factors Involved in Cold-Responsive Gene Expression in Transgenic Rice. Plant Cell Physiol. 2006, 47, 141–153. [Google Scholar] [CrossRef] [Green Version]
- Xie, K.; Chen, J.; Wang, Q.; Yang, Y. Direct Phosphorylation and Activation of a Mitogen-Activated Protein Kinase by a Calcium-Dependent Protein Kinase in Rice. Plant Cell 2014, 26, 3077–3089. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) Transcription Factor Enhances Drought Resistance and Salt Tolerance in Rice. Proc. Natl. Acad. Sci. USA. 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [Green Version]
- Bai, M.; Zhang, L.; Gampala, S.S.; Zhu, S.; Song, W.; Chong, K.; Wang, Z. Functions of OsBZR1 and 14-3-3 Proteins in Brassinosteroid Signaling in Rice. Proc. Natl. Acad. Sci. USA. 2007, 104, 13839–13844. [Google Scholar] [CrossRef] [Green Version]
- Hong, Z.; Ueguchi-Tanaka, M.; Shimizu-Sato, S.; Inukai, Y.; Fujioka, S.; Shimada, Y.; Takatsuto, S.; Agetsuma, M.; Yoshida, S.; Watanabe, Y. Loss-of-function of a Rice Brassinosteroid Biosynthetic Enzyme, C-6 Oxidase, Prevents the Organized Arrangement and Polar Elongation of Cells in the Leaves and Stem. Plant J. 2002, 32, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Hakata, M.; Kuroda, M.; Ohsumi, A.; Hirose, T.; Nakamura, H.; Muramatsu, M.; Ichikawa, H.; Yamakawa, H. Overexpression of a Rice TIFY Gene Increases Grain Size through Enhanced Accumulation of Carbohydrates in the Stem. Biosci. Biotechnol. Biochem. 2012, 76, 2129–2134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wuriyanghan, H.; Zhang, B.; Cao, W.; Ma, B.; Lei, G.; Liu, Y.; Wei, W.; Wu, H.; Chen, L.; Chen, H. The Ethylene Receptor ETR2 Delays Floral Transition and Affects Starch Accumulation in Rice. Plant Cell 2009, 21, 1473–1494. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, M.; Higo, K.; Takano, M. Circadian Clock-and Phytochrome-regulated Dof-like Gene, Rdd1, is Associated with Grain Size in Rice. Plant Cell Environ. 2009, 32, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhou, D. Rice jmjC Domain-Containing Gene JMJ706 Encodes H3K9 Demethylase Required for Floral Organ Development. Proc. Natl. Acad. Sci. USA 2008, 105, 13679–13684. [Google Scholar] [CrossRef] [Green Version]
- Ren, D.; Li, Y.; Zhao, F.; Sang, X.; Shi, J.; Wang, N.; Guo, S.; Ling, Y.; Zhang, C.; Yang, Z. MULTI-FLORET SPIKELET1, which Encodes an AP2/ERF Protein, Determines Spikelet Meristem Fate and Sterile Lemma Identity in Rice. Plant Physiol. 2013, 162, 872–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Yu, J.; Park, J.; Li, J.; Yoo, S.; Lee, N.; Lee, S.; Jeong, S.; Seo, H.S.; Koh, H. The Senescence-Induced Staygreen Protein Regulates Chlorophyll Degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [Green Version]
- Ding, B.; del Rosario Bellizzi, M.; Ning, Y.; Meyers, B.C.; Wang, G. HDT701, a Histone H4 Deacetylase, Negatively Regulates Plant Innate Immunity by Modulating Histone H4 Acetylation of Defense-Related Genes in Rice. Plant Cell 2012, 24, 3783–3794. [Google Scholar] [CrossRef] [Green Version]
- Sugio, A.; Yang, B.; Zhu, T.; White, F.F. Two Type III Effector Genes of Xanthomonas Oryzae Pv. Oryzae Control the Induction of the Host Genes OsTFIIAγ1 and OsTFX1 during Bacterial Blight of Rice. Proc. Natl. Acad. Sci. USA 2007, 104, 10720–10725. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Pei, Z.; Tian, Y.; He, C. OsLSD1, a Rice Zinc Finger Protein, Regulates Programmed Cell Death and Callus Differentiation. Mol. Plant Microbe Interact. 2005, 18, 375–384. [Google Scholar] [CrossRef] [Green Version]
- Park, C.; Sharma, R.; Lefebvre, B.; Canlas, P.E.; Ronald, P.C. The Endoplasmic Reticulum-Quality Control Component SDF2 is Essential for XA21-Mediated Immunity in Rice. Plant Sci. 2013, 210, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of Stress-Responsive CIPK Genes in Rice for Stress Tolerance Improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hudson, D.; Guevara, D.R.; Hand, A.J.; Xu, Z.; Hao, L.; Chen, X.; Zhu, T.; Bi, Y.; Rothstein, S.J. Rice Cytokinin GATA Transcription Factor1 Regulates Chloroplast Development and Plant Architecture. Plant Physiol. 2013, 162, 132–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Qin, F.; Huang, L.; Sun, Q.; Li, C.; Zhao, Y.; Zhou, D. Rice Histone Deacetylase Genes Display Specific Expression Patterns and Developmental Functions. Biochem. Biophys. Res. Commun. 2009, 388, 266–271. [Google Scholar]
- Rosa, S.B.; Caverzan, A.; Teixeira, F.K.; Lazzarotto, F.; Silveira, J.A.; Ferreira-Silva, S.L.; Abreu-Neto, J.; Margis, R.; Margis-Pinheiro, M. Cytosolic APx Knockdown Indicates an Ambiguous Redox Responses in Rice. Phytochemistry 2010, 71, 548–558. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, F.; Shen, G.; Gao, J.; Snustad, D.P.; Li, M.; Zhang, J.; Hong, M. The Amylose Content in Rice Endosperm is Related to the Post-transcriptional Regulation of the Waxy Gene. Plant J. 1995, 7, 613–622. [Google Scholar] [CrossRef]
- Xia, J.; Yamaji, N.; Ma, J.F. A Plasma Membrane-localized Small Peptide is Involved in Rice Aluminum Tolerance. Plant J. 2013, 76, 345–355. [Google Scholar] [CrossRef]
- Wei, X.; Xu, J.; Guo, H.; Jiang, L.; Chen, S.; Yu, C.; Zhou, Z.; Hu, P.; Zhai, H.; Wan, J. DTH8 Suppresses Flowering in Rice, Influencing Plant Height and Yield Potential Simultaneously. Plant Physiol. 2010, 153, 1747–1758. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Rahman, M.L.; Cho, S.; Kim, Y.; Koh, H.; Yoo, S.; Paek, N. The Rice Faded Green Leaf Locus Encodes Protochlorophyllide Oxidoreductase B and is Essential for Chlorophyll Synthesis Under High Light Conditions. Plant J. 2013, 74, 122–133. [Google Scholar] [CrossRef]
- Li, J.; Pandeya, D.; Nath, K.; Zulfugarov, I.S.; Yoo, S.; Zhang, H.; Yoo, J.; Cho, S.; Koh, H.; Kim, D. ZEBRA-NECROSIS, a Thylakoid-bound Protein, is Critical for the Photoprotection of Developing Chloroplasts during Early Leaf Development. Plant J. 2010, 62, 713–725. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for Inferring Cellular Functions from Protein Sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, C.; Wu, Y.; Shen, H.; Provart, N.J.; Geisler, M. A Predicted Protein Interactome for Rice. Rice 2012, 5, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirose, F.; Inagaki, N.; Hanada, A.; Yamaguchi, S.; Kamiya, Y.; Miyao, A.; Hirochika, H.; Takano, M. Cryptochrome and Phytochrome Cooperatively but Independently Reduce Active Gibberellin Content in Rice Seedlings Under Light Irradiation. Plant Cell Physiol. 2012, 53, 1570–1582. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A Bacterial-Type ABC Transporter is Involved in Aluminum Tolerance in Rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Ren, S.; Zhang, X.; Gao, M.; Ye, S.; Qi, Y.; Zheng, Y.; Wang, J.; Zeng, L.; Li, Q. BENT UPPERMOST INTERNODE1 Encodes the Class II Formin FH5 Crucial for Actin Organization and Rice Development. Plant Cell 2011, 23, 661–680. [Google Scholar] [CrossRef] [Green Version]
- Sunohara, H.; Kawai, T.; Shimizu-Sato, S.; Sato, Y.; Sato, K.; Kitano, H. A Dominant Mutation of TWISTED DWARF 1 Encoding an A-Tubulin Protein Causes Severe Dwarfism and Right Helical Growth in Rice. Genes Genet. Syst. 2009, 84, 209–218. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.; Cho, S.; Sugimoto, H.; Li, J.; Kusumi, K.; Koh, H.; Iba, K.; Paek, N. Rice Virescent3 and Stripe1 Encoding the Large and Small Subunits of Ribonucleotide Reductase are Required for Chloroplast Biogenesis during Early Leaf Development. Plant Physiol. 2009, 150, 388–401. [Google Scholar] [CrossRef] [Green Version]
- Chi, Y.H.; Moon, J.C.; Park, J.H.; Kim, H.; Zulfugarov, I.S.; Fanata, W.I.; Jang, H.H.; Lee, J.R.; Lee, Y.M.; Kim, S.T. Abnormal Chloroplast Development and Growth Inhibition in Rice Thioredoxin M Knock-Down Plants. Plant Physiol. 2008, 148, 808–817. [Google Scholar] [CrossRef] [Green Version]
- Bang, W.Y.; Chen, J.; Jeong, I.S.; Kim, S.W.; Kim, C.W.; Jung, H.S.; Lee, K.H.; Kweon, H.; Yoko, I.; Shiina, T. Functional Characterization of ObgC in Ribosome Biogenesis during Chloroplast Development. Plant J. 2012, 71, 122–134. [Google Scholar] [CrossRef]
- Park, J.; Jin, P.; Yoon, J.; Yang, J.; Jeong, H.J.; Ranathunge, K.; Schreiber, L.; Franke, R.; Lee, I.; An, G. Mutation in Wilted Dwarf and Lethal 1 (WDL1) Causes Abnormal Cuticle Formation and Rapid Water Loss in Rice. Plant Mol. Biol. 2010, 74, 91–103. [Google Scholar] [CrossRef]
- Higo, H.; Tahir, M.; Takashima, K.; Miura, A.; Watanabe, K.; Tagiri, A.; Ugaki, M.; Ishikawa, R.; Eiguchi, M.; Kurata, N. DDM1 (Decrease in DNA Methylation) Genes in Rice (Oryza Sativa). Mol. Genet. Genomics 2012, 287, 785–792. [Google Scholar] [CrossRef]
- Ashikari, M.; Wu, J.; Yano, M.; Sasaki, T.; Yoshimura, A. Rice Gibberellin-Insensitive Dwarf Mutant Gene Dwarf 1 Encodes the A-Subunit of GTP-Binding Protein. Proc. Natl. Acad. Sci. USA 1999, 96, 10284–10289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jan, A.; Nakamura, H.; Handa, H.; Ichikawa, H.; Matsumoto, H.; Komatsu, S. Gibberellin Regulates Mitochondrial Pyruvate Dehydrogenase Activity in Rice. Plant Cell Physiol. 2006, 47, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Kong, Z.; Omo-Ikerodah, E.; Xu, W.; Li, Q.; Xue, Y. Calcineurin B-Like Interacting Protein Kinase OsCIPK23 Functions in Pollination and Drought Stress Responses in Rice (Oryza sativa L.). J. Genet. Genomics 2008, 35, 531–543, S1–S2. [Google Scholar] [CrossRef]
- Cui, R.; Han, J.; Zhao, S.; Su, K.; Wu, F.; Du, X.; Xu, Q.; Chong, K.; Theißen, G.; Meng, Z. Functional Conservation and Diversification of Class E Floral Homeotic Genes in Rice (Oryza sativa). Plant J. 2010, 61, 767–781. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Attia, K.; Wei, C.; Li, K.; He, G.; Su, W.; Zhang, Q.; Qian, X.; Yang, J. Overexpression of the R FCA RNA Recognition Motif Affects Morphologies Modifications in Rice (Oryza sativa L.). Biosci. Rep. 2007, 27, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.; Yoo, S.; Park, J.; Kwon, C.; Lee, B.; An, G.; Zhang, Z.; Li, J.; Li, Z.; Paek, N. Natural Variation in OsPRR37 Regulates Heading Date and Contributes to Rice Cultivation at a Wide Range of Latitudes. Mol. Plant 2013, 6, 1877–1888. [Google Scholar] [CrossRef] [Green Version]
- Takano, M.; Inagaki, N.; Xie, X.; Yuzurihara, N.; Hihara, F.; Ishizuka, T.; Yano, M.; Nishimura, M.; Miyao, A.; Hirochika, H. Distinct and Cooperative Functions of Phytochromes A, B, and C in the Control of Deetiolation and Flowering in Rice. Plant Cell 2005, 17, 3311–3325. [Google Scholar] [CrossRef] [Green Version]
- Nallamilli, B.R.R.; Zhang, J.; Mujahid, H.; Malone, B.M.; Bridges, S.M.; Peng, Z. Polycomb Group Gene OsFIE2 Regulates Rice (Oryza Sativa) Seed Development and Grain Filling Via a Mechanism Distinct from Arabidopsis. PLoS Genet. 2013, 9, e1003322. [Google Scholar] [CrossRef] [Green Version]
- Yamagata, Y.; Yamamoto, E.; Aya, K.; Win, K.T.; Doi, K.; Ito, T.; Kanamori, H.; Wu, J.; Matsumoto, T.; Matsuoka, M. Mitochondrial Gene in the Nuclear Genome Induces Reproductive Barrier in Rice. Proc. Natl. Acad. Sci. USA 2010, 107, 1494–1499. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, N.; Itoh, H.; Sentoku, N.; Kojima, M.; Sakakibara, H.; Izawa, T.; Itoh, J.; Nagato, Y. The COP1 Ortholog PPS Regulates the Juvenile–adult and Vegetative–reproductive Phase Changes in Rice. Plant Cell 2011, 23, 2143–2154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Tang, D.; Luo, Q.; Jin, Y.; Shen, Y.; Wang, K.; Cheng, Z. BRK1, a Bub1-Related Kinase, is Essential for Generating Proper Tension between Homologous Kinetochores at Metaphase I of Rice Meiosis. Plant Cell 2012, 24, 4961–4973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukiyama, T.; Teramoto, S.; Yasuda, K.; Horibata, A.; Mori, N.; Okumoto, Y.; Teraishi, M.; Saito, H.; Onishi, A.; Tamura, K. Loss-of-Function of a Ubiquitin-Related Modifier Promotes the Mobilization of the Active MITE mPing. Mol. Plant 2013, 6, 790–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dreni, L.; Jacchia, S.; Fornara, F.; Fornari, M.; Ouwerkerk, P.B.; An, G.; Colombo, L.; Kater, M.M. The D-lineage MADS-box Gene OsMADS13 Controls Ovule Identity in Rice. Plant J. 2007, 52, 690–699. [Google Scholar] [CrossRef]
- Saito, H.; Okumoto, Y.; Yoshitake, Y.; Inoue, H.; Yuan, Q.; Teraishi, M.; Tsukiyama, T.; Nishida, H.; Tanisaka, T. Complete Loss of Photoperiodic Response in the Rice Mutant Line X61 is Caused by Deficiency of Phytochrome Chromophore Biosynthesis Gene. Theor. Appl. Genet. 2011, 122, 109–118. [Google Scholar] [CrossRef]
- Chen, R.; Zhao, X.; Shao, Z.; Wei, Z.; Wang, Y.; Zhu, L.; Zhao, J.; Sun, M.; He, R.; He, G. Rice UDP-Glucose Pyrophosphorylase1 is Essential for Pollen Callose Deposition and its Cosuppression Results in a New Type of Thermosensitive Genic Male Sterility. Plant Cell 2007, 19, 847–861. [Google Scholar] [CrossRef] [Green Version]
- Han, M.; Jung, K.; Yi, G.; An, G. Rice Importin Β1 Gene Affects Pollen Tube Elongation. Mol. Cells 2011, 31, 523–530. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Xia, H.; Yang, X.; Xu, T.; Si, H.J.; Cai, X.X.; Wang, F.; Su, J.; Snow, A.A.; Lu, B. A Novel 5-enolpyruvoylshikimate-3-phosphate (EPSP) Synthase Transgene for Glyphosate Resistance Stimulates Growth and Fecundity in Weedy Rice (O Ryza Sativa) without Herbicide. New Phytol. 2014, 202, 679–688. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Ren, Y.; Liu, X.; Jiang, L.; Chen, L.; Han, X.; Jin, M.; Liu, S.; Liu, F.; Lv, J. OsRab5a Regulates Endomembrane Organization and Storage Protein Trafficking in Rice Endosperm Cells. Plant J. 2010, 64, 812–824. [Google Scholar] [CrossRef]
- Lieberherr, D.; Thao, N.P.; Nakashima, A.; Umemura, K.; Kawasaki, T.; Shimamoto, K. A Sphingolipid Elicitor-Inducible Mitogen-Activated Protein Kinase is Regulated by the Small GTPase OsRac1 and Heterotrimeric G-Protein in Rice. Plant Physiol. 2005, 138, 1644–1652. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Yu, X.; An, C. Overexpression of Constitutively Active OsCPK10 Increases Arabidopsis Resistance Against Pseudomonas Syringae Pv. Tomato and Rice Resistance Against Magnaporthe Grisea. Plant Physiol. Biochem. 2013, 73, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Xiong, L.; Yang, Y. Rice SERK1 Gene Positively Regulates Somatic Embryogenesis of Cultured Cell and Host Defense Response Against Fungal Infection. Planta 2005, 222, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Bartley, L.E.; Chen, X.; Dardick, C.; Chern, M.; Ruan, R.; Canlas, P.E.; Ronald, P.C. OsWRKY62 is a Negative Regulator of Basal and Xa21-Mediated Defense Against Xanthomonas Oryzae Pv. Oryzae in Rice. Mol. Plant 2008, 1, 446–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Jiao, F.; Wu, Z.; Li, Y.; Wang, X.; He, X.; Zhong, W.; Wu, P. OsPHR2 is Involved in Phosphate-Starvation Signaling and Excessive Phosphate Accumulation in Shoots of Plants. Plant Physiol. 2008, 146, 1673–1686. [Google Scholar] [CrossRef] [Green Version]
- Qin, C.; Li, Y.; Gan, J.; Wang, W.; Zhang, H.; Liu, Y.; Wu, P. OsDGL1, a Homolog of an Oligosaccharyltransferase Complex Subunit, is Involved in N-Glycosylation and Root Development in Rice. Plant Cell Physiol. 2013, 54, 129–137. [Google Scholar] [CrossRef]
- Zang, A.; Xu, X.; Neill, S.; Cai, W. Overexpression of OsRAN2 in Rice and Arabidopsis Renders Transgenic Plants Hypersensitive to Salinity and Osmotic Stress. J. Exp. Bot. 2010, 61, 777–789. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Xie, K.; Hou, X.; Hu, H.; Xiong, L. Systematic Analysis of GT Factor Family of Rice Reveals a Novel Subfamily Involved in Stress Responses. Mol. Genet. Genomics 2010, 283, 157–169. [Google Scholar] [CrossRef]
- Lu, C.; Lin, C.; Lee, K.; Chen, J.; Huang, L.; Ho, S.; Liu, H.; Hsing, Y.; Yu, S. The SnRK1A Protein Kinase Plays a Key Role in Sugar Signaling during Germination and Seedling Growth of Rice. Plant Cell 2007, 19, 2484–2499. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Wang, Y.; Liu, X.; Jiang, L.; Ren, Y.; Liu, F.; Peng, C.; Li, J.; Jin, X.; Wu, F. The Failure to Express a Protein Disulphide Isomerase-Like Protein Results in a Floury Endosperm and an Endoplasmic Reticulum Stress Response in Rice. J. Exp. Bot. 2012, 63, 121–130. [Google Scholar] [CrossRef]
- Sheoran, I.S.; Koonjul, P.; Attieh, J.; Saini, H.S. Water-Stress-Induced Inhibition of A-Tubulin Gene Expression during Growth, and its Implications for Reproductive Success in Rice. Plant Physiol. Biochem. 2014, 80, 291–299. [Google Scholar] [CrossRef]
- Wang, G.; Ding, X.; Yuan, M.; Qiu, D.; Li, X.; Xu, C.; Wang, S. Dual Function of Rice OsDR8 Gene in Disease Resistance and Thiamine Accumulation. Plant Mol. Biol. 2006, 60, 437–449. [Google Scholar] [CrossRef]
- Heidarvand, L.; Amiri, R.M. What Happens in Plant Molecular Responses to Cold Stress? Acta Physiol. Plant 2010, 32, 419–431. [Google Scholar] [CrossRef]
- Webb, A.A. The Physiology of Circadian Rhythms in Plants. New Phytol. 2003, 160, 281–303. [Google Scholar] [CrossRef] [Green Version]
- Gil, K.; Park, C. Thermal Adaptation and Plasticity of the Plant Circadian Clock. New Phytol. 2019, 221, 1215–1229. [Google Scholar] [CrossRef] [Green Version]
- Gould, P.D.; Locke, J.C.; Larue, C.; Southern, M.M.; Davis, S.J.; Hanano, S.; Moyle, R.; Milich, R.; Putterill, J.; Millar, A.J. The Molecular Basis of Temperature Compensation in the Arabidopsis Circadian Clock. Plant Cell 2006, 18, 1177–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crepy, M.; Yanovsky, M.J.; Casal, J.J. Blue Rhythms between GIGANTEA and Phytochromes. Plant Signal. Behav. 2007, 2, 530–532. [Google Scholar] [CrossRef] [Green Version]
- Huq, E.; Tepperman, J.M.; Quail, P.H. GIGANTEA is a Nuclear Protein Involved in Phytochrome Signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 9789–9794. [Google Scholar] [CrossRef] [Green Version]
- Gould, P.D.; Ugarte, N.; Domijan, M.; Costa, M.; Foreman, J.; MacGregor, D.; Rose, K.; Griffiths, J.; Millar, A.J.; Finkenstädt, B. Network Balance Via CRY Signalling Controls the Arabidopsis Circadian Clock Over Ambient Temperatures. Mol. Syst. Biol. 2013, 9, 650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.; Fujiwara, S.; Suh, S.; Kim, J.; Kim, Y.; Han, L.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE is a Circadian Photoreceptor Stabilized by GIGANTEA in Blue Light. Nature 2007, 449, 356–360. [Google Scholar] [CrossRef]
- Miura, K.; Furumoto, T. Cold Signaling and Cold Response in Plants. Int. J. Mol. Sci. 2013, 14, 5312–5337. [Google Scholar] [CrossRef] [Green Version]
- Bendix, C.; Marshall, C.M.; Harmon, F.G. Circadian Clock Genes Universally Control Key Agricultural Traits. Mol Plant. 2015, 8, 1135–1152. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.T.; Leipner, J.; Stamp, P.; Guerra-Peraza, O. Low Temperature Stress in Maize (Zea mays L.) Induces Genes Involved in Photosynthesis and Signal Transduction as Studied by Suppression Subtractive Hybridization. Plant Physiol. Biochem. 2009, 47, 116–122. [Google Scholar] [CrossRef]
- Kathuria, H.; Giri, J.; Nataraja, K.N.; Murata, N.; Udayakumar, M.; Tyagi, A.K. Glycinebetaine-induced Water-stress Tolerance in codA-expressing Transgenic Indica Rice is Associated with Up-regulation of several Stress Responsive Genes. Plant Biotech. J. 2009, 7, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Kusakina, J.; Hall, A.; Gould, P.D.; Hanaoka, M. The Circadian Regulation of Photosynthesis. Photosynthesis Res. 2014, 119, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Kato, H.; Imai, R. Biochemical Identification of the OsMKK6–OsMPK3 Signalling Pathway for Chilling Stress Tolerance in Rice. Biochem. J. 2012, 443, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Jossier, M.; Bouly, J.; Meimoun, P.; Arjmand, A.; Lessard, P.; Hawley, S.; Grahame Hardie, D.; Thomas, M. SnRK1 (SNF1-related Kinase 1) has a Central Role in Sugar and ABA Signalling in Arabidopsis Thaliana. Plant J. 2009, 59, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Polge, C.; Thomas, M. SNF1/AMPK/SnRK1 Kinases, Global Regulators at the Heart of Energy Control? Trends Plant Sci. 2007, 12, 20–28. [Google Scholar] [CrossRef]
- Jangam, A.P.; Pathak, R.R.; Raghuram, N. Microarray Analysis of Rice D1 (RGA1) Mutant Reveals the Potential Role of G-Protein Alpha Subunit in Regulating Multiple Abiotic Stresses such as Drought, Salinity, Heat, and Cold. Front. Plant Sci. 2016, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Ferrero-Serrano, Á.; Assmann, S.M. The A-Subunit of the Rice Heterotrimeric G Protein, RGA1, Regulates Drought Tolerance during the Vegetative Phase in the Dwarf Rice Mutant D1. J. Exp. Bot. 2016, 67, 3433–3443. [Google Scholar] [CrossRef] [Green Version]
- Ferrero-Serrano, Á.; Su, Z.; Assmann, S.M. Illuminating the Role of the Gα Heterotrimeric G Protein Subunit, RGA1, in Regulating Photoprotection and Photoavoidance in Rice. Plant Cell Environ. 2018, 41, 451–468. [Google Scholar] [CrossRef]
- Kumar, M.; Gho, Y.; Jung, K.; Kim, S. Genome-Wide Identification and Analysis of Genes, Conserved between Japonica and Indica Rice Cultivars, that Respond to Low-Temperature Stress at the Vegetative Growth Stage. Front. Plant Sci. 2017, 8, 1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Lee, S.; Jeong, H.; An, G.; Jeon, J.; Jung, K. Crosstalk between Diurnal Rhythm and Water Stress Reveals an Altered Primary Carbon Flux into Soluble Sugars in Drought-Treated Rice Leaves. Sci. Reports 2017, 7, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M. NCBI GEO: Archive for Functional Genomics Data Sets—update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy—analysis of Affymetrix GeneChip Data at the Probe Level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions using R and Bioconductor; Anonymous, Ed.; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. Mev: Multiexperiment viewer. In Biomedical Informatics for Cancer Research; Anonymous, Ed.; Springer: New York, NY, USA, 2010; pp. 267–277. [Google Scholar]
- Cao, P.; Jung, K.; Choi, D.; Hwang, D.; Zhu, J.; Ronald, P.C. The Rice Oligonucleotide Array Database: An Atlas of Rice Gene Expression. Rice 2012, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.; Han, Y.; He, Q. clusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A User-driven Tool to Display Genomics Data Sets Onto Diagrams of Metabolic Pathways and Other Biological Processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological Network Exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8.13. 1–8.13. 24. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Luo, C.; Chen, Y.; Xiao, X.; Fu, C.; Yang, Y. Transcriptome-based Discovery of AP2/ERF tRanscription Factors and Expression Profiles Under Herbivore Stress Conditions in Bamboo (Bambusa emeiensis). J. Plant Biol. 2019, 62, 297–306. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of Housekeeping Genes as Internal Control for Studying Gene Expression in Rice by Quantitative Real-Time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef]
- Kim, E.J.; Kim, Y.J.; Hong, W.J.; Lee, C.; Jeon, J.S.; Jung, K.H. Genome-wide Analysis of Root Hair Preferred RBOH Genes Suggests that Three RBOH Genes are Associated with Auxin-mediated Root Hair Development in Rice. J. Plant Biol. 2019, 62, 229–238. [Google Scholar] [CrossRef]
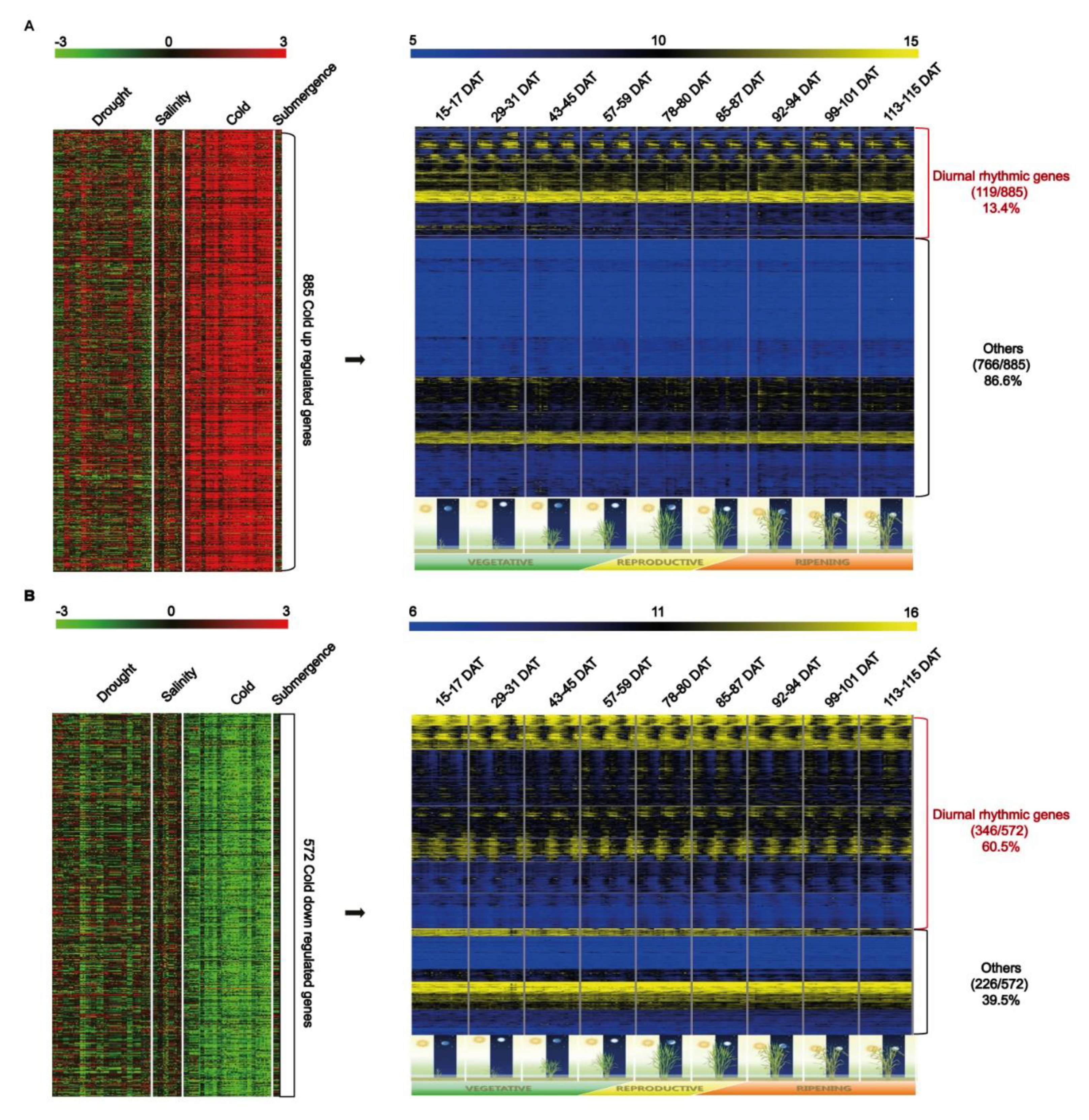

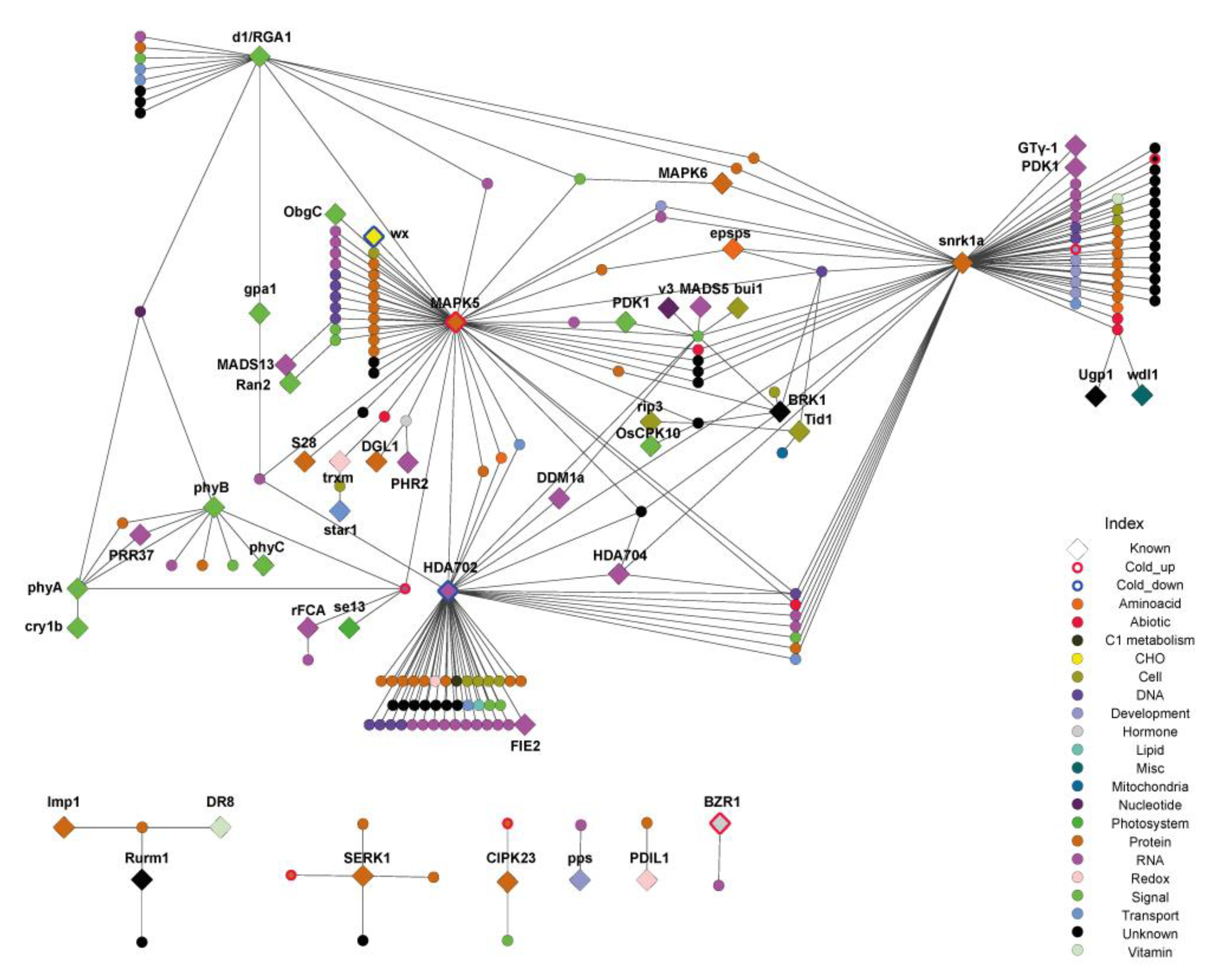

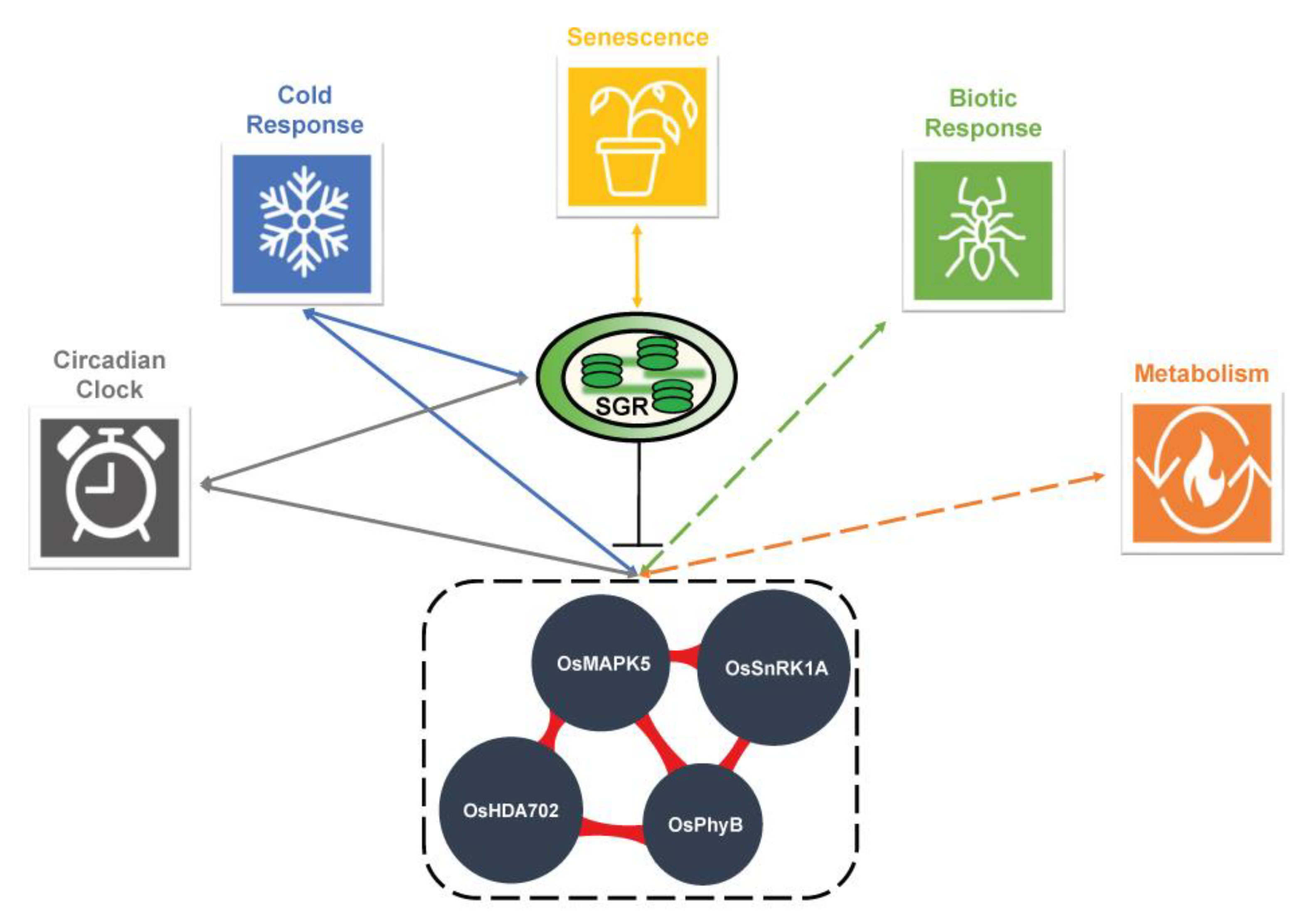
| Locus_ID | Gene_Symbol | Character_Minor | Method 1 | Detailed Functions | DOI 2 |
|---|---|---|---|---|---|
| Upregulated cold-responsive diurnal rhythmic genes (Upregulated CD genes) | |||||
| LOC_Os07g22710 | CPK18 | Blast resistance | Knockdown | Binds and phosphorylates MPK5, regulating resistant to blast (Magnaporthe oryzae) | [28] |
| LOC_Os06g45140 | OsbZIP52/RISBZ5 | Cold tolerance | Overexpression | Cold and drought tolerance | [24] |
| LOC_Os04g58810 | OsCAF1B | Cold tolerance | Others | Drought tolerance | [25] |
| LOC_Os09g35030 | OsDREB1A | Cold tolerance | Overexpression | Cold, drought and salinity tolerance. | [27] |
| LOC_Os03g17700 | OsMAPK5 | Cold tolerance | Knockdown Overexpression | Resistance to Magnaporthe grisea and Burkholderia glumae. Cold, drought and salinity tolerance. | [26] |
| LOC_Os03g60080 | SNAC1 | Drought tolerance | Overexpression | Drought and salinity tolerance. Stomatal control. | [29] |
| LOC_Os07g39220 | OsBZR1 | Dwarfism | Knockdown | Dwarfism. Leaf angle. Brassinosteroid sensitivity. | [30] |
| LOC_Os03g40540 | OsDWARF | Dwarf | Mutant | Dwarfism. Brassinosteroid biosynthesis. | [31] |
| LOC_Os03g08330 | TIFY11b | Dwarf | Overexpression | Grain size. Plant height | [32] |
| LOC_Os04g08740 | Etr2 | Flowering | Mutant | Flowering time. Ethylene sensitivity. Stem starch content. | [33] |
| LOC_Os01g15900 | Rdd1 | Flowering | Knockdown Overexpression | Grain length and width. 1000-grain weight. Flowering time. | [34] |
| LOC_Os10g42690 | Jmj6 | Panicle flower | Mutant | Number and morphology of floral organ. | [35] |
| LOC_Os05g41760 | MSF1 | Panicle flower | Mutant | Spikelet determinacy. Floral organ development. | [36] |
| LOC_Os09g36200 | SGR | Source activity | Mutant | Leaf senescence. Chlorophyll degradation. | [37] |
| Downregulated cold-responsive diurnal rhythmic genes (Downregulated CD genes) | |||||
| LOC_Os05g51830 | HDT701 | Bacterial blight resistance | Knockdown Overexpression | Resistance to Magnaporthe oryzae and Xanthomonas oryzae pv oryzae. | [38] |
| LOC_Os09g29820 | OsTFX1 | Bacterial blight resistance | Overexpression | Resistance to Xanthomonas oryzae pv. oryzae. | [39] |
| LOC_Os08g06280 | OsLSD1 | Blast resistance | Knockdown Overexpression | Lesion mimic. Resistance to Magnaporthe grisea. | [40] |
| LOC_Os08g17680 | SDF2-1 | Blast resistance | Knockdown | XA21-mediated resistance to Xanthomonas oryzae pv. oryzae | [41] |
| LOC_Os01g55450 | OsCIPK12 | Drought tolerance | Overexpression | Drought tolerance. | [42] |
| LOC_Os02g12790 | Cga1 | Dwarf | Knockdown Overexpression | Dwarfism. Tillering. Chlorophyll content. Grain filling rate. | [43] |
| LOC_Os06g38470 | HDA702 | Dwarf | Knockdown | Elongated uppermost internode. Fertility. | [44] |
| LOC_Os07g49400 | OsApx2 | Dwarf | Knockdown | Aluminum tolerance. Dwarfism. | [45] |
| LOC_Os06g04200 | Wx | Eating quality | Natural variation | Seed amylose content. | [46] |
| LOC_Os08g07740 | DTH8 | Flowering | Natural variation | Flowering time under long day condition. | [48] |
| LOC_Os01g08300 | OsCDT3 | Other soil stress tolerance | Knockdown | Aluminum tolerance. | [47] |
| LOC_Os10g35370 | Fgl | Source activity | Mutant | Chlorophyll synthesis under high light conditions. | [49] |
| LOC_Os06g02580 | Zn | Source activity | Mutant | Chloroplast biosynthesis. | [50] |
| Locus_ID | Gene_Symbol | Character_Minor | Methods 1 | Detailed Functions | DOI 2 |
|---|---|---|---|---|---|
| LOC_Os04g37920 | Cry1b | Shoot seedling | Mutant | Leaf sheath elongation during seedling stage. Gibberellin metabolism. | [53] |
| LOC_Os06g48060 | Star1 | Other soil stress tolerance | Mutant | Aluminum tolerance. | [54] |
| LOC_Os07g40510 | Bui1 | Dwarf | Mutant | Cell division and expansion. Actin organization. | [55] |
| LOC_Os11g14220 | Tid1 | Culm leaf | Mutant | Cell division and expansion. Twisted growth. Microtubule arrangement. | [56] |
| LOC_Os06g07210 | V3 | Source activity | Mutant | Chloroplast development during seedling stage. | [57] |
| LOC_Os12g08730 | Ostrxm | Source activity | Knockdown | Chloroplast development. Growth retardation | [58] |
| LOC_Os07g47300 | ObgC | Source activity | Mutant | Chloroplast development. Plastid ribosome biogenesis | [59] |
| LOC_Os11g48070 | Wdl1 | Drought tolerance | Mutant | Cuticle formation. Drought tolerance. | [60] |
| LOC_Os09g27060 | OsDDM1a | Dwarf | Knockdown | Dwarfism, DNA methylation | [61] |
| LOC_Os05g26890 | D1 | Dwarf | Mutant | Dwarfism. | [62] |
| LOC_Os07g44330 | OsPDK1 | Dwarf | Knockdown | Dwarfism. | [63] |
| LOC_Os07g06980 | HDA704 | Culm leaf | Knockdown | Dwarfism. Twisted flag leaf. | [44] |
| LOC_Os07g05620 | OsCIPK23 | Salinity tolerance | Knockdown Overexpression | Fertility. Salinity tolerance. | [64] |
| LOC_Os06g06750 | OsMADS5 | Panicle flower | Knockdown | Floral organ formation. | [65] |
| LOC_Os09g03610 | rFCA | Flowering | Overexpression | Flowering time. | [66] |
| LOC_Os07g49460 | OsPRR37 | Flowering | Natural variation | Flowering time. | [67] |
| LOC_Os03g51030 | OsPhyA | Flowering | Mutant | Flowering time. Deetiolation response. Sensitivity to red and far-red light. | [68] |
| LOC_Os03g19590 | OsPhyB | Flowering | Mutant | Flowering time. Deetiolation response. Sensitivity to red and far-red light. | [68] |
| LOC_Os03g54084 | OsPhyC | Flowering | Mutant | Flowering time. Deetiolation response. Sensitivity to red and far-red light. | [68] |
| LOC_Os08g04270 | OsFIE2 | Germination dormancy | Knockdown | Grain size. Grain filling rate. Seed dormancy. | [69] |
| LOC_Os04g25540 | S28 | Sterility | Natural variation | Hybrid sterility between Oryza sativa and Oryza glaberrima. Pollen development. Interaction with S27 | [70] |
| LOC_Os02g53140 | Pps | Dwarf | Mutant | Juvenile to adult phase change. Flowering time independent of daylength. Dwarfism. | [71] |
| LOC_Os07g32480 | BRK1 | Sterility | Mutant | Meiosis. | [72] |
| LOC_Os07g28280 | Rurm1 | Others | Mutant | Mobilization of the Active MITE mPing | [73] |
| LOC_Os12g10540 | OsMADS13 | Panicle flower | Mutant | Ovule identity. | [74] |
| LOC_Os01g72090 | Se13 | Flowering | Mutant | Photoperiodic response. | [75] |
| LOC_Os09g38030 | Ugp1 | Dwarf | Knockdown | Pollen callose deposition. Dwarfism. | [76] |
| LOC_Os05g28510 | OsImpβ1 | Sterility | Mutant | Pollen tube elongation. | [77] |
| LOC_Os06g04280 | Epsps | Panicle flower | Others | production of panicle | [78] |
| LOC_Os12g43550 | Gpa1 | Eating quality | Mutant | Pro-gultelin content in seed. Floury endosperm. | [79] |
| LOC_Os06g06090 | OsMAPK6 | Others | Knockdown | Regulation of stress response genes. | [80] |
| LOC_Os03g57450 | OsCPK10 | Blast resistance | Overexpression | Resistance to Magnaporthe grisea. | [81] |
| LOC_Os04g38480 | OsSERK1 | Blast resistance | Overexpression | Resistance to Magnaporthe grisea. | [82] |
| LOC_Os09g25070 | OsWRKY62 | Bacterial blight resistance | Overexpression | Resistance to Xanthomonas oryzae pv. oryzae. | [83] |
| LOC_Os07g25710 | OsPHR2 | Other soil stress tolerance | Overexpression | Response to phosphate starvation. | [84] |
| LOC_Os07g10830 | OsDGL1 | Root | Mutant | Root development. | [85] |
| LOC_Os05g49890 | OsRan2 | Other stress resistance | Knockdown Overexpression | Salinity and osmotic stress tolerance. ABA sensitivity. | [86] |
| LOC_Os02g33770 | OsGTgamma-1 | Salinity tolerance | Mutant | Salinity tolerance. | [87] |
| LOC_Os05g45420 | OsSnRK1a | Germination dormancy | Mutant | Seed germination. Seedling growth. | [88] |
| LOC_Os11g09280 | PDIL1 | Eating quality | Mutant | Starch biosynthesis. | [89] |
| LOC_Os03g51600 | Rip-3 (a-tubulin) | Panicle flower | Others | suppress panicle elongation during water deficit | [90] |
| LOC_Os07g34570 | OsDR8 | Bacterial blight resistance | Knockdown | Thiamine mediated resistance to Xanthomonas oryzae and Magnaporthe grisea. | [91] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, W.-J.; Jiang, X.; Ahn, H.R.; Choi, J.; Kim, S.-R.; Jung, K.-H. Systematic Analysis of Cold Stress Response and Diurnal Rhythm Using Transcriptome Data in Rice Reveals the Molecular Networks Related to Various Biological Processes. Int. J. Mol. Sci. 2020, 21, 6872. https://doi.org/10.3390/ijms21186872
Hong W-J, Jiang X, Ahn HR, Choi J, Kim S-R, Jung K-H. Systematic Analysis of Cold Stress Response and Diurnal Rhythm Using Transcriptome Data in Rice Reveals the Molecular Networks Related to Various Biological Processes. International Journal of Molecular Sciences. 2020; 21(18):6872. https://doi.org/10.3390/ijms21186872
Chicago/Turabian StyleHong, Woo-Jong, Xu Jiang, Hye Ryun Ahn, Juyoung Choi, Seong-Ryong Kim, and Ki-Hong Jung. 2020. "Systematic Analysis of Cold Stress Response and Diurnal Rhythm Using Transcriptome Data in Rice Reveals the Molecular Networks Related to Various Biological Processes" International Journal of Molecular Sciences 21, no. 18: 6872. https://doi.org/10.3390/ijms21186872





