Urinary Biomarkers for Diagnosis and Prediction of Acute Kidney Allograft Rejection: A Systematic Review
Abstract
1. Introduction
2. Results
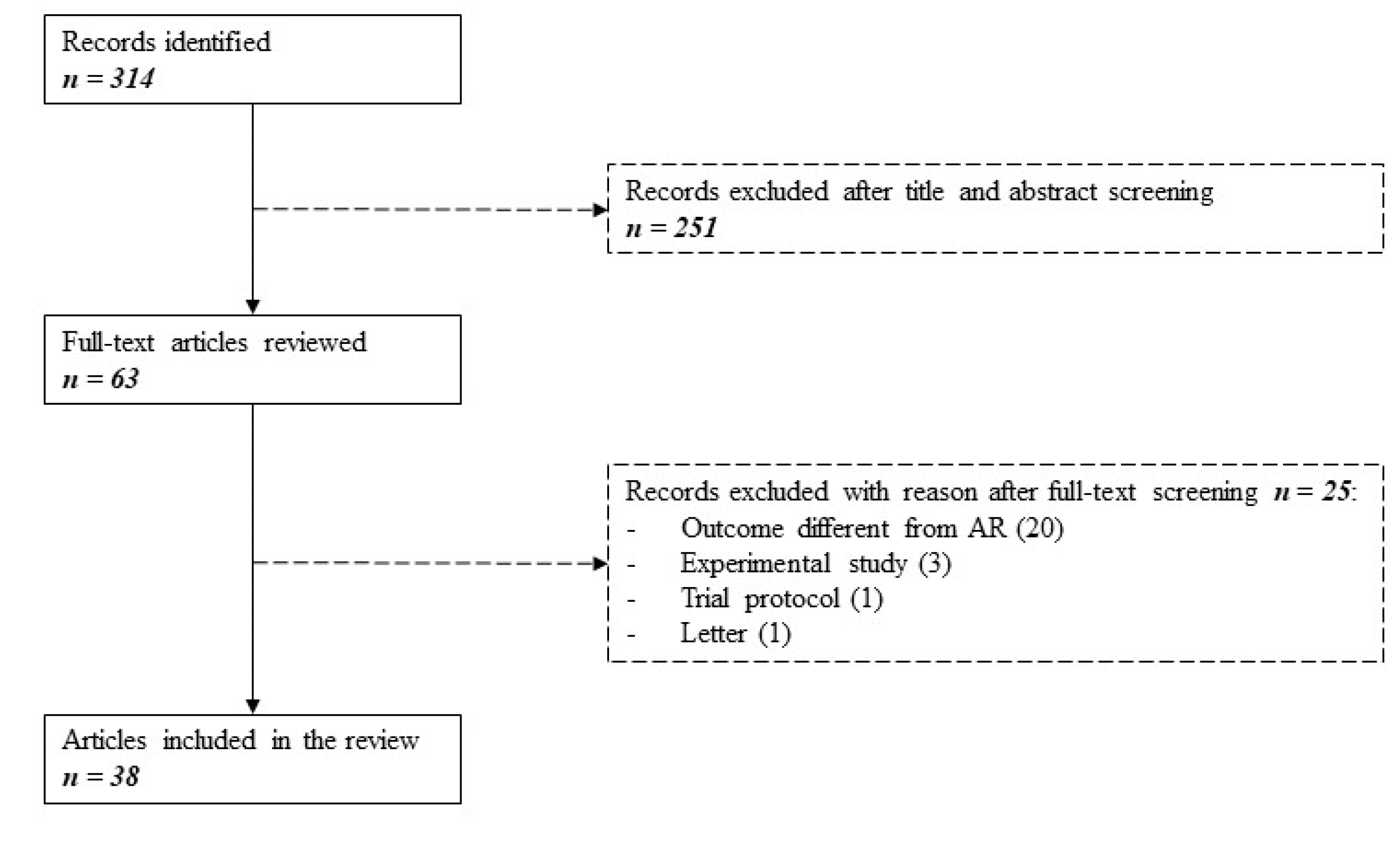
2.1. Included Studies
2.2. Study Characteristics
2.3. Biomarkers
2.4. Quality Assessment
2.5. Summary of the Results
2.5.1. Acute Rejection Diagnosis
2.5.2. T-Cell-Mediated Rejection Diagnosis
2.5.3. Antibody-Mediated Rejection Diagnosis
2.5.4. Acute Rejection, TCMR, and ABMR Prediction
3. Discussion
4. Materials and Methods
4.1. Literature Search
4.2. Selection Process
4.3. Data Collection and Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABMR | Antibody-mediated rejection |
| AKI | Acute kidney injury |
| AR | Acute rejection |
| ATN | Acute tubular necrosis |
| AUC | Area under the ROC curve |
| BKVN | BK virus nephropathy |
| CAN | Chronic allograft nephropathy |
| CCL2 | Chemokine ligand 2 |
| cfDNA | Cell free DNA |
| CKD | Chronic kidney disease |
| CTOT | Clinical trials in organ transplantation |
| CXCL | C-X-C motif chemokine ligands |
| DGF | Delayed graft function |
| DSA | Donor-specific antibodies |
| DTA | Diagnostic test accuracy |
| EPCAM | Epithelial cell adhesion molecule |
| FOXP3 | Forkhead box P3 |
| GABA | Gamma-aminobutyric acid |
| HE4 | Human epididymis protein 4 |
| HPX | Hemopexin |
| IFNγ | Interferon gamma |
| IFTA | Interstitial fibrosis and tubular atrophy |
| iKEA | Integrated kidney exosome analysis |
| IL | Interleukin |
| LC3 | Microtubule-associated protein 1A/1B-light chain 3 |
| LFABP | Liver-type fatty acid-binding protein |
| MMP7 | Matrix metalloproteinase 7 |
| MNA | 1-methylnicotinamide |
| NAD | Nicotinamide adenine dinucleotide |
| NADP | Nicotinamide adenine dinucleotide phosphate |
| NGAL | Neutrophil gelatinase-associated lipocalin |
| NPV | Negative predictive value |
| PB | Publication bias |
| PD1 | Programmed cell death protein 1 |
| PDX | Podocalyxin |
| PPV | Positive predictive value |
| PRISMA | Preferred reporting items for systematic reviews and meta-analysis |
| QUADAS | Quality assessment tool for diagnostic accuracy studies |
| Sens | Sensitivity |
| Spec | Specificity |
| sTIM3 | Soluble T cell immunoglobulin mucin domain 3 |
| TCMR | T-cell mediated rejection |
| TEC | Tubular epithelial cells |
| TNFα | Tumor necrosis factor alpha |
| TSPAN1 | Tetraspanin 1 |
| uCRM | Urinary common rejection module |
References
- Ronco, C.; Bellomo, R.; Kellum, J.A. Acute kidney injury. Lancet 2019, 394, 1949–1964. [Google Scholar] [CrossRef]
- Yang, J.Y.C.; Sarwal, R.D.; Fervenza, F.C.; Sarwal, M.M.; Lafayette, R.A. Noninvasive Urinary Monitoring of Progression in IgA Nephropathy. Int. J. Mol. Sci. 2019, 20, 4463. [Google Scholar] [CrossRef] [PubMed]
- Kwan, B.; Fuhrer, T.; Zhang, J.; Darshi, M.; Van Espen, B.; Montemayor, D.; de Boer, I.H.; Dobre, M.; Hsu, C.-Y.; Kelly, T.N.; et al. Metabolomic Markers of Kidney Function Decline in Patients With Diabetes: Evidence From the Chronic Renal Insufficiency Cohort (CRIC) Study. Am. J. Kidney Dis. 2020. S0272-6386(20)30572-2. [Google Scholar] [CrossRef] [PubMed]
- Naesens, M.; Anglicheau, D. Precision Transplant Medicine: Biomarkers to the Rescue. J. Am. Soc. Nephrol. 2018, 29, 24–34. [Google Scholar] [CrossRef]
- Guzzi, F.; Knight, S.R.; Ploeg, R.J.; Hunter, J.P. A systematic review to identify whether perfusate biomarkers produced during hypothermic machine perfusion can predict graft outcomes in kidney transplantation. Transpl. Int. 2020, 33, 590–602. [Google Scholar] [CrossRef]
- Jochmans, I.; Pirenne, J. Graft quality assessment in kidney transplantation: Not an exact science yet! Curr. Opin. Organ Transplant. 2011, 16, 174–179. [Google Scholar] [CrossRef]
- Jamshaid, F.; Froghi, S.; Cocco, P.D.; Dor, F.J. Novel non-invasive biomarkers diagnostic of acute rejection in renal transplant recipients: A systematic review. Int. J. Clin. Pract. 2018, 72, e13220. [Google Scholar] [CrossRef]
- Wiebe, C.; Ho, J.; Gibson, I.W.; Rush, D.N.; Nickerson, P.W. Carpe diem-Time to transition from empiric to precision medicine in kidney transplantation. Am. J. Transplant. 2018, 18, 1615–1625. [Google Scholar] [CrossRef]
- Thierry, A.; Thervet, E.; Vuiblet, V.; Goujon, J.-M.; Machet, M.-C.; Noel, L.-H.; Rioux-Leclercq, N.; Comoz, F.; Cordonnier, C.; François, A.; et al. Long-term impact of subclinical inflammation diagnosed by protocol biopsy one year after renal transplantation. Am. J. Transplant. 2011, 11, 2153–2161. [Google Scholar] [CrossRef]
- Haas, M.; Loupy, A.; Lefaucheur, C.; Roufosse, C.; Glotz, D.; Seron, D.; Nankivell, B.J.; Halloran, P.F.; Colvin, R.B.; Akalin, E.; et al. The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am. J. Transplant. 2018, 18, 293–307. [Google Scholar] [CrossRef]
- Singh, N.; Samant, H.; Hawxby, A.; Samaniego, M.D. Biomarkers of rejection in kidney transplantation. Curr Opin Organ Transplant 2019, 24, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Quaglia, M.; Merlotti, G.; Guglielmetti, G.; Castellano, G.; Cantaluppi, V. Recent Advances on Biomarkers of Early and Late Kidney Graft Dysfunction. Int. J. Mol. Sci. 2020, 21, 5404. [Google Scholar] [CrossRef] [PubMed]
- Anglicheau, D.; Naesens, M.; Essig, M.; Gwinner, W.; Marquet, P. Establishing Biomarkers in Transplant Medicine: A Critical Review of Current Approaches. Transplantation 2016, 100, 2024–2038. [Google Scholar] [CrossRef]
- Lo, D.J.; Kaplan, B.; Kirk, A.D. Biomarkers for kidney transplant rejection. Nat. Rev. Nephrol. 2014, 10, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Gwinner, W.; Metzger, J.; Husi, H.; Marx, D. Proteomics for rejection diagnosis in renal transplant patients: Where are we now? World J. Transplant 2016, 6, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Tinel, C.; Devresse, A.; Vermorel, A.; Sauvaget, V.; Marx, D.; Avettand-Fenoel, V.; Amrouche, L.; Timsit, M.-O.; Snanoudj, R.; Caillard, S.; et al. Development and validation of an optimized integrative model using urinary chemokines for noninvasive diagnosis of acute allograft rejection. Am. J. Transplant. 2020. [Google Scholar] [CrossRef]
- Yang, J.Y.C.; Sarwal, R.D.; Sigdel, T.K.; Damm, I.; Rosenbaum, B.; Liberto, J.M.; Chan-On, C.; Arreola-Guerra, J.M.; Alberu, J.; Vincenti, F.; et al. A urine score for noninvasive accurate diagnosis and prediction of kidney transplant rejection. Sci. Transl. Med. 2020, 12, eaba2501. [Google Scholar] [CrossRef]
- Kalantari, S.; Chashmniam, S.; Nafar, M.; Samavat, S.; Rezaie, D.; Dalili, N. A Noninvasive Urine Metabolome Panel as Potential Biomarkers for Diagnosis of T Cell-Mediated Renal Transplant Rejection. OMICS A J. Integr. Biol. 2020, 24, 140–147. [Google Scholar] [CrossRef]
- Verma, A.; Muthukumar, T.; Yang, H.; Lubetzky, M.; Cassidy, M.F.; Lee, J.R.; Dadhania, D.M.; Snopkowski, C.; Shankaranarayanan, D.; Salvatore, S.P.; et al. Urinary cell transcriptomics and acute rejection in human kidney allografts. JCI Insight 2020, 5, e131552. [Google Scholar] [CrossRef]
- Goerlich, N.; Brand, H.A.; Langhans, V.; Tesch, S.; Schachtner, T.; Koch, B.; Paliege, A.; Schneider, W.; Grützkau, A.; Reinke, P.; et al. Kidney transplant monitoring by urinary flow cytometry: Biomarker combination of T cells, renal tubular epithelial cells, and podocalyxin-positive cells detects rejection. Sci. Rep. 2020, 10, 796. [Google Scholar] [CrossRef]
- Banas, M.C.; Neumann, S.; Pagel, P.; Putz, F.J.; Krämer, B.K.; Böhmig, G.A.; Eiglsperger, J.; Schiffer, E.; Ruemmele, P.; Banas, B. A urinary metabolite constellation to detect acute rejection in kidney allografts. EBioMedicine 2019, 48, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Tajima, S.; Fu, R.; Shigematsu, T.; Noguchi, H.; Kaku, K.; Tsuchimoto, A.; Okabe, Y.; Masuda, S. Urinary Human Epididymis Secretory Protein 4 as a Useful Biomarker for Subclinical Acute Rejection Three Months after Kidney Transplantation. Int. J. Mol. Sci. 2019, 20, 4699. [Google Scholar] [CrossRef]
- Kölling, M.; Haddad, G.; Wegmann, U.; Kistler, A.; Bosakova, A.; Seeger, H.; Hübel, K.; Haller, H.; Mueller, T.; Wüthrich, R.P.; et al. Circular RNAs in Urine of Kidney Transplant Patients with Acute T Cell-Mediated Allograft Rejection. Clin. Chem. 2019, 65, 1287–1294. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Yang, J.Y.C.; Bestard, O.; Schroeder, A.; Hsieh, S.-C.; Liberto, J.M.; Damm, I.; Geraedts, A.C.M.; Sarwal, M.M. A urinary Common Rejection Module (uCRM) score for non-invasive kidney transplant monitoring. PLoS ONE 2019, 14, e0220052. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-Y.; Kim, B.K.; Gwon, M.-R.; Seong, S.J.; Ohk, B.; Kang, W.Y.; Lee, H.W.; Jung, H.-Y.; Cho, J.-H.; Chung, B.H.; et al. Urinary metabolomic profiling for noninvasive diagnosis of acute T cell-mediated rejection after kidney transplantation. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1118–1119, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Ciftci, H.S.; Tefik, T.; Savran, M.K.; Demir, E.; Caliskan, Y.; Ogret, Y.D.; Oktar, T.; Sanlı, O.; Kocak, T.; Ozluk, Y.; et al. Urinary CXCL9 and CXCL10 Levels and Acute Renal Graft Rejection. Int. J. Organ Transplant. Med. 2019, 10, 53–63. [Google Scholar]
- Banas, M.; Neumann, S.; Eiglsperger, J.; Schiffer, E.; Putz, F.J.; Reichelt-Wurm, S.; Krämer, B.K.; Pagel, P.; Banas, B. Identification of a urine metabolite constellation characteristic for kidney allograft rejection. Metabolomics 2018, 14, 116. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.-H.; Lee, C.-H.; Kim, K.Y.; Jung, H.-Y.; Choi, J.-Y.; Cho, J.-H.; Park, S.-H.; Kim, Y.-L.; Baek, M.-C.; Park, J.B.; et al. Novel urinary exosomal biomarkers of acute T cell-mediated rejection in kidney transplant recipients: A cross-sectional study. PLoS ONE 2018, 13, e0204204. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, J.; Peng, W.; Weng, C.; Chen, J. Urinary C-X-C motif chemokine 13 is a noninvasive biomarker of antibody-mediated renal allograft rejection. Mol. Med. Rep. 2018, 18, 2399–2406. [Google Scholar] [CrossRef]
- Barabadi, M.; Shahbaz, S.K.; Foroughi, F.; Hosseinzadeh, M.; Nafar, M.; Yekaninejad, M.S.; Amirzargar, A. High Expression of FOXP3 mRNA in Blood and Urine as a Predictive Marker in Kidney Transplantation. Prog. Transplant. 2018, 28, 134–141. [Google Scholar] [CrossRef]
- Mockler, C.; Sharma, A.; Gibson, I.W.; Gao, A.; Wong, A.; Ho, J.; Blydt-Hansen, T.D. The prognostic value of urinary chemokines at 6 months after pediatric kidney transplantation. Pediatr. Transplant. 2018, 22, e13205. [Google Scholar] [CrossRef] [PubMed]
- Senturk Ciftci, H.; Demir, E.; Savran Karadeniz, M.; Tefik, T.; Yazici, H.; Nane, I.; Savran Oguz, F.; Aydin, F.; Turkmen, A. Serum and Urinary Levels of Tumor Necrosis Factor-Alpha in Renal Transplant Patients. Exp. Clin. Transplant. 2018, 16, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lin, H.-Y.; Assaker, J.P.; Jeong, S.; Huang, C.-H.; Kurdi, A.; Lee, K.; Fraser, K.; Min, C.; Eskandari, S.; et al. Integrated Kidney Exosome Analysis for the Detection of Kidney Transplant Rejection. ACS Nano 2017, 11, 11041–11046. [Google Scholar] [CrossRef] [PubMed]
- Millán, O.; Budde, K.; Sommerer, C.; Aliart, I.; Rissling, O.; Bardaji, B.; Matz, M.; Zeier, M.; Silva, I.; Guirado, L.; et al. Urinary miR-155-5p and CXCL10 as prognostic and predictive biomarkers of rejection, graft outcome and treatment response in kidney transplantation. Br. J. Clin. Pharmacol. 2017, 83, 2636–2650. [Google Scholar] [CrossRef]
- Seo, J.-W.; Moon, H.; Kim, S.-Y.; Moon, J.-Y.; Jeong, K.H.; Lee, Y.-H.; Kim, Y.-G.; Lee, T.-W.; Ihm, C.-G.; Kim, C.-D.; et al. Both absolute and relative quantification of urinary mRNA are useful for non-invasive diagnosis of acute kidney allograft rejection. PLoS ONE 2017, 12, e0180045. [Google Scholar] [CrossRef]
- Gandolfini, I.; Harris, C.; Abecassis, M.; Anderson, L.; Bestard, O.; Comai, G.; Cravedi, P.; Cremaschi, E.; Duty, J.A.; Florman, S.; et al. Rapid Biolayer Interferometry Measurements of Urinary CXCL9 to Detect Cellular Infiltrates Noninvasively After Kidney Transplantation. Kidney Int. Rep. 2017, 2, 1186–1193. [Google Scholar] [CrossRef]
- Chen, D.; Peng, W.; Jiang, H.; Yang, H.; Wu, J.; Wang, H.; Chen, J. Noninvasive detection of acute renal allograft rejection by measurement of soluble Tim-3 in urine. Mol. Med. Rep. 2017, 16, 915–921. [Google Scholar] [CrossRef]
- Domenico, T.D.; Joelsons, G.; Montenegro, R.M.; Manfro, R.C. Upregulation of microRNA 142-3p in the peripheral blood and urinary cells of kidney transplant recipients with post-transplant graft dysfunction. Braz. J. Med. Biol. Res. 2017, 50, e5533. [Google Scholar] [CrossRef]
- Lee, H.; Park, Y.-M.; We, Y.-M.; Han, D.J.; Seo, J.-W.; Moon, H.; Lee, Y.-H.; Kim, Y.-G.; Moon, J.-Y.; Lee, S.-H.; et al. Evaluation of Digital PCR as a Technique for Monitoring Acute Rejection in Kidney Transplantation. Genomics Inform. 2017, 15, 2–10. [Google Scholar] [CrossRef]
- Seeman, T.; Vondrak, K.; Dusek, J.; Simankova, N.; Zieg, J.; Hacek, J.; Chadimova, M.; Sopko, B.; Fortova, M. Urinary Neutrophil Gelatinase-Associated Lipocalin Does Not Distinguish Acute Rejection from Other Causes of Acute Kidney Injury in Pediatric Renal Transplant Recipients. Clin. Lab. 2017, 63, 111–114. [Google Scholar] [CrossRef]
- Blydt-Hansen, T.D.; Sharma, A.; Gibson, I.W.; Wishart, D.S.; Mandal, R.; Ho, J.; Nickerson, P.; Rush, D. Urinary Metabolomics for Noninvasive Detection of Antibody-Mediated Rejection in Children After Kidney Transplantation. Transplantation 2017, 101, 2553–2561. [Google Scholar] [CrossRef] [PubMed]
- Belmar Vega, L.; Rodrigo Calabia, E.; Gómez Román, J.J.; Ruiz San Millán, J.C.; Martín Penagos, L.; Arias Rodríguez, M. Relationship Between Albuminuria During the First Year and Antibody-Mediated Rejection in Protocol Biopsies in Kidney Transplant Recipients. Transplant. Proc. 2016, 48, 2950–2952. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Firasat, S.; Khaliq, S.; Khan, A.R.; Mahmood, S.; Aziz, T.; Mubarak, M.; Naqvi, S.A.A.; Rizvi, S.A.H.; Abid, A. Monocyte Chemoattractant Protein-1 (MCP-1/CCL2) Levels and Its Association with Renal Allograft Rejection. Immunol. Investig. 2017, 46, 251–262. [Google Scholar] [CrossRef]
- Galichon, P.; Amrouche, L.; Hertig, A.; Brocheriou, I.; Rabant, M.; Xu-Dubois, Y.-C.; Ouali, N.; Dahan, K.; Morin, L.; Terzi, F.; et al. Urinary mRNA for the Diagnosis of Renal Allograft Rejection: The Issue of Normalization. Am. J. Transplant. 2016, 16, 3033–3040. [Google Scholar] [CrossRef]
- Sigdel, T.K.; Gao, Y.; He, J.; Wang, A.; Nicora, C.D.; Fillmore, T.L.; Shi, T.; Webb-Robertson, B.-J.; Smith, R.D.; Qian, W.-J.; et al. Mining the human urine proteome for monitoring renal transplant injury. Kidney Int. 2016, 89, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- García-Covarrubias, L.; Ventura, E.; Soto, V.; González, E.; García, A.; Aguilar, J.C.; Torres, J.M.; Hinojosa, H.; Fragoso, P.; De Los Santos, J.; et al. Lack of Association Between Elevated Urinary Levels of Interleukin-10 and Interferon Gamma With the Presence of Inflammation in Kidney Transplant Recipients. Transplant. Proc. 2016, 48, 583–587. [Google Scholar] [CrossRef]
- Ho, J.; Rush, D.N.; Krokhin, O.; Antonovici, M.; Gao, A.; Bestland, J.; Wiebe, C.; Hiebert, B.; Rigatto, C.; Gibson, I.W.; et al. Elevated Urinary Matrix Metalloproteinase-7 Detects Underlying Renal Allograft Inflammation and Injury. Transplantation 2016, 100, 648–654. [Google Scholar] [CrossRef]
- Abd Elaziz, M.M.; Bakry, S.; M Abd ElAal, A.E.; Rashed, L.; Hesham, D. Validation of Urinary PD-1 and FOXP3 mRNA in a Cohort of Egyptian Renal Allograft Recipients. Ann. Transplant. 2016, 21, 17–24. [Google Scholar] [CrossRef]
- Lorenzen, J.M.; Schauerte, C.; Kölling, M.; Hübner, A.; Knapp, M.; Haller, H.; Thum, T. Long Noncoding RNAs in Urine Are Detectable and May Enable Early Detection of Acute T Cell-Mediated Rejection of Renal Allografts. Clin. Chem. 2015, 61, 1505–1514. [Google Scholar] [CrossRef]
- Rabant, M.; Amrouche, L.; Morin, L.; Bonifay, R.; Lebreton, X.; Aouni, L.; Benon, A.; Sauvaget, V.; Le Vaillant, L.; Aulagnon, F.; et al. Early Low Urinary CXCL9 and CXCL10 Might Predict Immunological Quiescence in Clinically and Histologically Stable Kidney Recipients. Am. J. Transplant. 2016, 16, 1868–1881. [Google Scholar] [CrossRef]
- Rabant, M.; Amrouche, L.; Lebreton, X.; Aulagnon, F.; Benon, A.; Sauvaget, V.; Bonifay, R.; Morin, L.; Scemla, A.; Delville, M.; et al. Urinary C-X-C Motif Chemokine 10 Independently Improves the Noninvasive Diagnosis of Antibody-Mediated Kidney Allograft Rejection. J. Am. Soc. Nephrol. 2015, 26, 2840–2851. [Google Scholar] [CrossRef] [PubMed]
- Blydt-Hansen, T.D.; Gibson, I.W.; Gao, A.; Dufault, B.; Ho, J. Elevated urinary CXCL10-to-creatinine ratio is associated with subclinical and clinical rejection in pediatric renal transplantation. Transplantation 2015, 99, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Sigdel, T.K.; Ng, Y.W.; Lee, S.; Nicora, C.D.; Qian, W.-J.; Smith, R.D.; Camp, D.G., II; Sarwal, M.M. Perturbations in the urinary exosome in transplant rejection. Front. Med. (Lausanne) 2014, 1, 57. [Google Scholar] [CrossRef] [PubMed]
- Suthanthiran, M.; Schwartz, J.E.; Ding, R.; Abecassis, M.; Dadhania, D.; Samstein, B.; Knechtle, S.J.; Friedewald, J.; Becker, Y.T.; Sharma, V.K.; et al. Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. N. Engl. J. Med. 2013, 369, 20–31. [Google Scholar] [CrossRef] [PubMed]
- van Enst, W.A.; Ochodo, E.; Scholten, R.J.P.M.; Hooft, L.; Leeflang, M.M. Investigation of publication bias in meta-analyses of diagnostic test accuracy: A meta-epidemiological study. BMC Med. Res. Methodol. 2014, 14, 70. [Google Scholar] [CrossRef]
- Li, Y.M.; Li, Y.; Yan, L.; Wang, H.; Wu, X.J.; Tang, J.T.; Wang, L.L.; Shi, Y.Y. Comparison of urine and blood NGAL for early prediction of delayed graft function in adult kidney transplant recipients: A meta-analysis of observational studies. BMC Nephrol. 2019, 20, 291. [Google Scholar] [CrossRef]
- Tsuchimoto, A.; Shinke, H.; Uesugi, M.; Kikuchi, M.; Hashimoto, E.; Sato, T.; Ogura, Y.; Hata, K.; Fujimoto, Y.; Kaido, T.; et al. Urinary Neutrophil Gelatinase-Associated Lipocalin: A Useful Biomarker for Tacrolimus-Induced Acute Kidney Injury in Liver Transplant Patients. PLoS ONE 2014, 9, e110527. [Google Scholar] [CrossRef]
- Zununi Vahed, S.; Omidi, Y.; Ardalan, M.; Samadi, N. Dysregulation of urinary miR-21 and miR-200b associated with interstitial fibrosis and tubular atrophy (IFTA) in renal transplant recipients. Clin. Biochem. 2017, 50, 32–39. [Google Scholar] [CrossRef]
- Naesens, M.; Lerut, E.; Emonds, M.-P.; Herelixka, A.; Evenepoel, P.; Claes, K.; Bammens, B.; Sprangers, B.; Meijers, B.; Jochmans, I.; et al. Proteinuria as a Noninvasive Marker for Renal Allograft Histology and Failure: An Observational Cohort Study. J. Am. Soc. Nephrol. 2016, 27, 281–292. [Google Scholar] [CrossRef]
- Bertrand, D.; Gatault, P.; Jauréguy, M.; Garrouste, C.; Sayegh, J.; Bouvier, N.; Caillard, S.; Lanfranco, L.; Galinier, A.; Laurent, C.; et al. Protocol Biopsies in Patients with Subclinical De Novo DSA After Kidney Transplantation: A multicentric study. Transplantation 2020, 104, 1726–1737. [Google Scholar] [CrossRef]
- Diena, D.; Messina, M.; De Biase, C.; Fop, F.; Scardino, E.; Rossetti, M.M.; Barreca, A.; Verri, A.; Biancone, L. Relationship between early proteinuria and long term outcome of kidney transplanted patients from different decades of donor age. BMC Nephrol. 2019, 20, 443. [Google Scholar] [CrossRef] [PubMed]
- Rotondi, M.; Rosati, A.; Buonamano, A.; Lasagni, L.; Lazzeri, E.; Pradella, F.; Fossombroni, V.; Cirami, C.; Liotta, F.; La Villa, G.; et al. High pretransplant serum levels of CXCL10/IP-10 are related to increased risk of renal allograft failure. Am. J. Transplant. 2004, 4, 1466–1474. [Google Scholar] [CrossRef] [PubMed]
- Lazzeri, E.; Rotondi, M.; Mazzinghi, B.; Lasagni, L.; Buonamano, A.; Rosati, A.; Pradella, F.; Fossombroni, V.; La Villa, G.; Gacci, M.; et al. High CXCL10 expression in rejected kidneys and predictive role of pretransplant serum CXCL10 for acute rejection and chronic allograft nephropathy. Transplantation 2005, 79, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Rotondi, M.; Netti, G.S.; Lazzeri, E.; Stallone, G.; Bertoni, E.; Chiovato, L.; Grandaliano, G.; Gesualdo, L.; Salvadori, M.; Schena, F.P.; et al. High pretransplant serum levels of CXCL9 are associated with increased risk of acute rejection and graft failure in kidney graft recipients. Transpl. Int. 2010, 23, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.A.; Kim, E.J.; Begley, B.; Cheeseman, J.; Harden, T.; Perez, S.D.; Thomas, S.; Warshaw, B.; Kirk, A.D. Urinary chemokines CXCL9 and CXCL10 are noninvasive markers of renal allograft rejection and BK viral infection. Am. J. Transplant. 2011, 11, 2228–2234. [Google Scholar] [CrossRef] [PubMed]
- Hricik, D.E.; Nickerson, P.; Formica, R.N.; Poggio, E.D.; Rush, D.; Newell, K.A.; Goebel, J.; Gibson, I.W.; Fairchild, R.L.; Riggs, M.; et al. Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am. J. Transplant. 2013, 13, 2634–2644. [Google Scholar] [CrossRef] [PubMed]
- Oetting, W.S.; Rogers, T.B.; Krick, T.P.; Matas, A.J.; Ibrahim, H.N. Urinary beta2-microglobulin is associated with acute renal allograft rejection. Am. J. Kidney Dis. 2006, 47, 898–904. [Google Scholar] [CrossRef]
- Li, B.; Hartono, C.; Ding, R.; Sharma, V.K.; Ramaswamy, R.; Qian, B.; Serur, D.; Mouradian, J.; Schwartz, J.E.; Suthanthiran, M. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N. Engl. J. Med. 2001, 344, 947–954. [Google Scholar] [CrossRef]
- Muthukumar, T.; Dadhania, D.; Ding, R.; Snopkowski, C.; Naqvi, R.; Lee, J.B.; Hartono, C.; Li, B.; Sharma, V.K.; Seshan, S.V.; et al. Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N. Engl. J. Med. 2005, 353, 2342–2351. [Google Scholar] [CrossRef]
- Manfro, R.C.; Aquino-Dias, E.C.; Joelsons, G.; Nogare, A.L.; Carpio, V.N.; Gonçalves, L.F.S. Noninvasive Tim-3 messenger RNA evaluation in renal transplant recipients with graft dysfunction. Transplantation 2008, 86, 1869–1874. [Google Scholar] [CrossRef]
- Lorenzen, J.M.; Volkmann, I.; Fiedler, J.; Schmidt, M.; Scheffner, I.; Haller, H.; Gwinner, W.; Thum, T. Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am. J. Transplant. 2011, 11, 2221–2227. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.E. Evaluation and Treatment of Acute Rejection in Kidney Allografts. CJASN 2020, 15, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Haas, M. Evolving criteria for the diagnosis of antibody-mediated rejection in renal allografts. Curr. Opin. Nephrol. Hypertens. 2018, 27, 137–143. [Google Scholar] [CrossRef]
- Sis, B.; Jhangri, G.S.; Bunnag, S.; Allanach, K.; Kaplan, B.; Halloran, P.F. Endothelial gene expression in kidney transplants with alloantibody indicates antibody-mediated damage despite lack of C4d staining. Am. J. Transplant. 2009, 9, 2312–2323. [Google Scholar] [CrossRef]
- Haas, M.; Sis, B.; Racusen, L.C.; Solez, K.; Glotz, D.; Colvin, R.B.; Castro, M.C.R.; David, D.S.R.; David-Neto, E.; Bagnasco, S.M.; et al. Banff 2013 meeting report: Inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am. J. Transplant. 2014, 14, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ 2009, 339, b2535. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.G.; Sterne, J.A.C.; Bossuyt, P.M.M. QUADAS-2 Group QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
| Ref | Study Design | Single/Multicenter | Patients (n) | Enrolment (years) | Urinary Biomarker(s) | Ref. Standard | Outcome |
|---|---|---|---|---|---|---|---|
| Tinel [16] | Cross-sectional | Single center | 329 | 2011–2016 | CXCL9, CXCL10 | Banff ‘15 | TCMR, ABMR |
| Yang [17] | Cross-sectional | Multicenter | 364 | 2010–2018 | Q score | Banff ‘17 | TCMR, ABMR |
| Kalantari [18] | Case-control | Single center | 22 | 2016–2018 | Unbiased metabolomics 1 | Banff ‘97 | TCMR |
| Verma [19] | Case-control | Single center | 53 | N/R | RNA-Seq signature | Banff ‘17 | TCMR |
| Goerlich [20] | Case-control | Single center | 39 | 2016–2017 | T cells, TEC, PDX | Banff ‘13 | TCMR, ABMR |
| Banas [21] | Cross-sectional | Single center | 109 | 2011–2012 | Unbiased metabolomics 2 | Banff ‘09 | TCMR, ABMR |
| Tajima [22] | Cross-sectional | Single center | 80 | 2014–2016 | LC3, CCL2, LFABP, NGAL, HE4 | Banff ‘09 | TCMR, ABMR |
| Kolling [23] | Case-control | Single center | 93 | N/R | Circular RNAs | Banff ‘09 | TCMR |
| Sigdel [24] | Cross-sectional | Multicenter | 150 | 2000–2016 | uCRM score | Banff ‘09 | TCMR, ABMR |
| Kim [25] | Case-control | Multicenter | 23 | N/R | Unbiased metabolomics 3 | Banff ‘07 | TCMR |
| Ciftci [26] * | Prospective | Single center | 85 | 2014–2017 | CXCL9, CXCL10 | Banff ‘13 | TCMR, ABMR |
| Banas [27] | Case-control | Single center | 358 | 2008–2010; 2015–2016 | Unbiased metabolomics 2 | Banff ‘97 | TCMR |
| Lim [28] | Case-control | Multicenter | 47 | 2013–2015 | Exosome proteins | Banff ‘07 | TCMR |
| Chen [29] | Case-control | Single center | 49 | 2006–2009 | CXCL13 | Banff ‘97 | TCMR, ABMR |
| Barabadi [30] § | Cross-sectional | Single center | 91 | 2013–2015 | FOXP3 | Banff ‘13 | AR |
| Mockler [31] *§ | Prospective | Single center | 38 | N/R | CCL2 | Banff ‘13 | TCMR |
| Ciftci [32] * | Prospective | Single center | 65 | 2013–2015 | TNFα | Banff ‘97 | AR |
| Park [33] | Case-control | Single center | 44 | N/R | Exosome proteins | Banff (N/R) | TCMR |
| Millan [34] * | Prospective | Multicenter | 80 | N/R | miR-155-5p, CXCL10 | Banff ‘97 | TCMR |
| Seo [35] | Case-control | Multicenter | 88 | 2013–2015 | CTOT4 formula | Banff (N/R) | TCMR, ABMR |
| Gandolfini [36] § | Case-control | Multicenter | 56 | N/R | CXCL9 | Banff ‘13 | TCMR |
| Chen [37] | Case-control | Single center | 156 | 2006–2009 | sTim3 | Banff ‘97 | TCMR, ABMR |
| Domenico [38] § | Case-control | Single center | 49 | N/R | miRNA-142-3p | Banff ‘07 | AR |
| Lee [39] § | Case-control | Single center | 34 | N/R | Donor-derived cfDNA | unclear | AR |
| Seeman [40] § | Case-control | Single center | 15 | 2013–2014 | NGAL | Banff ‘09 | TCMR, ABMR |
| Blydt-H. [41] | Cross-sectional | Multicenter | 59 | 2002–N/R | ABMR score | Banff ‘13 | ABMR |
| Belmar V. [42] * | Retrospective | Single center | 86 | 2012–2015 | Albumin | Banff (N/R) | ABMR |
| Raza [43] | Cross-sectional | Single center | 300 | 2009–2014 | CCL2 | Banff ‘97 | TCMR |
| Galichon [44] | Cross-sectional | Multicenter | 108 | N/R | CTOT4 formula | Banff ‘09 | TCMR, ABMR |
| Sigdel [45] | Cross-sectional | Single center | 396 | 2000–2011 | Unbiased proteomics | Banff ‘07 | TCMR, ABMR |
| Garcìa-C. [46] § | Cross-sectional | Single center | 50 | N/R | IL10, IFNγ | Banff ‘09 | TCMR, ABMR |
| Ho [47] § | Cross-sectional | Single center | 133 | N/R | MMP7, CXCL10 | Banff ‘07 | TCMR |
| A. Elaziz [48] | Cross-sectional | Single center | 54 | 2011–2014 | PD1, FOXP3 | Banff ‘07 | TCMR |
| Lorenzen [49] | Cross-sectional | Single center | 93 | N/R | LncRNAs | Banff ‘09 | TCMR |
| Rabant [50] * | Prospective | Single center | 300 | 2010–2012 | CXCL9, CXCL10 | Banff ‘07 | TCMR, ABMR |
| Rabant [51] | Cross-sectional | Single center | 244 | 2011–2013 | CXCL9, CXCL10 | Banff ‘07 | TCMR, ABMR |
| Blydt-H. [52] | Cross-sectional | Single center | 51 | 2002–N/R | CXCL10 | Banff ‘07 | TCMR |
| Sigdel [53] § | Case-control | Single center | 30 | 2000–2009 | Exosome proteins | Banff ‘07 | AR |
| Category | Biomarkers |
|---|---|
| Cytokines | |
| Chemokines Other | CCL2, CXCL9, CXCL10, CXCL13 IFNγ, IL10, TNFα |
| Metabolites | |
| Nucleotides Amino acids and Organic acids Other small molecules | NAD, NADP Alanine, Citrate, GABA, 4-Guanidinobutyric Acid, Guanidoacetic Acid, Homocysteine, Lactate, Methylimidazoleacetic Acid, Nicotinic Acid, l-Tryptophan Cholesterol Sulfate, Dopamine, MNA, Urea |
| Proteins | Albumin, LFAPB, HE4, LC3, MMP7, NGAL, sTIM3, Urinary extracellular vesicle (exosome) proteins (HPX, TSPAN1) |
| RNAs micro RNAs | Circular RNAs, FOXP3 mRNA, LncRNAs, PD1 mRNA, RNA-seq miR-142-3p, miR-155-5p |
| Urinary Cells | CD4+/CD8+ T cells, CD10+/EPCAM+ cells, PDX+ cells, TEC |
| Scores and Formulas | |
| ABMR score [41] CTOT-4 formula [54] Q score [17] uCRM score [24] | Signature of 133 unique metabolites CD3ε mRNA + CXCL10 mRNA + 18S rRNA Cell-free DNA + Clusterin + Creatinine + CXCL10 + Methylated Cell-free DNA + Total Urinary Protein 11 genes expression score on urinary cell pellet (including CXCL9 and CXCL10) |
| Ref | Risk of Bias | Applicability Concerns | |||||
|---|---|---|---|---|---|---|---|
| Patient Selection | Index Test | Reference Standard | Flow and Timing | Patient Selection | Index Test | Reference Standard | |
| Tinel [16] | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Yang [17] | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Kalantari [18] | ☹ | ☹ | ☺ | ? | ☹ | ☹ | ☺ |
| Verma [19] | ☹ | ☹ | ☺ | ☺ | ☹ | ☺ | ☺ |
| Goerlich [20] | ☹ | ☹ | ☺ | ☺ | ☹ | ☺ | ☺ |
| Banas [21] | ☺ | ☺ | ☺ | ? | ☺ | ☺ | ☺ |
| Tajima [22] | ☺ | ☹ | ☺ | ☺ | ☹ | ☺ | ☺ |
| Kolling [23] | ☹ | ☹ | ☺ | ☺ | ☹ | ☺ | ☺ |
| Sigdel [24] | ☺ | ☹ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Kim [25] | ☹ | ☹ | ☺ | ☺ | ☹ | ☺ | ☺ |
| Ciftci [26] * | ☹ | ☹ | ☺ | ? | ☹ | ☺ | ☺ |
| Banas [27] | ☹ | ☹ | ☺ | ☺ | ☹ | ☺ | ☺ |
| Lim [28] | ☹ | ☹ | ☺ | ☺ | ☹ | ☺ | ☺ |
| Chen [29] | ☹ | ☹ | ? | ☺ | ☹ | ☺ | ☺ |
| Ciftci [31] * | ☹ | ☹ | ? | ? | ☹ | ☺ | ☺ |
| Park [33] | ☹ | ☺ | ☺ | ☺ | ☹ | ☺ | ☺ |
| Millan [34] * | ? | ☹ | ☺ | ? | ☺ | ☺ | ☺ |
| Seo [35] | ☹ | ☹ | ☺ | ☹ | ☹ | ☺ | ☺ |
| Chen [37] | ☹ | ☹ | ? | ☺ | ☹ | ☺ | ☺ |
| Blydt-H. [41] | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Belm.V. [42] * | ☹ | ☹ | ? | ☺ | ☹ | ☺ | ☺ |
| Raza [43] | ☺ | ☹ | ☺ | ☹ | ☹ | ☺ | ☺ |
| Galichon [44] | ☺ | ☹ | ? | ☺ | ☺ | ☺ | ☺ |
| Sigdel [45] | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ | ☺ |
| A. Elaziz [48] | ? | ☹ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Lorenzen [49] | ☺ | ☹ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Rabant [50] * | ☺ | ☹ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Rabant [51] | ☺ | ☹ | ☺ | ☺ | ☺ | ☺ | ☺ |
| Blydt-H. [52] | ☺ | ☹ | ☺ | ☹ | ☺ | ☺ | ☺ |
| Ref. | Outcome (n) | Control Group (n) | Test Design, Biomarkers, Thresholds | Diagnostic Test Accuracy (95%CI) | ||||
|---|---|---|---|---|---|---|---|---|
| Sens. | Spec. | PPV | NPV | AUC–Accuracy(%) | ||||
| Tinel [16] | TCMR (17), ABMR (64), mixed (14) | ALL-B (normal, 21; IFTA, 154; BKVN, 23; ATN, 11; recurrent disease, 9; other, 78) | CXCL9 + CXCL10 for AR | 62% | 72% | 41% | 86% | 0.70 (0.64–0.76) |
| CXCL9 + CXCL10 for TCMR | 79% | 74% | 21% | 98% | 0.81 (0.73–0.89) | |||
| CXCL9 + CXCL10 for ABMR | 72% | 54% | 28% | 88% | 0.67 (0.61–0.74) | |||
| Yang [17] | TCMR + ABMR (103) | ALL-B (normal, 170; bAR, 50; | Training: AR vs normal (Q score ≥ 32) | 95% | 100% | - | - | 0.99 (0.99–1.00) |
| BKVN, 9) | Validation 1: AR vs normal | 91% | 92% | - | - | 0.98 (0.96–1.00) | ||
| Validation 2: AR vs normal | 100% | 96% | - | - | 1.00 (1.00–1.00) | |||
| All AR vs All normal | 95% | 96% | 87% | 98% | 0.99(0.98–0.99) | |||
| All AR vs ALL-B | - | - | - | - | 0.96 (0.94–0.98) | |||
| Kalantari [18] | TCMR (7) | DYS-B (normal, 15) | Unbiased metab.1 | 67–71% | 40–100% | - | - | 0.51–0.71 |
| Verma [19] | TCMR (22) | ALL-B (normal, 28) | 13-gene urinary cell signature | - | - | - | - | 0.92 (0.85–0.99) |
| Goerlich [20] | TCMR (14) + ABMR (7) | DYS-B (normal, 18) | T cells + total TEC | - | - | - | - | 0.90 |
| T cells + CD10+ TEC | - | - | - | - | 0.89 | |||
| T cells + ECPAM+ TEC | - | - | - | - | 0.91 | |||
| T cells + PDX+ cells | - | - | - | - | 0.89 | |||
| Banas [21] | TCMR + ABMR + mixed | ALL-B (normal) + STA | Unbiased metab.2 | - | - | - | - | 0.75 (0.68–0.83) |
| Score = 3.0 | 91% (79–98) | 34% (30–38) | - | - | - | |||
| Score = 13.0 | 48% (33–63) | 89% (86–91) | - | - | - | |||
| + bAR | + (IFTA + other) | - | - | - | - | 0.71 (0.64–0.79) | ||
| Tajima [22] | TCMR + ABMR (subclinical, 11) | STA-B (normal or borderline AR, 69) | LC3 (517.9 pg/mg) | 64% (31–89) | 78% (67–87) | 32% | 93% | 0.73 (0.55–0.90) |
| CCL2 (226.0 pg/mg) | 82% (48–98) | 57% (44–68) | 23% | 95% | 0.69 (0.54–0.84) | |||
| L-FABP (7.6 ng/mg) | 9% (0–41) | 88% (78–94) | 15% | 100% | 0.61 (0.45–0.77) | |||
| NGAL (12.8 ng/mg) | 100% (72–100) | 48% (36–60) | 23% | 100% | 0.72 (0.59–0.84) | |||
| HE4 (789.1 ng/mg) | 100% (72–100) | 54% (41–66) | 26% | 100% | 0.81 (0.70–0.92) | |||
| Kolling [23] | TCMR (11; subclinical, 51) | STA-B (normal, 31) | hsa_circ_0001334 (2.41) | 70% (59–80) | 92% (64–100) | 98% | 32% | 0.85 (p < 0.0001) |
| Sigdel [24] | TCMR + ABMR (45) | ALL-B (normal, 43; bAR, 19; BKVN, 43) | AR vs normal (uCRM score = 3.63) | 95% | 98% | - | - | 0.99, p < 0.0001 |
| AR vs normal + bAR | 87% | 98% | - | - | - | |||
| AR vs normal + bAR + BKVN | 77% | 98% | - | - | 96.6% | |||
| Kim [25] | TCMR (14) | STA-B (normal, 17) | Unbiased metab.3 | - | - | - | - | - |
| Training: TCMR (10) vs STA-B (13) | 90% | 85% | - | - | 0.93 (0.72–1.00) - 87% | |||
| Validation: TCMR (4) vs STA-B (4) | - | - | - | - | 62.5% | |||
| Banas [27] | TCMR | ALL-B (normal) + STA (extended) | Unbiased metab.2, train (180) | - | - | - | - | 0.76 (0.69–0.82) |
| Test (178) strict/extended cohort | - | - | - | - | 0.72 (0.58–0.86)/ 0.74 (0.62–0.86) | |||
| Lim [28] | TCMR (25) | STA-B (normal, 22) | TSPAN1 + HPX | 64% | 73% | - | - | 0.74 |
| Chen [29] | TCMR (37) + ABMR (12) | ALL-B (normal, 58; CAN, 29; ATN, 10) | CXCL13 for AR vs. normal | 84% | 79% | - | - | 0.82 (0.73–0.90) |
| CXCL13 for AR vs. CAN + ATN | - | - | - | - | 0.63 (0.52–0.75) | |||
| Park [33] | TCMR (22) | DYS-B (normal, 22) | iKEA | |||||
| Training: TCMR (15) vs normal (15) | 93% | 88% | - | - | 0.91 ± 0.02 - 90% | |||
| Validation: TCMR (7) vs normal (7) | 64% | 100% | - | - | 0.84 ± 0.11 - 71% | |||
| Seo [35] | TCMR (27) + ABMR (13) | STA-B (normal, 17); STA (22) | CTOT4 formula | - | - | - | - | 0.72 (0.60–0.83) |
| CXCL10 mRNA | - | - | - | - | 0.72 (0.60–0.83) | |||
| CD3ε mRNA | - | - | - | - | 0.71 (0.60–0.83) | |||
| 18S rRNA | - | - | - | - | 0.47 (0.33–0.60) | |||
| Chen [37] | TCMR (37) + ABMR (12) | STA-B (normal, 58) | sTim-3 (1.836 ng/mmol) | 90% | 83% | - | - | 0.88 (0.81–0.95) |
| Blydt-H. [41] | ABMR (10) | ALL-B (normal, TCMR, transplant glomerulopathy, IFTA, other, 49) | ABMR score = 0.23 | 78% | 83% | 40% | 96% | 0.84 (0.77–0.91) |
| ABMR score with top 10 metabolites | - | - | - | - | 0.80 (0.73–0.88) | |||
| Validation | - | - | - | - | 0.76 (0.67–0.84) | |||
| Raza [43] | TCMR (acute, 101; borderline, 47; vascular, 17) | DYS-B (normal, 47; IFTA, 46) + STA (42) | CCL2 (198 pg/mL) | 87% | 62% | - | - | 0.81 (0.76–0.86) |
| Galichon [44] | TCMR (11) + bAR (3) + ABMR (28) + mixed (9) | ALL-B (56) | CTOT4 formula | - | - | - | - | 0.72 (0.61–0.82) |
| CXCL10 mRNA | - | - | - | - | 0.76 (0.66–0.86) | |||
| CD3ε mRNA | - | - | - | - | 0.67 (0.56–0.78) | |||
| 18S rRNA | - | - | - | - | 0.63 (0.53–0.74) | |||
| Sigdel [45] | TCMR + ABMR (42) | ALL-B (normal, 47; CAN, 46; BKVN, 16) | Unbiased proteomics (11 peptides) | |||||
| Validation: AR (20) vs normal (27), CAN (15), BKVN (16) | - | - | - | - | 0.94 (0.93–0.95) | |||
| A. Elaziz [48] | TCMR (31) | STA-B (normal, 23) | PD1 mRNA (2.6) | 80% | 84% | - | - | 0.81 |
| FOXP3 mRNA (1.5) | 83% | 90% | - | - | 0.91 | |||
| PD1 + FOXP3 mRNA | 94% | 97% | - | - | 0.98 | |||
| Lorenzen [49] | TCMR (11; subclinical 51) | STA-B (normal, 31) | RNA L328 (9.556) | 49% | 96% | 49% | 93% | 0.76 (p < 0.001) |
| Rabant [51] | TCMR (10) + ABMR (37) + mixed (31) | DYS-B (203) | CXCL9 | 58% | 85% | 59% | 84% | 0.71 (0.64–0.78) |
| CXCL10 | 59% | 83% | 58% | 84% | 0.74 (0.68–0.80) | |||
| Blydt-H. [52] | TCMR (subclinical, 17; clinical, 9) | ALL-B (normal, 21; IFTA, 31) | CXCL10, subclinical (4.82 ng/mL) | 59% | 67% | - | - | 0.81 (0.70–0.92) |
| Clinical (4.72 ng/mL) | 77% | 60% | - | - | 0.88 (0.73–1.0) | |||
| Ref. | Outcome (n) | Control Group (n) | Biomarkers, Thresholds and Time Post-Transplant | Diagnostic Test Accuracy (95%CI) | ||||
|---|---|---|---|---|---|---|---|---|
| Sens. | Spec. | PPV | NPV | AUC | ||||
| Ciftci [26] | TCMR (9) + ABMR (6) | STA (70) | CXCL9, 1 day - 3 months | 70–85% | 37–88% | 60–71% | 71–90% | 0.71–0.95 |
| CXCL10, 1 day - 3 months | 78–82% | 58–85% | 59–73% | 74–87% | 0.75–0.97 | |||
| Ciftci [32] | AR (9) | STA (56) | TNF-α (12.08 pg/mL), 1 day | 71% | 57% | - | - | 0.74 (0.51–0.97) |
| TNF-α (11.03), 7 days | 100% | 84% | - | - | 0.95 (0.88–1.00) | |||
| TNF-α (9.85), 1 month | 100% | 83% | - | - | 0.91 (0.81–1.00) | |||
| TNF-α (9.13), 3 months | 100% | 71% | - | - | 0.83 (0.75–0.98) | |||
| TNF-α (7.42), 6 months | 100% | 62% | - | - | 0.82 (0.69–0.95) | |||
| Millan [34] | TCMR (8) | STA (72) | miR-155-5p (0.51), 1wk-6m | 85% | 86% | 88% | 100% | 0.88 (0.78–0.97) |
| CXCL10 (84.73 pg/mL),1wk-6m | 84% | 80% | 90% | 85% | 0.87 (0.81–0.92) | |||
| CXCL10:Cr (0.43), 1wk-6m | 72% | 73% | 90% | 96% | 0.75 (0.67–0.83) | |||
| Belm.V. [42] | ABMR (subclinical) | ALL-B | Albuminuria (> 30 mg/g), 6m | - | - | - | - | 0.75 (0.55–0.95) |
| Rabant [50] | AR (TCMR + ABMR + mixed, 76) | ALL-B | CXCL9:Cr (1.78 ng/mmoL),10d | 61% | 50% | 24% | 84% | 0.58 (0.47–0.68) |
| CXCL9:Cr (0.96), 1 month | 81% | 35% | 23% | 89% | 0.50 (0.37–0.62) | |||
| CXCL9:Cr (1.67), 3 months | 57% | 62% | 18% | 91% | 0.57 (0.39–0.75) | |||
| CXCL10:Cr (4.80), 10 days | 57% | 52% | 23% | 83% | 0.54 (0.43–0.65) | |||
| CXCL10:Cr (2.79), 1 month | 83% | 51% | 29% | 93% | 0.72 (0.61–0.80) | |||
| CXCL10:Cr (5.32), 3 months | 54% | 77% | 25% | 92% | 0.68 (0.55–0.80) | |||
| Ref. | Outcome (n) | Control Group (n) | Biomarkers, Thresholds and Main Results |
|---|---|---|---|
| Barabadi [30] | AR (27) | ALL-B (normal, 45; CAN, 19) | FOXP3 mRNA expression was significantly higher in AR (p < 0.001) |
| Mockler [31] * | TCMR (5; borderline, 3) | STA-B | There was no significant association between 6 months post-transplant CCL2 and TCMR changes (p = 0.46) |
| Gandolfini [36] | TCMR (22) | ALL-B (normal, 19) | CXCL9 > 200 pg/mL in TCMR, 100-200 in dysfunction graft, and < 100 pg/mL in stable graft (p < 0.01) |
| Domenico [38] | AR (23) | ALL-B (ATN, 18; normal, 8) | mirRNA 142-3p was significantly higher in AR compared to stable graft (p < 0.001); not compared to ATN (p = 0.079) |
| Lee [39] | AR (8) | STA (8); DYS-B (ATN, 8; other, 4) | Donor-derived cfDNA was not significantly different between groups (p = 0.95) |
| Seeman [40] | TCMR (2) + ABMR (2) | DYS-B (11) | NGAL was not significantly different between groups (p = 0.48) |
| Garcìa-C. [46] | AR (9) | ALL-B (fibrosis, 31; other, 10) | IL10 and IFNγ were not significantly different between groups (p = 0.95, p = 0.1) |
| Ho [47] | TCMR (17; subclinical, 17) | ALL-B (normal, 22) | MMP7 and CXCL10 were significantly elevated in subclinical (p = 0.01, p < 0.0001) and clinical (p < 0.001) TCMR |
| Sigdel [53] | AR (10) | DYS-B (IFTA, BKVN, 20) | Ten urinary exosomal proteins were significantly increased in AR (p < 0.05) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guzzi, F.; Cirillo, L.; Buti, E.; Becherucci, F.; Errichiello, C.; Roperto, R.M.; Hunter, J.P.; Romagnani, P. Urinary Biomarkers for Diagnosis and Prediction of Acute Kidney Allograft Rejection: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 6889. https://doi.org/10.3390/ijms21186889
Guzzi F, Cirillo L, Buti E, Becherucci F, Errichiello C, Roperto RM, Hunter JP, Romagnani P. Urinary Biomarkers for Diagnosis and Prediction of Acute Kidney Allograft Rejection: A Systematic Review. International Journal of Molecular Sciences. 2020; 21(18):6889. https://doi.org/10.3390/ijms21186889
Chicago/Turabian StyleGuzzi, Francesco, Luigi Cirillo, Elisa Buti, Francesca Becherucci, Carmela Errichiello, Rosa Maria Roperto, James P. Hunter, and Paola Romagnani. 2020. "Urinary Biomarkers for Diagnosis and Prediction of Acute Kidney Allograft Rejection: A Systematic Review" International Journal of Molecular Sciences 21, no. 18: 6889. https://doi.org/10.3390/ijms21186889
APA StyleGuzzi, F., Cirillo, L., Buti, E., Becherucci, F., Errichiello, C., Roperto, R. M., Hunter, J. P., & Romagnani, P. (2020). Urinary Biomarkers for Diagnosis and Prediction of Acute Kidney Allograft Rejection: A Systematic Review. International Journal of Molecular Sciences, 21(18), 6889. https://doi.org/10.3390/ijms21186889





