Signalling Pathways Implicated in Alzheimer′s Disease Neurodegeneration in Individuals with and without Down Syndrome
Abstract
:1. Introduction
2. Amyloid Plaques
3. Neurofibrillary Tangles
4. Cholinergic Neurodegeneration
5. Changes in Energy Consumption and Accumulation: Oxidative Stress, Mitochondrial Alterations, and Energy Metabolism
5.1. Oxidative Stress
5.2. Mitochondria
5.3. Energy Metabolism
6. Cellular Senescence
7. Immune Response/Inflammation
8. Changes in Cell Proliferation/Differentiation and Migration
9. Alterations in Intercellular Signalling: Neurotransmitter Release, Synapses, and Receptors
9.1. Neurotransmitter Release
9.2. Synapses
9.3. Receptors
10. Therapies Targeting Different Pathways Implicated in AD Pathology
11. Concluding Remarks
12. Key Summary Points
- AD, in individuals with or without DS, is a disease with a complex set of neuropathological signs.
- Numerous signalling pathways are implicated in the onset and aggravation of this neuropathology.
- The same signalling pathway often plays a role in the appearance or progression of different signs of AD.
- In many cases, synergic effects and feedback loops exist between these pathways.
- Because of the complex etiopathology of AD and the interrelation between the factors responsible for the symptoms of the disease, therapeutic approaches should combine different targets.
Author Contributions
Funding
Conflicts of Interest
References
- Alzheimer’s Association Home Page. Available online: http://Alzh.org (accessed on 18 September 2020).
- Antonarakis, S.E.; Skotko, B.G.; Rafii, M.S.; Strydom, A.; Pape, S.E.; Bianchi, D.W.; Sherman, S.L.; Reeves, R.H. Down syndrome. Nat. Rev. Dis. Primers 2020, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Lott, I.T. Neurological phenotypes for Down syndrome across the life span. Prog. Brain Res. 2012, 197, 101–121. [Google Scholar] [PubMed] [Green Version]
- Haydar, T.F.; Reeves, R.H. Trisomy 21 and early brain development. Trends Neurosci. 2012, 35, 81–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teipel, S.J.; Hampel, H. Neuroanatomy of Down syndrome in vivo: A model of preclinical Alzheimer´s disease. Behav. Genet. 2006, 36, 405–415. [Google Scholar] [CrossRef]
- Cenini, G.; Dowling, A.L.; Beckett, T.L.; Barone, E.; Mancuso, C.; Murphy, M.P.; LeVine III, H.; Lott, I.T.; Schmitt, F.A.; Butterfield, D.A.; et al. Association between frontal cortex oxidative damage and beta-amyloid as a function of age in Down syndrome. Biochim. Biophys. Acta 2012, 1822, 130–138. [Google Scholar] [CrossRef]
- Wilcock, D.M.; Griffin, W.S. Down’s syndrome, neuroinflammation, and Alzheimer neuropathogenesis. J. Neuroinflammation 2013, 10, 84. [Google Scholar] [CrossRef] [Green Version]
- Sabbagh, M.N.; Fleisher, A.; Chen, K.; Rogers, J.; Berk, C.; Reiman, E.; Pontecorvo, M.; Mintun, M.; Skovronsky, D.; Jacobson, S.A.; et al. Positron emission tomography and neuropathologic estimates of fibrillar amyloid-β in a patient with Down syndrome and Alzheimer disease. Arch. Neurol. 2011, 68, 1461–1466. [Google Scholar] [CrossRef]
- Moreno-Jiménez, E.P.; Flor-García, M.; Terreros-Roncal, J.; Rábano, A.; Cafini, F.; Pallas-Bazarra, N.; Ávila, J.; Llorens-Martín, M. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 2019, 25, 554–560. [Google Scholar] [CrossRef]
- Kim, J.; Basak, J.M.; Holtzman, D.M. The role of apolipoprotein E in Alzheimer’s disease. Neuron 2009, 63, 287–303. [Google Scholar] [CrossRef] [Green Version]
- Chang, K.T.; Min, K.T. Drosophila melanogaster homolog of Down syndrome critical region 1 is critical for mitochondrial function. Nat. Neurosci. 2005, 8, 1577–1585. [Google Scholar] [CrossRef]
- Coskun, P.E.; Wyrembak, J.; Derbereva, O.; Melkonian, G.; Doran, E.; Lott, I.T.; Head, E.; Cotman, C.W.; Wallace, D.C. Systemic mitochondrial dysfunction and the etiology of Alzheimer’s disease and down syndrome dementia. J. Alzheimers Dis. 2010, 20, 293–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butterfield, D.A.; Di Domenico, F.; Swomley, A.M.; Head, E.; Perluigi, M. Redox proteomics analysis to decipher the neurobiology of Alzheimer-like neurodegeneration: Overlaps in Down’s syndrome and Alzheimer’s disease brain. Biochem. J. 2014, 463, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesini, A.; Iglesias, E.; Bayona-Bafaluy, M.P.; Garrido-Pérez, N.; Meade, P.; Gaudó, P.; Jiménez-Salvador, I.; Andrés-Benito, P.; Montoya, J.; Ferrer, I.; et al. Brain pyrimidine nucleotide synthesys and Alzheimer disease. Aging 2019, 11, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cué, C.; Rueda, N. Cellular senescence in neurodegenerative diseases. Front. Cell. Neurosci. 2020, 14, 16. [Google Scholar] [CrossRef]
- Hard, J. Has the amyloid cascade hypothesis for Alzheimer’s disease been proved? Curr. Alzheimer Res. 2006, 3, 71–73. [Google Scholar] [CrossRef]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef]
- Gitter, B.D.; Cox, L.M.; Rydel, R.E.; May, P.C. Amyloid beta peptide potentiates cytokine secretion by interleukin-1 beta-activated human astrocytoma cells. Proc. Natl. Acad. Sci. USA 1995, 92, 10738–10741. [Google Scholar] [CrossRef] [Green Version]
- Chong, Y. Effect of a carboxy-terminal fragment of the Alzheimer’s amyloid precursor protein on expression of proinflammatory cytokines in rat glial cells. Life Sci. 1997, 61, 2323–2333. [Google Scholar] [CrossRef]
- Weldon, D.T.; Rogers, S.D.; Ghilardi, J.R.; Finke, M.P.; Cleary, J.P.; O’Hare, E.; Esler, W.P.; Maggio, J.E.; Mantyh, P.W. Fibrillar beta-amyloid induces microglial phagocytosis, expression of inducible nitric oxide synthase, and loss of a select population of neurons in the rat CNS in vivo. J. Neurosci. 1998, 18, 2161–2173. [Google Scholar] [CrossRef] [Green Version]
- Eikelenboom, P.; Veerhuis, R.; Scheper, W.; Rozemuller, A.J.M.; van Gool, W.A.; Hoozemans, J.J.M. He significance of neuroinflammation in understanding Alzheimer’s disease. J. Neural. Transm. 2006, 113, 1685–1695. [Google Scholar] [CrossRef]
- Sipos, E.; Kurunczi, A.; Kasza, A.; Horváth, J.; Felszeghy, K.; Laroche, S.; Toldi, J.; Párducz, A.; Penke, B.; Penke-Verdier, Z. Beta-amyloid pathology in the entorhinal cortex of rats induces memory deficits: Implications for Alzheimer’s disease. Neuroscience 2007, 147, 28–36. [Google Scholar] [CrossRef]
- Wilcock, D.M. Neuroinflammation in the aging Down syndrome brain; Lessons from Alzheimer’s disease. Curr. Gerontol. Geriatr. Res. 2012, 51, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, S.J.; Hill, W.D.; Marioni., R.E.; Davies, G.; Hagenaars, S.P.; Harris, S.E.; Cox, S.R.; Taylor, A.M.; Corley, J.; Pattie, A.; et al. Polygenic predictors of age-related decline in cognitive ability. Mol. Psychiatry 2019, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, M.S.C.; Ojinaga Alfageme, O.; D’Souza, H.; Patkee, P.A.; Rutherford, M.A.; Mok, K.Y.; Hardy, J.; Karmiloff-Smith, A.; the LonDownS Consortium. A multi-level developmental approach to exploring individual differences in Down syndrome: Genes, brain, behaviour, and environment. Res. Dev. Disabil. 2020, 104, 103638. [Google Scholar] [CrossRef] [PubMed]
- Bu, G. Apolipoprotein E and its receptors in Alzheimer’s disease: Pathways, pathogenesis and therapy. Nat. Rev. Neurosci. 2009, 10, 333–344. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Mucke, L. Alzheimer mechanisms and therapeutic strategies. Cell 2012, 148, 1204–1222. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef] [Green Version]
- Prasher, V.P.; Sajith, S.G.; Rees, S.D.; Patel, A.; Tewari, S.; Schupf, N.; Zigman, W.B. Significant effect of APOE epsilon 4 genotype on the risk of dementia in Alzheimer’s disease and mortality in persons with Down syndrome. Int. J. Geriatr. Psychiatry 2008, 23, 1134–1140. [Google Scholar] [CrossRef] [Green Version]
- Tan, C.C.; Yu, J.T.; Tan, M.S.; Jiang, T.; Zhu, X.C.; Tan, L. Autophagy in aging and neurodegenerative diseases: Implications for pathogenesis and therapy. Neurobiol. Aging 2014, 35, 941–957. [Google Scholar] [CrossRef]
- Ryoo, S.R.; Cho, H.J.; Lee, H.W.; Jeong, H.K.; Radnaabazar, C.; Kim, Y.S.; Kim, M.-J.; Son, M.-Y.; Seo, H.; Chung, S.-H.; et al. Dual-specificity tyrosine(Y)-phosphorylation regulated kinase 1A-mediated phosphorylation of amyloid precursor protein: Evidence for a functional link between Down syndrome and Alzheimer’s disease. J. Neurochem. 2008, 104, 1333–1344. [Google Scholar] [CrossRef]
- Kentrup, H.; Becker, W.; Heukelbach, J.; Wilmes, A.; Schurmann, A.; Huppertz, C.; Kainulainen, K.; Joost, H.G. Dyrk, a dual specificity protein kinase with unique structural features whose activity is dependent on tyrosine residues between subdomains VII and VIII. J. Biol. Chem. 1996, 271, 3488–3495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.J.; Zhu, Y.; Zhang, J.; Cheng, J.F.; Rubin, E.M. Construction of a panel of transgenic mice containing a contiguous 2-Mb set of YAC/P1 clones from human chromosome 21q22.2. Genomics 1995, 27, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Stevens, M.E.; Sudanagunta, S.P.; Bronson, R.T.; Makhinson, M.; Watabe, A.M.; O’Dell, T.J.; Fung, J.; Weier, H.U.; Cheng, J.F.; et al. Functional screening of 2Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nat. Genet. 1997, 16, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Altafaj, X.; Dierssen, M.; Baamonde, C.; Martí, E.; Visa, J.; Guimerà, J.; Oset, M.; Gonzáles, J.R.; Flórez, J.; Fillat, C.; et al. Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down’s syndrome. Hum. Mol. Genet. 2001, 10, 1915–1923. [Google Scholar] [CrossRef] [Green Version]
- Ahn, K.J.; Jeong, H.K.; Choi, H.S.; Ryoo, S.R.; Kim, Y.J.; Goo, J.S.; Choi, S.-Y.; Han, J.-S.; Ha, I.; Song, W.J. DYRK1A BAC transgenic mice show altered synaptic plasticity with learning and memory defects. Neurobiol. Dis. 2006, 22, 463–472. [Google Scholar] [CrossRef]
- Kimura, R.; Kamino, K.; Yamamoto, M.; Nuripa, A.; Kida, T.; Kazui, H.; Hashimoto, R.; Tanaka, T.; Kudo, T.; Yamagata, H.; et al. The DYRK1A gene, encoded in chromosome 21 Down syndrome critical region, bridges between beta-amyloid production and tau phosphorylation in Alzheimer disease. Hum. Mol. Genet. 2007, 16, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Wegiel, J.; Gong, C.-X.; Hwang, Y.-W. The role of DYRK1A in neurodegenerative diseases. FEBS J. 2011, 278, 236–245. [Google Scholar] [CrossRef]
- Ryu, Y.S.; Park, S.Y.; Jung, M.S.; Yoon, S.H.; Kwen, M.Y.; Lee, S.Y.; Choi, S.-H.; Radnaabazar, C.; Kim, M.-K.; Kim, H.; et al. Dyrk1A-mediated phosphorylation of Presenilin 1: A functional link between Down syndrome and Alzheimer’s disease. J. Neurochem. 2010, 115, 574–584. [Google Scholar] [CrossRef]
- Ferrer, I.; Gomez-Isla, T.; Puig, B.; Freixes, M.; Ribe, E.; Dalfo, E.; Avila, J. Current advances on different kinases involved in tau phosphorylation, and implications in Alzheimer’s disease and tauopathies. Curr. Alzheimer Res. 2005, 2, 3–18. [Google Scholar] [CrossRef]
- Jung, M.S.; Park, J.H.; Ryu, Y.S.; Choi, S.H.; Yoon, S.H.; Kwen, M.Y.; Oh, J.Y.; Song, W.J.; Chung, S.H. Regulation of RCAN1 protein activity by Dyrk1A protein-mediated phosphorylation. J. Biol. Chem. 2011, 286, 40401–40412. [Google Scholar] [CrossRef] [Green Version]
- Mattson, M.P. Oxidative stress, perturbed calcium homeostasis, and immune dysfunction in Alzheimer’s disease. J. Neurovirol. 2002, 8, 539–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asai, M.; Kinjo, A.; Kimura, S.; Mori, R.; Kawajubo, T.; Shirotani, K.; Yagishita, S.; Maruyama, K.; Iwata, N. Perturbed calcineurin-NFAT signaling is associated with the development of Alzheimer’s disease. Biol. Pharm. Bull. 2016, 39, 1646–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, K.J.; Ermak, G.; Rothermel, B.A.; Pritchard, M.; Heitman, J.; Ahnn, J.; Henrique-Silva, F.; Crawford, D.; Canaider, S.; Strippoli, P.; et al. Renaming the DSCR1/Adapt78 gene family as RCAN: Regulators of calcineurin. FASEB J. 2007, 21, 3023–3028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdul, H.M.; Furman, J.L.; Sama, M.A.; Mathis, D.M.; Norris, C.M. NFATs and Alzheimer’s Disease. Mol. Cell Pharm. 2010, 2, 7–14. [Google Scholar]
- Ermak, G.; Morgan, T.E.; Davies, K.J. Chronic overexpression of the calcineurin inhibitory gene DSCR1 (Adapt78) is associated with Alzheimer’s disease. J. Biol. Chem. 2001, 276, 38787–38794. [Google Scholar] [CrossRef] [Green Version]
- Arron, J.R.; Winslow, M.M.; Polleri, A.; Chang, C.P.; Wu, H.; Gao, X.; Neilson, J.R.; Chen, L.; Heit, J.J.; Kim, S.K.; et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 2006, 441, 595–600. [Google Scholar] [CrossRef]
- Di Domenico, F.; Tramutola, A.; Foppoli, C.; Head, E.; Perluigi, M.; Butterfield, A. mTOR in Down syndrome: Role in Aβ and tau neuropathology and transition to Alzheimer disease-like dementia. Free Radic. Biol. Med. 2018, 114, 94–101. [Google Scholar] [CrossRef]
- Cho, H.J.; Jin, S.M.; Youn, H.D.; Huh, K.; Mook-Jung, I. Disrupted intracellular calcium regulates BACE1 gene expression via nuclear factor of activated T cells 1 (NFAT 1) signaling. Aging Cell. 2008, 7, 137–147. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, C. PI3-kinase/Akt/mTOR signaling: Impaired on/off switches in aging, cognitive decline and Alzheimer’s disease. Exp. Gerontol. 2013, 48, 647–653. [Google Scholar] [CrossRef]
- Holtzman, D.M.; Morris, J.C.; Goate, A.M. Alzheimer’s disease: The challenge of the second century. Sci. Transl. Med. 2011, 3, 77sr1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, J.J.; Braak, H.; An, W.L.; Winblad, B.; Cowburn, R.F.; Iqbal, K.; Grundke-Iqbal, I. Up-regulation ofmitogen-activated protein kinases ERK1/2 andMEK1/2 is associated with the progression of neurofibrillary degeneration in Alzheimer’s disease. Mol. Brain Res. 2002, 109, 45–55. [Google Scholar] [CrossRef]
- Swatton, J.E.; Sellers, L.A.; Faull, R.L.M.; Holland, A.; Iritani, S.; Bahn, S. Increased MAP kinase activity in Alzheimer’s and Down syndrome but not in schizophrenia human brain. Eur. J. Neurosci. 2004, 19, 2711–2719. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Hoeffer, C.A.; Capetillo-Zarate, E.; Yu, F.; Wong, H.; Lin, M.T.; Tampellini, D.; Klann, E.; Blitzer, R.D.; Gouras, G.K. Dysregulation of the mTOR pathway mediates impairment of synaptic plasticity in a mouse model of Alzheimer’s disease. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Iyer, A.M.; van Scheppingen, J.; Milenkovic, I.; Anink, J.J.; Adle- Biassette, H.; Kovacs, G.G.; Aronica, E. mTOR Hyperactivation in down syndrome hippocampus appears early during development. J. Neuropathol. Exp. Neurol. 2014, 73, 671–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Troca-Marin, J.A.; Casanas, J.J.; Benito, I.; Montesinos, M.L. The Akt-mTOR pathway in Down’s syndrome: The potential use of rapamycin/rapalogs for treating cognitive deficits. CNS Neurol. Disord.-Drug Targets 2014, 13, 34–40. [Google Scholar] [CrossRef]
- Griffin, R.J.; Moloney, A.; Kelliher, M.; Johnston, J.A.; Ravid, R.; Dockery, P.; O’Connor, R.; O’Neill, C. Activation of Akt/PKB, increased phosphorylation of Akt substrates and loss and altered distribution of Akt and PTEN are features of Alzheimer’s disease pathology. J. Neurochem. 2005, 93, 105–117. [Google Scholar] [CrossRef]
- Li, X.; Alafuzoff, I.; Soininen, H.; Winblad, B.; Pei, J.J. Levels of mTOR and its downstream targets 4E-BP1, eEF2, and eEF2 kinase in relationships with tau in Alzheimer’s disease brain. FEBS J. 2005, 272, 4211–4220. [Google Scholar] [CrossRef]
- Peil, J.J.; Hugon, J. mTOR-dependent signalling in Alzheimer’s disease. J. Cell Mol. Med. 2008, 12, 2525–2532. [Google Scholar]
- Sun, Y.X.; Ji, X.; Mao, X.; Xie, L.; Jia, J.; Galvan, V.; Greenberg, D.A.; Jin, K. Differential activation of mTOR complex 1 signaling in human brain with mild to severe Alzheimer’s disease. J. Alzheimers Dis. 2014, 38, 437–444. [Google Scholar] [CrossRef]
- Martin, D.; Salinas, M.; Lopez-Valdaliso, R.; Serrano, E.; Recuero, M.; Cuadrado, A. Effect of the Alzheimer amyloid fragment Abeta(25-35) on Akt/PKB kinase and survival of PC12 cells. J. Neurochem. 2001, 78, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.L.; Norton, D.D.; Wang, X.T.; Kusiak, J.W. A beta 17-42 in Alzheimer’s disease activates JNK and caspase-8 leading to neuronal apoptosis. Brain 2002, 125, 2036–2043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perluigi, M.; Pupo, G.; Tramutola, A.; Cini, C.; Coccia, R.; Barone, E.; Head, E.; Butterfield, D.A.; Di Domenico, F. Neuropathological role of PI3K/Akt/mTOR axis in Down syndrome brain. Biochim. Biophys. Acta 2014, 1842, 1144–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Z.; Zhao, B.; Li, K.; Zhang, L.; Li, C.; Quazi, S.H.; Tan, Y. Mammalian target of rapamycin: A valid therapeutic target through the autophagy pathway for Alzheimer’s disease? J. Neurosci. Res. 2012, 90, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Chen, G.; He, W.; Xiao, M.; Yan, L.J. Activation of mTOR: A culprit of Alzheimer’s disease? Neuropsychiatr. Dis. Treat. 2015, 11, 1015–1030. [Google Scholar] [CrossRef] [Green Version]
- Caccamo, A.; Majumder, S.; Richardson, A.; Strong, R.; Oddo, S. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and Tau: Effects on cognitive impairments. J. Biol. Chem. 2010, 285, 13107–13120. [Google Scholar] [CrossRef] [Green Version]
- Di Domenico, F.; Head, E.; Butterfield, D.A.; Perluigi, M. Oxidative Stress and Proteostasis Network: Culprit and Casualty of Alzheimer’s-Like Neurodegeneration. Adv. Geriatr. 2014, 14. [Google Scholar] [CrossRef] [Green Version]
- Funderburk, S.F.; Wang, Q.J.; Yue, Z. The Beclin 1–VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell. Biol. 2010, 20, 355–362. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef] [Green Version]
- Nixon, R.A. Autophagy, amyloidogenesis and Alzheimer disease. J. Cell Sci. 2007, 120, 4081–4091. [Google Scholar] [CrossRef] [Green Version]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A.; Ojala, J.; Haapasalo, A.; Soininen, H.; Hiltunen, M. Impaired autophagy and APP processing in Alzheimer’s disease: The potential role of Beclin 1 interactome. Prog. Neurobiol. 2013, 106–107, 33–54. [Google Scholar] [CrossRef] [PubMed]
- Caccamo, A.; Maldonado, M.A.; Majumder, S.; Medina, D.X.; Holbein, W.; Magri, A.; Oddo, S. Naturally secreted amyloid-beta increases mammalian target of rapamycin (mTOR) activity via a PRAS40-mediated mechanism. J. Biol. Chem. 2011, 286, 8924–8932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majumder, S.; Richardson, A.; Strong, R.; Oddo, S. Inducing autophagy by rapamycin before, but not after, the formation of plaques and tangles ameliorates cognitive deficits. PLoS ONE 2011, 6, e25416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.Q.; De Felice, F.G.; Fernandez, S.; Chen, H.; Lambert, M.P.; Quon, M.J.; Krafft, G.A.; Klein, W.L. Amyloid beta oligomers induce impairment of neuronal insulin receptors. FASEB J. 2008, 22, 246–260. [Google Scholar] [CrossRef] [Green Version]
- Pláteník, J.; Fišar, Z.; Buchal, R.; Jirák, R.; Kitzlerová, E.; Zvěřová, M.; Raboch, J. GSK3β, CREB, and BDNF in peripheral blood of patients with Alzheimer’s disease and depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 2014, 50, 83–93. [Google Scholar] [CrossRef]
- Walton, M.R.; Dragunow, M. Is CREB a key to neuronal survival? Trends Neurosci. 2000, 23, 48–53. [Google Scholar] [CrossRef]
- Grimes, C.A.; Jope, R.S. CREB DNA binding activity is inhibited by glycogen synthase kinase-3β and facilitated by lithium. J. Neurochem. 2001, 78, 1219–1232. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.M.; Roach, P.J.; Fiol, C.J. Use of a synthetic peptide as a selective substrate for glycogen synthase kinase 3. Anal. Biochem. 1994, 220, 397–402. [Google Scholar] [CrossRef]
- DaRocha-Souto, B.; Coma, M.; Perez-Nievas, B.; Scotton, T.; Siao, M.; Sánchez-Ferrer, P.; Hashimoto, T.; Fan, Z.; Hudry, E.; Barroeta, I. Activation of glycogen synthase kinase-3 beta mediates β-amyloid induced neuritic damage in Alzheimer’s disease. Neurobiol. Dis. 2012, 45, 425–437. [Google Scholar] [CrossRef] [Green Version]
- Tong, T.; Thornton, P.L.; Balazs, R.; Cotman, C.W. β-amyloid-(1–42) impairs activity-dependent cAMPresponse element-binding protein signaling in neurons at concentrations in which cell survival is not compromised. J. Biol. Chem. 2001, 276, 17301–17306. [Google Scholar] [CrossRef] [Green Version]
- Vitolo, O.V.; Sant’Angelo, A.; Costanzo, V.; Battaglia, F.; Arancio, O.; Shelanski, M. Amyloid β-peptide inhibition of the PKA/CREB pathway and long-term potentiation: Reversibility by drugs that enhance cAMP signaling. Proc. Natl. Acad. Sci. USA 2002, 99, 13217–13221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowburn, R.F.; O’Neill, C.; Ravid, R.; Alafuzoff, I.; Winblad, B.; Fowler, C.J. Adenylyl cyclase activity in postmortem human brain: Evidence of altered G protein mediation in Alzheimer’s disease. J. Neurochem. 1992, 58, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Schnecko, A.; Witte, K.; Bohl, J.; Ohm, T.; Lemmer, B. Adenylyl cyclase activity in Alzheimer’s disease brain: Stimulatory and inhibitory signal transduction pathways are differently affected. Brain Res. 1994, 644, 291–296. [Google Scholar] [CrossRef]
- Gong, B.; Cao, Z.; Zheng, P.; Vitolo, O.V.; Liu, S.; Staniszewski, A.; Moolman, D.; Zhang, H.; Shelanski, M.; Arancio, O. Ubiquitin hydrolase Uch-L1 rescues β-amyloid-induced decreases in synaptic function and contextual memory. Cell 2006, 126, 775–788. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.L.; Pozueta, J.; Gong, B.; Arancio, O.; Shelanski, M. Reversal of long-term dendritic spine alterations in Alzheimer disease models. Proc. Natl. Acad. Sci. USA 2009, 106, 16877–16882. [Google Scholar] [CrossRef] [Green Version]
- Rosa, E.; Fahnestock, M. CREB expression mediates amyloid β-induced basal BDNF downregulation. Neurobiol. Aging 2015, 36, 2406–2413. [Google Scholar] [CrossRef]
- Connor, B.; Young, D.; Yan, Q.; Faull, R.; Synek, B.; Dragunow, M. Brain-derived neurotrophic factor is reduced in Alzheimer’s disease. Mol. Brain Res. 1997, 49, 71–81. [Google Scholar] [CrossRef]
- Hock, C.; Heese, K.; Hulette, C.; Rosenberg, C.; Otten, U. Region-specific neurotrophin imbalances in Alzheimer disease: Decreased levels of brain-derived neurotrophic factor and increased levels of nerve growth factor in hippocampus and cortical areas. Arch. Neurol. 2000, 57, 846–851. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-G.; Zhu, X.; Casadesus, G.; Pallas, M.; Camins, A.; O’Neill, M.J.; Nakanishi, S.; Perry, G.; Smith, M.A. The effect of mGluR2 activation on signal transduction pathways and neuronal cell survival. Brain Res. 2009, 1249, 244–250. [Google Scholar] [CrossRef] [Green Version]
- Cowansage, K.K.; LeDoux, J.E.; Monfils, M.-H. Brain-derived neurotrophic factor: A dynamic gatekeeper of neural plasticity. Curr. Mol. Pharmacol. 2010, 3, 12–29. [Google Scholar] [CrossRef]
- Fahnestock, M. Brain-derived neurotrophic factor: The link between amyloid-β and memory loss. Future Neurol. 2011, 6, 627–639. [Google Scholar] [CrossRef]
- Peng, S.; Wuu, J.; Mufson, E.J.; Fahnestock, M. Precursor form of brain-derived neurotrophic factor and mature brain-derived neurotrophic factor are decreased in the pre-clinical stages of Alzheimer’s disease. J. Neurochem. 2005, 93, 1412–1421. [Google Scholar] [CrossRef] [PubMed]
- Garzon, D.J.; Fahnestock, M. Oligomeric amyloid decreases basal levels of brain-derived neurotrophic factor (BDNF) mRNA via specific downregulation of BDNF transcripts IV and V in differentiated human neuroblastoma cells. J. Neurosci. 2007, 27, 2628–2635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaves, J.C.S.; Machado, F.T.; Almeida, M.F.; Bacovsky, T.B.; Ferrari, M.F.R. microRNAs expression correlates with levels of APP, DYRK1A, hyperphosphorylated Tau and BDNF in the hippocampus of a mouse model for Down syndrome during ageing. Neurosci. Lett. 2020, 714, 134541. [Google Scholar] [CrossRef]
- Andrade-Talavera, Y.; Benito, I.; Casañas, J.J.; Rodríguez-Moreno, A.; Montesinos, M.L. Rapamycin restores BDNF-LTP and the persistence of long-term memory in a model of Down’s syndrome. Neurobiol. Dis. 2015, 82, 16–525. [Google Scholar] [CrossRef] [PubMed]
- Budni, J.; Bellettini-Santos, T.; Mina, F.; Garcez, M.L.; Zugno, A.I. The involvement of BDNF, NGF and GDNF in aging and Alzheimer’s disease. Aging Dis. 2015, 6, 331. [Google Scholar] [CrossRef] [Green Version]
- Bruno, M.A.; Mufson, E.J.; Wuu, J.; Cuello, A.C. Increased matrix metalloproteinase 9 activity in mild cognitive impairment. J. Neuropathol. Exp. Neurol. 2009, 68, 1309–1318. [Google Scholar] [CrossRef]
- Iulita, M.F.; Do Carmo, S.; Ower, A.K.; Fortress, A.M.; Flores Aguilar, L.; Hanna, M.; Wisniewski, T.; Granholm, A.C.; Buhusi, M.; Busciglio, J.; et al. Nerve growth factor metabolic dysfunction in Down’s syndrome brains. Brain 2014, 137, 860–872. [Google Scholar] [CrossRef] [Green Version]
- Matrone, C.; Ciotti, M.T.; Mercanti, D.; Marolda, R.; Calissano, P. NGF and BDNF signaling control amyloidogenic route and Abeta production in hippocampal neurons. Proc. Natl. Acad. Sci. USA 2008, 105, 13139–13144. [Google Scholar] [CrossRef] [Green Version]
- España, J.; Valero, J.; Miñano-Molina, A.J.; Masgrau, R.; Martín, E.; Guardia-Laguarta, C.; Lleó, A.; Giménez-Llort, L.; Rodríguez-Alvarez, J.; Saura, C.A. Amyloid disrupts activity-dependent gene transcription required for memory through the CREB coactivator CRTC1. J. Neurosci. 2010, 30, 9402–9410. [Google Scholar]
- Pugazhenthi, S.; Wang, M.; Pham, S.; Sze, C.-I.; Eckman, C.B. Downregulation of CREB expression in Alzheimer’s brain and in Aβ-treated rat hippocampal neurons. Mol. Neurodegener. 2011, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Allen, S.J.; Watson, J.J.; Dawbarn, D. The neurotrophins and their role in Alzheimer’s disease. Curr. Neuropharmacol. 2011, 9, 559–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, B. BDNF and activity-dependent synaptic modulation. Learn. Mem. 2003, 10, 86–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dineley, K.T.; Westerman, M.; Bui, D.; Bell, K.; Ashe, K.H.; Sweatt, J.D. β-Amyloid activates the mitogenactivated protein kinase cascade via hippocampal α7 nicotinic acetylcholine receptors: In vitro and in vivo mechanisms related to Alzheimer’s disease. J. Neurosci. 2001, 21, 4125–4133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dineley, K.T.; Xia, X.; Bui, D.; Sweatt, J.D.; Zheng, H. Accelerated plaque accumulation, associative learning deficits, and up-regulation of α7 nicotinic receptor protein in transgenic mice co-expressing mutant human presenilin 1 and amyloid precursor proteins. J. Biol. Chem. 2002, 277, 22768–22780. [Google Scholar] [CrossRef] [Green Version]
- Gong, B.; Vitolo, O.V.; Trinchese, F.; Liu, S.; Shelanski, M.; Arancio, O. Persistent improvement in synaptic and cognitive functions in an Alzheimer mouse model after rolipram treatment. J. Clin. Investig. 2004, 114, 1624–1634. [Google Scholar] [CrossRef] [Green Version]
- Bancher, C.; Grundke-Iqbal, I.; Iqbal, K.; Fried, V.A.; Smith, H.T.; Wisniewski, H.M. Abnormal phosphorylation of tau precedes ubiquitination in neurofibrillary pathology of Alzheimer disease. Brain Res. 1991, 539, 11–18. [Google Scholar] [CrossRef]
- Grundke-Iqbal, I.; Iqbal, K.; Tung, Y.C.; Quinlan, M.; Wisniewski, H.M.; Binder, L.I. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. USA 1986, 83, 4913–4917. [Google Scholar] [CrossRef] [Green Version]
- Hanger, D.P.; Brion, J.P.; Gallo, J.M.; Cairns, N.J.; Luthert, P.J.; Anderton, B.H. Tau in Alzheimer’s disease and Down’s syndrome is insoluble and abnormally phosphorylated. Biochem. J. 1991, 275, 99–104. [Google Scholar] [CrossRef] [Green Version]
- Cardenas, A.M.; Ardiles, A.O.; Barraza, N.; Baez-Matus, X.; Caviedes, P. Role of tau protein in neuronal damage in Alzheimer’s disease and Down syndrome. Arch. Med. Res. 2012, 43, 645–654. [Google Scholar] [CrossRef]
- Ksiezak-Reding, H.; Liu, W.-K.; Yen, S.-H. Phosphate analysis and dephosphorylation of modified tau associated with paired helical filaments. Brain Res. 1992, 597, 209–219. [Google Scholar] [CrossRef]
- Kimura, T.; Yamashita, S.; Fukuda, T.; Park, J.M.; Murayama, M.; Mizoroki, T.; Yoshiike, Y.; Sahara, N.; Takashima, A. Hyperphosphorylated tau in parahippocampal cortex impairs place learning in aged mice expressing wild-type human tau. EMBO J. 2007, 26, 5143–5152. [Google Scholar] [CrossRef] [Green Version]
- Head, E.; Lott, I.T.; Hof, P.R.; Bouras, C.; Su, J.H.; Kim, R.; Haier, R.; Cotman, C.W. Parallel compensatory and pathological events associated with tau pathology in middle aged individuals with Down syndrome. J. Neuropathol. Exp. Neurol. 2003, 62, 917–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Lian, Z.; Wegiel, J.; Hwang, Y.W.; Iqbal, K.; Grundke-Iqbal, I.; Ramakrishna, N.; Gong, C.-X. Overexpression of Dyrk1A contributes to neurofibrillary degeneration in Down syndrome. FASEB J. 2008, 22, 3224–3233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukkur, E.A.; Shimohata, A.; Akagi, T.; Yu, W.; Yamaguchi, M.; Murayama, M.; Chui, D.; Takeuchi, T.; Amano, K.; Subramhanya, K.H.; et al. Mitochondrial dysfunction and tau hyperphosphorylation in Ts1Cje, a mouse model for Down syndrome. Hum. Mol. Genet. 2006, 15, 2752–2762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woods, Y.L.; Cohen, P.; Becker, W.; Jakes, R.; Goedert, M.; Wang, X.; Proud, C.G. The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2Bepsilon at Ser539 and the microtubule-associated protein tau at Thr212: Potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem. J. 2001, 355, 609–615. [Google Scholar] [CrossRef]
- Azorsa, D.O.; Robeson, R.H.; Frost, D.; Meec Hoovet, B.; Brautigam, G.R.; Dickey, C.; Beaudry, C.; Basu, G.D.; Holz, D.R.; Hernandez, J.A.; et al. High-content siRNA screening of the kinome identifies kinases involved in Alzheimer’s disease-related tau hyperphosphorylation. BMC Genom. 2010, 11, 25. [Google Scholar] [CrossRef] [Green Version]
- Frost, D.; Meechoovet, B.; Wang, T.; Gately, S.; Giorgetti, M.; Shcherbakova, I.; Dunckley, T. beta-carboline compounds, including harmine, inhibit DYRK1A and tau phosphorylation at multiple Alzheimer’s disease-related sites. PLoS ONE 2011, 6, e19264. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.; Jin, N.; Gu, J.; Shi, J.; Zhou, J.; Gong, C.X.; Iqbal, K.; Grundke-Iqbal, I.; Liu, F. Dual-specificity tyrosine phosphorylation-regulated kinase 1A (Dyrk1A) modulates serine/arginine-rich protein 55 (SRp55)-promoted Tau exon 10 inclusion. J. Biol. Chem. 2012, 287, 30497–30506. [Google Scholar] [CrossRef] [Green Version]
- Mancini, M.; Toker, A. NFAT proteins: Emerging roles in cancer progression. Nat. Rev. Cancer 2009, 9, 810–820. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Peisley, A.; Graef, I.A.; Crabtree, G.R. NFAT signaling and the invention of vertebrates. Trends Cell Biol. 2007, 17, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G.; Jakes, R.; Rutherford, D.; Crowther, R.A. Multiple isoforms of humanmicrotubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 1989, 3, 519–526. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, T.; Zhou, C.; Chohan, M.O.; Gu, X.; Wegiel, J.; Zhou, J.; Hwang, Y.-W.; Iqbal, K.; Drundke-Iqbal, I.; et al. Increased dosage of Dyrk1A alters alternative splicing factor (ASF)-regulated alternative splicing of tau in Down syndrome. J. Biol. Chem. 2008, 283, 28660–28669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wegiel, J.; Kaczmarski, W.; Barua, M.; Kuchna, I.; Nowicki, K.; Wang, K.C.; Wegiel, J.; Ma, S.Y.; Franckowiak, J.; Mazur-Kolecka, B.; et al. Link between DYRK1A overexpression and several-fold enhancement of neurofibrillary degeneration with 3-repeat tau protein in Down syndrome. J. Neuropathol. Exp. Neurol. 2011, 70, 36–50. [Google Scholar] [CrossRef] [Green Version]
- Ermak, G.; Pritchard, M.A.; Dronjak, S.; Niu, B.; Davies, K.J. Do RCAN1 proteins link chronic stress with neurodegeneration? FASEB J. 2011, 25, 3306–3311. [Google Scholar] [CrossRef] [Green Version]
- Poppek, D.; Keck, S.; Ermak, G.; Jung, T.; Stolzing, A.; Ullrich, O.; Davies, K.J.A.; Grune, T. Phosphorylation inhibits turnover of the tau protein by the proteasome: Influence of RCAN1 and oxidative stress. Biochem. J. 2006, 400, 511–520. [Google Scholar] [CrossRef] [Green Version]
- Lloret, A.; Badia, M.C.; Giraldo, E.; Ermak, G.; Alonso, M.-D.; Pallardó, F.V.; Davies, K.J.A.; Viña, J. Amyloid-beta toxicity and tau hyperphosphorylation are linked via RCAN1 in Alzheimer’s Disease. J. Alzheimers Dis. 2011, 27, 701–709. [Google Scholar] [CrossRef] [Green Version]
- Ermak, G.; Harris, C.D.; Battocchio, D.; Davies, K.J. RCAN1 (DSCR1 or Adapt78) stimulates expression of GSK-3beta. FEBS J. 2006, 273, 2100–2109. [Google Scholar] [CrossRef]
- Reynolds, C.H.; Betts, J.C.; Blackstock, W.P.; Nebreda, A.R.; Anderton, B.H. Phosphorylation sites on tau identified by nanoelectrospray mass spectrometry: Differences in vitro between the mitogen-activated protein kinases ERK2, c-Jun Nterminal kinase and P38, and glycogen synthase kinase-3b. J. Neurochem. 2000, 74, 1587–1595. [Google Scholar] [CrossRef]
- Takashima, A.; Honda, T.; Yasutake, K.; Michel, G.; Murayama, O.; Murayama, M.; Ishiguro, K.; Yamaguchi, H. Activation of tau protein kinaseI/glycogen synthase kinase-3beta by amyloid beta peptide enhances phosphorylation of tau in hippocampal neurons. Neurosci. Res. 1998, 31, 317–323. [Google Scholar] [CrossRef]
- Qing, H.; He, G.; Ly, P.T.; Fox, C.J.; Staufenbiel, M.; Cai, F.; Zhang, Z.; Wei, S.; Sun, X.; Chen, C.-H.; et al. Valproic acid inhibits Abeta production, neuritic plaque formation, and behavioral deficits in Alzheimer’s disease mouse models. J. Exp. Med. 2008, 205, 2781–2789. [Google Scholar] [CrossRef] [PubMed]
- Pollonini, G.; Gao, V.; Rabe, A.; Palminiello, S.; Albertini, G.; Alberini, C.M. Abnormal expression of synaptic proteins and neurotrophin-3 in the Down syndrome mouse model Ts65Dn. Neuroscience 2008, 156, 99–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz, J.C.; Tseng, H.-C.; Goldman, J.A.; Shih, H.; Tsai, L.-H. Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron 2003, 40, 471–483. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Hawkes, C.; Qureshi, H.Y.; Kar, S.; Paudel, H.K. Cyclindependent protein kinase 5 primes microtubule-associated protein tau site-specifically for glycogen synthase kinase 3b. Biochemistry 2006, 45, 3134–3145. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Liu, F.; Iqbal, K.; Grundke-Iqbal, I.; Wegiel, J.; Gong, C.X. Decrease of protein phosphatase 2A and its association with accumulation and hyperphosphorylation of tau in Down syndrome. J. Alzheimers Dis. 2008, 13, 295–302. [Google Scholar] [CrossRef] [Green Version]
- Sontag, J.M.; Sontag, E. Protein phosphatase 2A dysfunction in Alzheimer’s disease. Front. Mol. Neurosci. 2014, 7, 16. [Google Scholar] [CrossRef]
- Chohan, M.O.; Khatoon, S.; Iqbal, I.G.; Iqbal, K. Involvement of I2PP2A in the abnormal hyperphosphorylation of tau and its reversal by Memantine. FEBS Lett. 2006, 580, 3973–3979. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Grundke-Iqbal, I.; Iqbal, K.; Gong, C.X. Contributions of protein phosphatases PP1, PP2A, PP2B and PP5 to the regulation of tau phosphorylation. Eur. J. Neurosci. 2005, 22, 1942–1950. [Google Scholar] [CrossRef]
- Gong, C.X.; Singh, T.J.; Grundkeiqbal, I.; Iqbal, K. Phosphoprotein phosphatase-Activities in alzheimer-disease brain. J. Neurochem. 1993, 61, 921–927. [Google Scholar] [CrossRef]
- Gong, C.X.; Shaikh, S.; Wang, J.Z.; Zaidi, T.; Grundkeiqbal, I.; Iqbal, K. Phosphataseactivity toward abnormally phosphorylated-tau - decrease in alzheimer-disease brain. J. Neurochem. 1995, 65, 732–738. [Google Scholar] [CrossRef]
- Sun, L.; Liu, S.Y.; Zhou, X.W.; Wang, X.C.; Liu, R.; Wang, Q.; Wang, J.C. Inhibition of protein phosphatase 2A- and protein phosphatase 1-induced tau hyperphosphorylation and impairment of spatial memory retention in rats. Neuroscience 2003, 118, 1175–1182. [Google Scholar] [CrossRef]
- Julien, C.; Tremblay, C.; Emond, V.; Lebbadi, M.; Salem, N.; Bennett, D.A.; Calon, F. Sirtuin 1 reduction parallels the accumulation of tau in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2009, 68, 48–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tramutola, A.; Pupo, G.; Di Domenico, F.; Barone, E.; Arena, A.; Lanzillotta, C.; Brokeaart, D.; Blarzino, C.; Head, E.; Butterfiels, D.A.; et al. Activation of p53 in Down Syndrome and in the Ts65Dn Mouse Brain is Associated with a Pro-Apoptotic Phenotype. J. Alzheimers Dis. 2016, 52, 359–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Williams, J.G.; Schug, T.T.; Li, X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J. Biol. Chem. 2010, 285, 13223–13232. [Google Scholar] [CrossRef] [Green Version]
- Counts, S.E.; Mufson, E.J. The role of nerve growth factor receptors in cholinergic basal forebrain degeneration in prodromal Alzheimer disease. J. Neuropathol. Exp. Neurol. 2005, 64, 263–272. [Google Scholar] [CrossRef] [Green Version]
- Forman, M.S.; Mufson, E.J.; Leurgans, S.; Pratico, D.; Joyce, S.; Leight, S.; Lee, M.-Y.; Trojanowski, Q. Cortical biochemistry in MCI and Alzheimer disease: Lack of correlation with clinical diagnosis. Neurology 2007, 68, 757–763. [Google Scholar] [CrossRef]
- Ginsberg, S.D.; Che, S.; Counts, S.E.; Mufson, E.J. Single cell gene expression profiling in Alzheimer’s disease. NeuroRx 2006, 3, 302–318. [Google Scholar] [CrossRef]
- Ginsberg, S.D.; Che, S.; Counts, S.E.; Mufson, E.J. Shift in the ratio of three-repeat tau and four-repeat tau mRNAs in individual cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer’s disease. J. Neurochem. 2006, 96, 1401–1408. [Google Scholar] [CrossRef]
- Mesulam, M.M. The cholinergic lesion of Alzheimerˇıs disease: Pivotal factor or side show. Learn. Mem. 2004, 11, 43–49. [Google Scholar] [CrossRef] [Green Version]
- Geula, C.; Nagykery, N.; Nicholas, A.; Wu, C.K. Cholinergic neuronal and axonal abnormalities are present early in aging and in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2008, 67, 309–318. [Google Scholar] [CrossRef] [Green Version]
- Sassin, I.; Schultz, C.; Thal, D.R.; Rüb, U.; Arai, K.; Braak, E. Evolution of Alzheimer’s disease-related cytoskeletal changes in the basal nucleus of Meynert. Acta Neuropathol. 2000, 100, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Belarbi, K.; Schindowski, K.; Burnouf, S.; Caillierez, R.; Grosjean, M.E.; Demeyer, D.; Hamdane, M.; Sergeant, N.; Blum, D.; Buée, L. Early Tau pathology involving the septo-hippocampal pathway in a Tau transgenic model: Relevance to Alzheimer’s disease. Curr. Alzheimer Res. 2009, 6, 152–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellström-Lindahl, E.; Moore, H.; Nordberg, A. Increased levels of tau protein in SH-SY5Y cells after treatment with cholinesterase inhibitors and nicotinic agonists. J. Neurochem. 2000, 74, 777–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckingham, S.D.; Jones, A.K.; Brown, L.A.; Sattelle, D.B. Nicotinic acetylcholine receptor signaling: Roles in Alzheimer’s disease and amyloid neuroprotection. Pharm. Rev. 2009, 61, 39–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bencherif, M.; Lippiello, P.M. Alpha7 neuronal nicotinic receptors: The missing link to understanding Alzheimer’s etiopathology? Med. Hypotheses 2010, 74, 281–285. [Google Scholar] [CrossRef]
- Wang, H.Y.; Li, W.; Benedetti, N.J.; Lee, D.H. α7 nicotinic acetylcholine receptors mediate β-amyloid peptide-induced tau protein phosphorylation. J. Biol. Chem. 2003, 278, 31547–31553. [Google Scholar] [CrossRef] [Green Version]
- Caccamo, A.; Oddo, S.; Billings, L.M.; Green, K.N.; Martinez-Coria, H.; Fisher, A.; LaFerla, F.M. M1 receptors play a central role in modulating AD-like pathology in transgenic mice. Neuron 2006, 49, 671–682. [Google Scholar] [CrossRef] [Green Version]
- Ferreira-Vieira, T.H.; Guimaraes, M.; Silva, F.R.; Ribeiro, F.M. Alzheimer’s disease: Targeting the cholinergic system. Curr. Neuropharmacol. 2016, 14, 101–115. [Google Scholar] [CrossRef] [Green Version]
- Goekoop, R.; Scheltens, P.; Barkhoh, F.; Rombouts, S.A.R.B. Cholinergic challenge in Alzheimer patients and mild cognitive impairment differentially affects hippocampal activation – a pharmacological fMRI study. Brain 2006, 129, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Bierer, L.M.; Haroutunian, V.; Gabriel, S.; Knott, P.J.; Carlin, L.S.; Purohit, D.P.; Perl, D.P.; Schmeidler, J.; Kanof, P.; Davis, K.L. Neurochemical correlates of dementia severity in Alzheimer’s disease: Relative importance of the cholinergic deficits. J. Neurochem. 1995, 64, 749–760. [Google Scholar] [CrossRef]
- Gsell, W.; Jungkunz, G.; Riederer, P. Functional neurochemistry of Alzheimer’s disease. Curr. Pharm. Des. 2004, 10, 265–293. [Google Scholar] [CrossRef] [PubMed]
- Contestabile, A.; Ciani, E.; Contestabile, A. The place of choline acetyltransferase activity measurement in the “cholinergic hypothesis” of neurodegenerative diseases. Neurochem. Res. 2008, 33, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Mufson, E.J.; Counts, S.E.; Perez, S.E.; Ginsberg, S.D. Cholinergic system during the progression of Alzheimer’s disease: Therapeutic implications. Exp. Rev. Neurother. 2008, 8, 1703–1718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burghaus, L.; Schütz, U.; Krempel, U.; de Vos, R.A.; Jansen Steur, E.N.; Wevers, A.; Lindstrom, J.; Schröder, H. Quantitative assessment of nicotinic acetylcholine receptor proteins in the cerebral cortex of Alzheimer patients. Mol. Brain Res. 2000, 76, 385–388. [Google Scholar] [CrossRef]
- Mousavi, M.; Hellström-Lindahl, E.; Guan, Z.Z.; Shan, K.R.; Ravid, R.; Nordberg, A. Protein and mRNA levels of nicotinic receptors in brain of tobacco using controls and patients with Alzheimer’s disease. Neuroscience 2003, 122, 515–520. [Google Scholar] [CrossRef]
- Nordberg, A. Nicotinic receptor abnormalities of Alzheimer’s disease: Therapeutic implications. Biol. Psychiatry 2001, 49, 200–210. [Google Scholar] [CrossRef]
- Wilcock, G.K.; Esiri, M.M.; Bowen, D.M.; Smith, C.C. Alzheimer’s disease. Correlation of cortical choline acetyltransferase activity with the severity of dementia and histological abnormalities. J. Neurol. Sci. 1982, 57, 407–417. [Google Scholar] [CrossRef]
- Perry, E.K.; Morris, C.M.; Court, J.A.; Cheng, A.; Fairbairn, A.F.; McKeith, I.G.; Irving, D.; Brown, A.; Perry, R.H. Alteration in nicotine binding sites in Parkinson’s disease. Lewy body dementia and Alzheimer’s disease: Possible index of early neuropathology. Neuroscience 1995, 64, 385–395. [Google Scholar] [CrossRef]
- Mufson, E.J.; Counts, S.E.; Fahnestock, M.; Ginsberg, S.D. Cholinotrophic molecular substrates of mild cognitive impairment in the elderly. Curr. Alzheimer Res. 2007, 4, 340–350. [Google Scholar] [CrossRef]
- Cuello, A.C.; Bruno, M.A.; Bell, K.F. NGF-cholinergic dependency in brain aging. MCI and Alzheimer’s disease. Curr. Alzheimer Res. 2007, 4, 351–358. [Google Scholar] [CrossRef]
- Auld, D.S.; Kornecook, T.J.; Bastianetto, S.; Quirion, R. Alzheimer’s disease and the basal forebrain cholinergic system: Relations to β-amyloid peptides, cognition, and treatment strategies. Prog. Neurobiol. 2002, 68, 209–245. [Google Scholar] [CrossRef]
- Yan, Z.; Feng, J. Alzheimer’s disease: Interactions between cholinergic functions and β-amyloid. Curr. Alzheimer Res. 2004, 1, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Apelt, J.; Kumar, A.; Schliebs, R. Impairment of cholinergic neurotransmission in adult and aged transgenic Tg2576 mouse brain expressing the Swedish mutation of human beta-amyloid precursor protein. Brain Res. 2002, 953, 17–30. [Google Scholar] [CrossRef]
- Di Domenico, F.; Tramutola, A.; Butterfield, D.A. Role of 4-hydroxy-2-nonenal (HNE) in the pathogenesis of alzheimer disease and other selected age-related neurodegenerative disorders. Free Radic. Biol. Med. 2017, 111, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Rueda, N.; Martínez-Cué, C. Antioxidants in Down Syndrome: From Preclinical Studies to Clinical Trials. Antioxidants 2020, 9, 692. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Boyd-Kimball, D. Redox proteomics and amyloid β-peptide: Insights into Alzheimer disease. J. Neurochem. 2019, 151, 459–487. [Google Scholar] [CrossRef] [Green Version]
- Di Carlo, M.; Giacomazza, D.; Picone, P.; Nuzzo, D.; San Biagio, P.L. Are oxidative stress and mitochondrial dysfunction the key players in the neurodegenerative diseases? Free Radic. Res. 2012, 46, 1327–1338. [Google Scholar] [CrossRef]
- Lott, I.T. Antioxidants in Down syndrome. Biochim. Biophys. Acta 2012, 1822, 657–663. [Google Scholar] [CrossRef] [Green Version]
- Perluigi, M.; Butterfield, D.A. Oxidative Stress and Down Syndrome: A Route toward Alzheimer-Like Dementia. Curr. Gerontol. Geriatr. Res. 2012, 724904. [Google Scholar] [CrossRef] [Green Version]
- Wiseman, F.K.; Al-Janabi, T.; Hardy, J.; Karmiloff-Smith, A.; Nizetic, D.; Tybulewicz, V.L.; Fisher, E.M.; Strydom, A. A genetic cause of Alzheimer disease: Mechanistic insights from Down syndrome. Nat. Rev. Neurosci. 2015, 16, 564–574. [Google Scholar] [CrossRef] [Green Version]
- De Haan, J.B.; Cristiano, F.; Iannello, R.C.; Kola, I. Cu/Zn-superoxide dismutase and glutathione peroxidase during aging. Biochem. Mol. Biol. Int. 1995, 35, 1281–1297. [Google Scholar] [PubMed]
- Ermak, G.; Sojitra, S.; Yin, F.; Cadenas, E.; Cuervo, A.M.; Davies, K.J. Chronic expression of RCAN1-1L protein induces mitochondrial autophagy and metabolic shift from oxidative phosphorylation to glycolysis in neuronal cells. J. Biol. Chem. 2012, 287, 14088–14098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Wu, Y.; Herculano, B.; Song, W. RCAN1 overexpression exacerbates calcium overloading-induced neuronal apoptosis. PLoS ONE 2014, 9, e95471. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Jang, H.; Cho, E.J.; Youn, H.D. Down syndrome critical region 1 enhances the proteolytic cleavage of calcineurin. Exp. Mol. Med. 2009, 41, 471–477. [Google Scholar] [CrossRef]
- Celsi, F.; Svedberg, M.; Unger, C.; Cotman, C.W.; Carrì, M.T.; Ottersen, O.P.; Nordberg, A.; Torp, R. Beta-amyloid causes downregulation of calcineurin in neurons through induction of oxidative stress. Neurobiol. Dis. 2007, 26, 342–352. [Google Scholar] [CrossRef]
- Crawford, D.R.; Leahy, K.P.; Abramova, N.; Lan, L.; Wang, Y.; Davies, K.J. Hamster adapt78 mRNA is a Down syndrome critical region homologue that is inducible by oxidative stress. Arch. Biochem. Biophys. 1997, 342, 6–12. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Boyd-Kimball, D. Oxidative stress, amyloid-b peptide, and altered key molecular pathways in the pathogenesis and progression of Alzheimer’s disease. J. Alzheimers Dis. 2018, 62, 1345–1367. [Google Scholar] [CrossRef] [Green Version]
- Olney, J.W. Excitotoxicity: An overview. Can. Dis. Wkly. Rep. 1990, 16, S47–S57. [Google Scholar]
- Butterfield, D.A.; Pocernich, C.B. The glutamatergic system and Alzheimer’s disease: Therapeutic implications. CNS Drugs 2003, 17, 641–652. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Lauderback, C.M. Lipid peroxidation and protein oxidation in Alzheimer’s disease brain: Potential causes and consequences involving amyloid b-peptide-associated free radical oxidative stress. Free Radic. Biol. Med. 2002, 32, 1050–1060. [Google Scholar] [CrossRef]
- Di Domenico, F.; Barone, E.; Perluigi, M.; Butterfield, D.A. The triangle of death in Alzheimer’s disease brain: The aberrant cross-talk among energy metabolism, mammalian target of rapamycin signaling, and protein homeostasis revealed by redox proteomics. Antioxid. Redox Signal 2017, 26, 364–387. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, F.; Tramutola, A.; Barone, E.; Lanzillotta, C.; Defever, O.; Arena, A.; Zuliani, I.; Foppoli, C.; Iavarone, F.; Vincenzoni, F.; et al. Restoration of aberrant mTOR signaling by intranasal rapamycing reduces oxidative damage: Focus on HNE-modified proteins in a mouse model of Down syndrome. Redox Biol. 2019, 23, 101162. [Google Scholar] [CrossRef] [PubMed]
- D i Domenico, F.; Coccia, R.; Cocciolo, A.; Murphy, M.P.; Cenini, G.; Head, E.; Butterfield, D.A.; Giorgi, A.; Schinina, M.E.; Mancuso, C.; et al. Impairment of proteostasis network in Down syndrome prior to the development of Alzheimer’s disease neuropathology: Redox proteomics analysis of human brain. Biochim. Biophys. Acta 2013, 1832, 1249–1259. [Google Scholar] [CrossRef]
- Di Domenico, F.; Pupo, G.; Tramutola, A.; Giorgi, A.; Schinina, M.E.; Coccia, R.; Head, E.; Butterfield, D.A.; Perluigi, M. Redox proteomics analysis of HNE-modified proteins in Down syndrome brain: Clues for understanding the development of Alzheimer disease. Free Radic. Biol. Med. 2014, 71, 270–280. [Google Scholar] [CrossRef] [Green Version]
- Tramutola, A.; Lanzillotta, C.; Arena, A.; Barone, E.; Perluigi, M.; Di Domenico, F. Increased Mammalian Target of Rapamycin Signaling Contributes to the Accumulation of Protein Oxidative Damage in a Mouse Model of Down’s Syndrome. Neurodegener. Dis. 2016, 16, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Perluigi, M.; Di Domenico, F.; Butterfield, D.A. mTOR signaling in aging and neurodegeneration: At the crossroad between metabolism dysfunction and impairment of autophagy. Neurobiol. Dis. 2015, 84, 39–49. [Google Scholar] [CrossRef]
- Tramutola, A.; Lanzillotta, C.; Barone, E.; Arena, A.; Zuliani, I.; Mosca, L.; Blarzino, C.; Butterfield, D.A.; Perluigi, M.; Di Domenico, F. Intranasal rapamycin ameliorates Alzheimer-like cognitive decline in a mouse model of Down syndrome. Transl. Neurodegener. 2018, 7, 28. [Google Scholar] [CrossRef] [Green Version]
- Godoy, J.A.; Rios, J.A.; Zolezzi, J.M.; Braidy, N.; Inestrosa, N.C. Signaling pathway cross talk in Alzheimer’s disease. Cell Commun. Signal. 2014, 12, 23. [Google Scholar] [CrossRef] [Green Version]
- Dröge, W.; Schipper, H. Oxidative stress and aberrant signaling in aging and cognitive decline. Aging Cell 2007, 6, 361–370. [Google Scholar] [CrossRef]
- Brand, M.D. The role of mitochondria in longevity and healthspan. Longev. Healthspan 2014, 3, 7. [Google Scholar] [CrossRef] [Green Version]
- Hall, C.N.; Klein-Flügge, M.C.; Howarth, C.; Attwell, D. Oxidative phosphorylation, not glycolysis, powers presynaptic and postsynaptic mechanisms underlying brain information processing. J. Neurosci. 2012, 32, 8940–8951. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, R.H.; Khan, S.M. A “mitochondrial cascade hypothesis” for sporadic Alzheimer’s disease. Med. Hypotheses 2004, 63, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, H.M.; Swerdlow, R.H. Amyloid precursor protein processing and bioenergetics. Brain Res. Bull. 2017, 133, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Pesini, A.; Iglesias, E.; Garrido, N.; Bayona-Bafaluy, M.P.; Montoya, J.; Ruiz-Pesini, E. OXPHOS, pyrimidine nucleotides, and Alzheimer’s disease: A pharmacogenomics approach. J. Alzheimers Dis. 2014, 42, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Alzheimer’s disease is a synaptic failure. Science 2002, 298, 789–791. [Google Scholar] [CrossRef] [Green Version]
- Krapfenbauer, K.; Yoo, B.C.; Cairns, N.; Lubec, G. Differential display reveals deteriorated mRNA levels of NADH3 (complex I) in cerebellum of patients with Down syndrome. J. Neural. Transm. Suppl. 1999, 57, 211–220. [Google Scholar]
- Salemi, M.; Barone, C.; Salluzzo, M.G.; Giambirtone, M.; Scillato, F.; Rando, R.G.; Romano, C.; Morale, M.C.; Ridolfi, F.; Romano, C. A polymorphism (rs1042522) in TP53 gene is a risk factor for Down Syndrome in Sicilian mothers. J. Matern. Fetal Neonatal Med. 2017, 30, 2752–2754. [Google Scholar] [CrossRef]
- Conti, A.; Fabbrini, F.; D’Agostino, P.; Negri, R.; Greco, D.; Genesio, R.; D’Armiento, M.; Olla, C.; Paladini, D.; Zannini, M.; et al. Altered expression of mitochondrial and extracellular matrix genes in the heart of human fetuses with chromosome 21 trisomy. BMC Genom. 2007, 8, 268. [Google Scholar] [CrossRef] [Green Version]
- Piccoli, C.; Izzo, A.; Scrima, R.; Bonfiglio, F.; Manco, R.; Negri, R.; Quarato, G.; Cela, O.; Ripoli, M.; Prisco, M.; et al. Chronic pro-oxidative state and mitocondrial dysfunctions are more pronounced in fibroblasts from Down syndrome foeti with congenital heart defects. Hum. Mol. Genet. 2013, 22, 1218–1232. [Google Scholar] [CrossRef] [Green Version]
- Qiu, J.J.; Liu, Y.N.; Ren, Z.R.; Yan, J.B. Dysfunctions of mitochondria in close association with strong perturbation of long noncoding RNAs expression in Down syndrome. Int. J. Biochem. Cell. Biol. 2017, 92, 115–120. [Google Scholar] [CrossRef]
- Kim, S.H.; Vlkolinsky, R.; Cairns, N.; Lubec, G. Decreased levels of complex III core protein 1 and complex V beta chain in brains from patients with Alzheimer’s disease and Down syndrome. Cell. Mol. Life Sci. 2000, 57, 1810–1816. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Dierssen, M.; Ferreres, J.C.; Fountoulakis, M.; Lubec, G. Increased protein levels of heterogeneous nuclear ribonucleoprotein A2/B1 in fetal Down syndrome brains. J. Neural Transm. Suppl. 2001, 57, 273–280. [Google Scholar]
- Kim, S.H.; Vlkolinsky, R.; Cairns, N.; Fountoulakis, M.; Lubec, G. The reduction of NADH ubiquinone oxidoreductase 24- and 75-kDa subunits in brains of patients with Down syndrome and Alzheimer’s disease. Life Sci. 2001, 68, 2741–2750. [Google Scholar] [CrossRef]
- Valenti, D.; Tullo, A.; Caratozzolo, M.F.; Merafina, R.S.; Scartezzini, P.; Marra, E.; Vacca, R.A. Impairment of F1F0-ATPase, adenine nucleotide translocator and adenylate kinase causes mitochondrial energy deficit in human skin fibroblasts with chromosome 21 trisomy. Biochem. J. 2010, 431, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Valenti, D.; Manente, G.A.; Moro, L.; Marra, E.; Vacca, R.A. Deficit of complex I activity in human skin fibroblasts with chromosome 21 trisomy and overproduction of reactive oxygen species by mitochondria: Involvement of the cAMP/PKA signalling pathway. Biochem. J. 2011, 435, 679–688. [Google Scholar] [CrossRef] [Green Version]
- Valenti, D.; De Rasmo, D.; Signorile, A.; Rossi, L.; de Bari, L.; Scala, I.; Granese, B.; Papa, S.; Vacca, R.A. Epigallocatechin-3-gallate prevents oxidative phosphorylation deficit and promotes mitochondrial biogenesis in human cells from subjects with Down’s syndrome. Biochim. Biophys. Acta 2013, 1832, 542–552. [Google Scholar] [CrossRef] [Green Version]
- Izzo, A.; Nitti, M.; Mollo, N.; Paladino, S.; Procaccini, C.; Faicchia, D.; Cali, G.; Genesio, R.; Bonfiglio, F.; Cicatiello, R.; et al. Metformin restores the mitochondrial network and reverses mitochondrial dysfunction in Down syndrome cells. Hum. Mol. Genet. 2017, 26, 1056–1069. [Google Scholar] [CrossRef] [Green Version]
- Panagaki, T.; Randi, E.B.; Augsburger, F.; Szabo, C. Overproduction of H2S, generated by CBS, inhibits mitochondrial Complex IV and suppresses oxidative phosphorylation in Down syndrome. Proc. Natl. Acad. Sci. USA 2019, 116, 18769–18771. [Google Scholar] [CrossRef] [Green Version]
- Helguera, P.; Seiglie, J.; Rodriguez, J.; Hanna, M.; Helguera, G.; Busciglio, J. Adaptive downregulation of mitochondrial function in Down syndrome. Cell Metab. 2013, 17, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Izzo, A.; Mollo, N.; Nitti, M.; Paladino, S.; Calì, G.; Genesio, R.; Bonfiglio, F.; Cicatiello, R.; Barbato, M.; Sarnataro, V.; et al. Mitochondrial dysfunction in down syndrome: Molecular mechanisms and therapeutic targets. Mol. Med. 2018, 24, 2. [Google Scholar] [CrossRef] [Green Version]
- Izzo, A.; Manco, R.; Bonfiglio, F.; Calì, G.; De Cristofaro, T.; Patergnani, S.; Cicatiello, R.; Scrima, R.; Zannini, M.; Pinton, P.; et al. NRIP1/RIP140 siRNA-mediated attenuation counteracts mitochondrial dysfunction in Down syndrome. Hum. Mol. Genet. 2014, 23, 4406–4419. [Google Scholar] [CrossRef] [PubMed]
- Schieke, S.M.; Finkel, T. Mitochondrial signaling, TOR, and life span. Biol. Chem. 2006, 387, 1357–1361. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunnane, S.; Nugent, S.; Roy, M.; Courchesne-Loye, A.; Croteau, E.; Tremblay, S.; Castellano, A.; Pifferi, F.; Bocti, C.; Paquet, N.; et al. Brain fuel metabolism, aging, and Alzheimer’s disease. Nutrition 2011, 27, 3–20. [Google Scholar] [CrossRef] [Green Version]
- Anstey, K.J.; Cherbuin, N.; Budge, M.; Young, J. Body mass index in midlife and late-life as a risk factor for dementia: A metaanalysis of prospective studies. Obes. Rev. 2011, 12, e426–e437. [Google Scholar] [CrossRef]
- Ronnemaa, E.; Zethelius, B.; Lannfelt, L.; Kilander, L. Vascular risk factors and dementia: 40-year follow-up of a population-based cohort. Dement. Geriatr. Cogn. Disord. 2011, 31, 460–466. [Google Scholar] [CrossRef]
- Loef, M.; Walach, H. Midlife obesity and dementia: Metaanalysis and adjusted forecast of dementia prevalence in the United States and China. Obesity 2013, 21, E51–E55. [Google Scholar] [CrossRef]
- Gottesman, R.F.; Schneider, A.L.; Zhou, Y.; Coresh, J.; Green, E.; Gupta, N.; Knopman, D.S.; Mintz, A.; Rahmim, A.; Scharrett, A.R.; et al. Association between midlife vascular risk factors and estimated brain amyloid deposition. JAMA 2017, 317, 1443–1450. [Google Scholar] [CrossRef]
- Hayden, M.R. Type 2 Diabetes Mellitus Increases The Risk of Late-Onset Alzheimer’s Disease: Ultrastructural Remodeling of the Neurovascular Unit and Diabetic Gliopathy. Brain Sci. 2019, 9, 262. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, S.; Mudher, A. Alzheimer’s disease and type 2 diabetes: A critical assessment of the shared pathological traits. Front. Neurosci. 2018, 12, 383. [Google Scholar] [CrossRef] [Green Version]
- Tumminia, A.; Vinciguerra, F.; Parisi, M.; Frittitta, L. Type 2 diabetes mellitus and Alzheimer’s disease: Role of insulin signalling and therapeutic implications. Int. J. Mol. Sci. 2018, 19, 3306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, B.L. Glucose, glycolysis, and neurodegenerative diseases. J. Cell. Physiol. 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Soloman, A.; Kivipelto, M.; Wolozin, B.; Zhou, J.; Whitmer, R.A. Midlife serum cholesterol and increased risk of Alzheimer’s and vascular dementia three decades later. Dement. Geriatr. Cogn. Disord. 2009, 28, 75–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.F.; Yu, J.T.; Wang, H.F.; Tan, M.S.; Wang, C.; Tan, C.C.; Tan, L. Midlife vascular risk factors and the risk of Alzheimer’s disease: A systematic review and meta-analysis. J. Alzheimers Dis. 2014, 42, 1295–1310. [Google Scholar] [CrossRef]
- Lee, H.J.; Seo, H.I.; Cha, H.Y.; Yang, Y.J.; Kwon, S.H.; Yang., S.J. Diabetes and Alzheimer’s disease: Mechanisms and nutritional aspects. Clin. Nutr. Res. 2018, 7, 229–240. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Huang, E.; Gao, S. Type 1 diabetes mellitus and cognitive impairments: A systematic review. J. Alzheimers Dis. 2017, 57, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Desai, G.S.; Zheng, C.; Geetha, T.; Mathews, S.T.; White, B.D.; Huggins, K.W.; Zizza, C.A.; Broderick, T.L.; Babu, J.R. The pancreas-brain axis: Insight into disrupted mechanisms associating type 2 diabetes and Alzheimer’s disease. J. Alzheimers Dis. 2014, 42, 347–356. [Google Scholar] [CrossRef]
- Arnold, S.E.; Arvanitakis, Z.; Macauley-Rambach, S.L.; Koenig, A.M.; Wang, H.-Y.; Ahima, R.S.; Craft, S.; Gandy, S.; Buettner, C.; Stoeckel, L.E.; et al. Brain insulin resistance in type 2 diabetes and Alzheimer disease: Concepts and conundrums. Nat. Rev. Neurol. 2018, 14, 168–181. [Google Scholar] [CrossRef]
- Tramutola, A.; Lanzillotta, C.; Di Domenico, F.; Head, E.; Butterfield, D.A.; Perluigi, M.; Barone, E. Brain insulin resistance triggers early onset Alzheimer disease in Dowon syndrome. Neurobiol. Dis. 2020, 137, 104772. [Google Scholar] [CrossRef]
- Biessels, G.J.; Reagan, L.P. Hippocampal insulin resistance and cognitive dysfunction. Nat. Rev. Neurosci. 2015, 16, 660–671. [Google Scholar] [CrossRef]
- De Felice, F.G. Alzheimer’s disease and insulin resistance: Translating basic science into clinical applications. J. Clin. Invest. 2013, 123, 531–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tramutola, A.; Triplett, J.C.; Di Domenico, F.; Niedowicz, D.M.; Murphy, M.P.; Coccia, R.; Perluigi, M.; Butterfield, D.A. Alteration of mTOR signaling occurs early in the progression of Alzheimer disease (AD): Analysis of brain from subjects with pre-clinical AD, amnestic mild cognitive impairment and late-stage AD. J. Neurochem. 2015, 133, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, E.; Velázquez, E.; Hurtado-Carneiro, V.; Ruiz-Albusac, J.M. Insulin in the brain: Its pathophysiological implications for States related with central insulin resistance, type 2 diabetes and Alzheimer’s disease. Front. Endocrinol. 2014, 5, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson, I.A.; Chundu, K.R.; Davies-Hill, T.; Honer, W.G.; Davies, P. Decreased concentrations of GLUT1 and GLUT3 glucose transporters in the brains of patients with Alzheimer’s disease. Ann. Neurol. 1994, 35, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Bergau, N.; Maul, S.; Rujescu, D.; Simm, A. Reduction of glycolysis intermediate concentrations in the cerebrospinal fluid of Alzheimer’s disease patients. Front. Neurosci. 2019, 13, 871. [Google Scholar] [CrossRef] [PubMed]
- Croteau, E.; Castellano, C.A.; Fortier, M.; Bocti, C.; Fulop, T.; Paquet, N.; Cunnane, S.C. A cross-sectional comparison of brain glucose and ketone metabolism in cognitively healthy older adults, mild cognitive impairment and early Alzheimer’s disease. Exp. Gerontol. 2018, 107, 18–26. [Google Scholar] [CrossRef]
- Theurey, P.; Connolly, N.M.C.; Fortunati, I.; Basso, E.; Lauwen, S.; Ferrante, C.; Pinho, C.M.; Joselin, A.; Gioran, A.; Bano, D.; et al. Systems biology identifies preserved integrity but impaired metabolism of mitochondria due to a glycolytic defect in Alzheimer’s disease neurons. Aging Cell 2019, 18, e12924. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, S.; Meguro, K.; Itoh, M.; Hayasaka, C.; Shimada, M.; Yamazaki, H.; Yamadori, A. Decreased cortical glucose metabolism correlates with hippocampal atrophy in Alzheimer’s disease as shown by MRI and PET. J. Neurol. Neurosurg. Psychiatry 1997, 62, 596–600. [Google Scholar] [CrossRef] [Green Version]
- Kato, T.; Inui, Y.; Nakamura, A.; Ito, K. Brain fluordeoxyglucose (FDG) PET in dementia. Ageing Res. Rev. 2016, 30, 73–84. [Google Scholar] [CrossRef]
- Hertz, L.; Chen, Y. Integration between glycolysis and glutamate-glutamine cycle flux may explain preferential glycolytic increase during brain activation, requiring glutamate. Front. Integr Neurosci. 2017, 11, 18. [Google Scholar] [CrossRef] [Green Version]
- Mergenthaler, P.; Lindauer, U.; Dienel, G.A.; Meisel, A. Sugar for the brain: The role of glucose in physiological and pathological brain function. Trends Neurosci. 2013, 36, 587–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueno, M.; Carvalheira, J.B.; Tambascia, R.C.; Bezerra, R.M.; Amaral, M.E.; Carneiro, E.M.; Folli, F.; Franchini, K.G.; Saad, M.J. Regulation of insulin signalling by hyperinsulinaemia: Role of IRS-1/2 serine phosphorylation and the mTOR/p70 S6K pathway. Diabetologia 2005, 48, 506–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alayev, A.; Holz, M.K. mTOR signaling for biological control and cancer. J. Cell Physiol. 2013, 228, 1658–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell. Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spilman, P.; Podlutskaya, N.; Hart, M.J.; Debnath, J.; Gorostiza, O.; Bredesen, D.; Richardson, A.; Strong, R.; Galvan, V. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS ONE 2010, 5, e9979. [Google Scholar] [CrossRef] [Green Version]
- Vicencio, J.M.; Galluzzi, L.; Tajeddine, N.; Ortiz, C.; Criollo, A.; Tasdemir, E.; Morselli, E.; Younes, A.B.; Maiuri, M.C.; Lavendero, S.; et al. Senescence, apoptosis or autophagy? When a damaged cell must decide its path-a mini-review. Gerontology 2008, 54, 92–99. [Google Scholar] [CrossRef]
- Faragher, R.G.A.; McArdle, A.; Willows, A.; Ostler, E.L. Senescence in the aging process. F1000Res 2017, 6, 1219. [Google Scholar] [CrossRef] [Green Version]
- Stein, G.H.; Drullinger, L.F.; Soulard, A.; Dulić, V. Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol. Cell Biol. 1999, 19, 2109–2117. [Google Scholar] [CrossRef] [Green Version]
- Krenning, L.; Feringa, F.M.; Shaltiel, I.A.; Van den Berg, J.; Medema, R.H. Transient activation of p53 in G2 phase is sufficient to induce senescence. Mol. Cell 2014, 55, 59–72. [Google Scholar] [CrossRef] [Green Version]
- Özcan, S.; Alessio, N.; Acar, M.B.; Mert, E.; Omerli, F.; Peluso, G.; Galderisi, U. Unbiased analysis of senescence associated secretory phenotype (SASP) to identify common components following different genotoxic stresses. Aging 2016, 8, 1316–1329. [Google Scholar] [CrossRef] [Green Version]
- Correia-Melo, C.; Passos, J.F. Mitochondria: Are they causal players in cellular senescence? Biochim. Biophys. Acta 2015, 1847, 1373–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weichhart, T. mTOR as regulator of lifespan, aging, and cellular senescence: A mini-review. Gerontology 2018, 64, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.J.; Chiang, Y.J.; Hathcock, K.S.; Horikawa, I.; Sedelnikova, O.A.; Hodes, R.J.; Bonner, W.M. Both telomeric and non-telomeric DNA damage are determinants of mammalian cellular senescence. Epigenetics Chromatin 2008, 1, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freund, A.; Laberge, R.-M.; Demaria, M.; Campisi, J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 2012, 23, 2066–2075. [Google Scholar] [CrossRef] [PubMed]
- Cormenier, J.; Martin, N.; Deslé, J.; Salazar-Cardozo, C.; Pourtier, A.; Abbadie, C.; Pluquet, O. The ATF6 arm of the Unfolded Protein Response mediates replicative senescence in human fibroblasts through a COX2/prostaglandin E2 intracrine pathway. Mech. Ageing Dev. 2018, 170, 82–91. [Google Scholar] [CrossRef]
- Zhang, P.; Kishimoto, Y.; Grammatikakis, I.; Gottimukkala, K.; Cutler, R.G.; Zhang, S.; Abdelmohsen, K.; Bohr, V.A.; Sen, J.M.; Gorospe, M.; et al. Senolytic therapy alleviates Aβ-associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer’s disease model. Nat. Neurosci. 2019, 22, 719–728. [Google Scholar] [CrossRef]
- Musi, N.; Valentine, J.M.; Sickora, K.R.; Baeuerle, E.; Thompson, C.S.; Shen, Q.; Orr, M.E. Tau protein aggregation is associated with cellular senescence in the brain. Aging Cell. 2018, 17, e12840. [Google Scholar] [CrossRef]
- Roberson, R.; Kuddo, T.; Horowitz, K.; Caballero, M.; Spong, C.Y. Cytokine and chemokine alterations in Down syndrome. Am. J. Perinatol. 2012, 29, 705–708. [Google Scholar] [CrossRef]
- Rueda, N.; Vidal, V.; García-Cerro, S.; Narcís, J.O.; Llorens-Martín, M.; Corrales, A.; Lantigua, S.; Iglesias, M.; Merino, J.; Merino, R.; et al. Anti-IL17 treatment ameliorates Down syndrome phenotypes in mice. Brain Behav. Immun. 2018, 73, 235–251. [Google Scholar] [CrossRef]
- Yang, S.; Wang, J.; Guo, S.; Huang, D.; Bestard Lorigados, I.; Nie, X.; Lou, D.; Li, Y.; Liu, M.; Kang, Y.; et al. Transcriptional activation of UPS16 gene expression by NFkB signaling. Mol. Brain 2019, 12, 120. [Google Scholar] [CrossRef] [Green Version]
- Adorno, M.; Sikandar, S.; Mitra, S.S.; Kuo, A.; Di Robilant, B.N.; Haro-Acosta, V.; Ouadah, Y.; Quarta, M.; Rodriguez, J.; Qian, D.; et al. Usp16 contributes to somatic stem-cell defects in Down’s syndrome. Nature 2013, 501, 380–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adorno, M.; di Robilant, B.N.; Sikandar, S.S.; Haro-Acosta, V.; Antony, J.; Heller, C.H.; Clarke, M.F. Usp16 modulates Wnt signaling in primary tissues through Cdkn2a regulation. Sci. Rep. 2018, 8, 17506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Chang, J.; Shao, L.; Feng, W.; Luo, Y.; Chow, M.; Du, W.; Meng, A.; Zhou, D. Hematopoietic Stem Cells from Ts65Dn Mice Are Deficient in the Repair of DNA Double-Strand Breaks. Radiat Res. 2016, 185, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Prattichizzo, F.; Grillari, J.; Balistreri, C.R. Cellular senescence and inflammaging in age-related diseases. Med. Inflamm. 2018, 9076485. [Google Scholar] [CrossRef] [PubMed]
- Fuster-Matanzo, A.; Llorens-Martín, M.; Hernández, F.; Avila, J. Role of neuroinflammation in adult neurogenesis and Alzheimer disease: Therapeutic approaches. Mediat. Inflamm. 2013, 260925. [Google Scholar] [CrossRef] [Green Version]
- Lyman, M.; Lloyd, D.G.; Ji, X.; Vizcaychipi, M.P.; Ma, D. Neuroinflammation: The role and consequences. Neurosci. Res. 2014, 79, 1–12. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [Green Version]
- Guerreiro, R.; Brás, J.; Hardy, J. Snapshot: Genetics of Alzheimer’s disease. Cell 2013, 155, 968. [Google Scholar] [CrossRef] [Green Version]
- Rawji, K.S.; Mishra, M.K.; Michaels, N.J.; Rivest, S.; Stys, P.K.; Yong, V.W. Immunosenescence of microglia and macrophages: Impact on the ageing central nervous system. Brain 2016, 139, 653–661. [Google Scholar] [CrossRef] [Green Version]
- Bauer, J.; Strauss, S.; Schreiter-Gasser, U.; Ganter, U.; Schlegel, P.; Witt, I.; Yolk, B.; Berger, M. Interleukin-6 and α-2-macroglobulin indicate an acute-phase state in Alzheimer’s disease cortices. FEBS Lett. 1991, 285, 111–114. [Google Scholar] [CrossRef] [Green Version]
- Huell, M.; Strauss, S.; Volk, B.; Berger, M.; Bauer, J. Interleukin-6 is present in early stages of plaque formation and is restricted to the brains of Alzheimer’s disease patients. Acta Neuropathol. 1995, 89, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Liu, M.; Nguyen, X.V.; Bing, G. P38 MAP kinase is activated at early stages in Alzheimer’s disease brain. Exp. Neurol. 2003, 183, 394–405. [Google Scholar] [CrossRef]
- Lai, K.S.P.; Liu, C.S.; Rau, A.; Lanctôt, K.L.; Köhler, C.A.; Pakosh, M.; Carvalho, A.F.; Herrmann, N. Peripheral inflammatory markers in Alzheimer’s disease: A systematic review and meta-analysis of 175 studies. J. Neurol. Neurosurg. Psychiatry 2017, 88, 876–882. [Google Scholar] [CrossRef] [PubMed]
- Rea, I.M.; Gibson, D.S.; McGilligan, V.; McNerlan, S.E.; Alexander, H.D.; Ross, O.A. Age and age-related diseases: Role of inflammation triggers and cytokines. Front. Immunol. 2018, 9, 586. [Google Scholar] [CrossRef] [PubMed]
- Blurton-Jones, M.; Laferla, F.M. Pathways by which Aβ facilitates tau pathology. Curr. Alzheimer Res. 2006, 3, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Minter, M.R.; Taylor, J.M.; Crack, P.J. The contribution of neuroinflammation to amyloid toxicity in Alzheimer’s disease. J. Neurochem. 2016, 136, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Barger, S.W.; Harmon, A.D. Microglial activation by alzhelmer amyloid precursor protein and modulation by apolipoprotein E. Nature 1997, 388, 878–881. [Google Scholar] [CrossRef]
- Ho, G.J.; Drego, R.; Hakimian, E.; Masliah, E. Mechanisms of cell signaling and inflammation in Alzheimer’s disease. Curr. Drug Targets Inflamm. Allergy 2004, 4, 247–256. [Google Scholar] [CrossRef]
- Sastre, M.; Dewachter, I.; Landreth, G.E.; Willson, T.M.; Klockgether, T.; van Leuven, F.; Heneka, M.T. Nonsteroidal anti-inflammatory drugs and peroxisome proliferator-activated receptor-γ agonists modulate immunostimulated processing of amyloid precursor protein through regulation of β-secretase. J. Neurosci. 2003, 23, 9796–97804. [Google Scholar] [CrossRef] [Green Version]
- Sastre, M.; Dewachter, I.; Rossner, S.; Bogdanovic, N.; Rosen, E.; Borghgraef, P.; Evert, B.O.; Dumitrescu-Ozimek, L.; Thal, D.R.; Landreth, G.; et al. Nonsteroidal anti-inflammatory drugs repress β- secretase gene promoter activity by the activation of PPARgamma. Proc. Natl. Acad. Sci. USA 2006, 103, 443–448. [Google Scholar] [CrossRef] [Green Version]
- Mrak, R.E.; Griffin, W.S. Potential inflammatory biomarkers in Alzheimer’s disease. J. Alzheimers Dis. 2005, 8, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Barger, S.; Barnum, S.; Bradt, B.; Bauer, J.; Cole, G.M.; Cooper, N.R.; Eikelenboom, P.; Emmerling, M.; Fiebich, B.L.; et al. Inflammation and Alzheimer’s disease. Neurobiol. Aging 2000, 21, 383–421. [Google Scholar] [CrossRef]
- Sapolsky, R.M. Glucocorticoid toxicity in the hippocampus: Reversal by supplementation with brain fuels. J. Neurosci. 1986, 6, 2240–2244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, D.; Rodrigues, B. Glucocorticoids produce whole body insulin resistance with changes in cardiac metabolism. Am. J. Physiol. Endocrinol. Metab. 2007, 292, 654–667. [Google Scholar] [CrossRef] [Green Version]
- Lim, G.P.; Yang, F.; Chu, T.; Gahtan, E.; Ubeda, O.; Beech, W.; Overmier, J.B.; Hsiao Ashe, K.; Frautschy, S.A.; Cole, G.M. Ibuprofen effects on Alzheimer pathology and open field activity in APPsw transgenic mice. Neurobiol. Aging 2001, 22, 983–991. [Google Scholar] [CrossRef]
- Vogel, A.; Upadhya, R.; Shetty, A.K. Neural stem cell derived extracellular vesicles: Attributes and prospects for treating neurodegenerative disorders. EBioMedicine 2018, 38, 273–282. [Google Scholar] [CrossRef]
- Wheeler, D.L.; Sariol, A.; Meyerholz, D.K.; Perlman, S. Microglia are required for protection against lethal coronavirus encephalitis in mice. J. Clin. Invest. 2018, 128, 931–943. [Google Scholar] [CrossRef]
- Halle, A.; Hornung, V.; Petzold, G.C.; Stewart, C.R.; Monks, B.G.; Reinheckel, T.; Gitzgerald, K.A.; Latz, E.; Moore, K.J.; Golenbock, D.T. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat. Immunol. 2008, 9, 857–865. [Google Scholar] [CrossRef] [Green Version]
- Johansson, J.U.; Woodling, N.S.; Wang, Q.; Panchal, M.; Liang, X.; Trueba-Saiz, A.; Brown, H.D.; Mhatre, S.D.; Loui, T.; Andreasson, K.I. Prostaglandin signaling suppresses beneficial microglial function in Alzheimer’s disease models. J. Clin. Invest. 2015, 125, 350–364. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Walter, S.; Stagi, M.; Cherny, D.; Letiembre, M.; Schulz-Schaeffer, W.; Heine, H.; Penke, B.; Neumann, H.; Fassbender, K. LPS receptor (CD14): A receptor for phagocytosis of Alzheimer’s amyloid peptide. Brain 2005, 128, 1778–1789. [Google Scholar] [CrossRef] [Green Version]
- Martin, E.; Boucher, C.; Fontaine, B.; Delarasse, C. Distinct inflammatory phenotypes of microglia and monocyte-derived macrophages in Alzheimer’s disease models: Effects of aging and amyloid pathology. Aging Cell 2017, 16, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Stancu, I.C.; Cremers, N.; Vanrusselt, H.; Couturier, J.; Vanoosthuyse, A.; Kessels, S.; Lodder, C.; Brône, B.; Huaux, F.; Octave, J.-N.; et al. Aggregated Tau activates NLRP3-ASC inflammasome exacerbating exogenously seeded and non-exogenously seeded Tau pathology in vivo. Acta Neuropathol. 2019, 137, 599–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Zhang, Z. Microglia and Wnt pathways: Prospects for inflammation in Alzheimer’s disease. Front. Aging Neurosci. 2020, 12, 120. [Google Scholar] [CrossRef]
- Contestabile, A.; Fila, T.; Ceccarelli, A.; Bonasoni, P.; Bonapace, L.; Santini, D.; Bartesaghi, R.; Ciani, E. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with Down syndrome and in Ts65Dn mice. Hippocampus 2007, 17, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Stagni, F.; Giacomini, A.; Emili, M.; Guidi, S.; Bartesaghi, R. Neurogenesis impairment: An early developmental defect in Down syndrome. Free Radic. Biol. Med. 2018, 114, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Hammerle, B.; Ulin, E.; Guimera, J.; Becker, W.; Guillemot, F.; Tejedor, F.J. Transient expression ofMnb/Dyrk1a couples cell cycle exit and differentiation of neuronal precursors by inducing p27KIP1 expression and suppressing NOTCH signaling. Development 2011, 138, 2543–2554. [Google Scholar] [CrossRef] [Green Version]
- Sadasiva, S.; DeCaprio, J.A. The DREAM complex: Master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer 2013, 13, 585–595. [Google Scholar] [CrossRef] [Green Version]
- Litovchick, L.; Florens, L.A.; Swanson, S.K.; Washburn, M.P.; DeCaprio, J.A. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 2011, 25, 801–813. [Google Scholar] [CrossRef] [Green Version]
- Yabut, O.; Domogauer, J.; D’Arcangelo, G. Dyrk1A overexpression inhibits proliferation and induces premature neuronal differentiation of neural progenitor cells. J. Neurosci. 2010, 30, 4004–4014. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Lin, J.-R.; Feng-Chiao, T.; Meyer, T. Dosage of Dyrk1A shifts cells within a p21-cyclin D1 signalling map to control the decision to enter the cell cycle. Mol. Cell 2013, 52, 87–100. [Google Scholar] [CrossRef] [Green Version]
- Kageyama, R.; Ohtsuka, T.; Shimojo, H.; Imayoshi, I. Dynamic regulation of Notch signaling in neural progenitor cells. Curr. Opin. Cell. Biol. 2009, 21, 733–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagarsheth, M.H.; Viehman, A.; Lippa, S.M.; Lippa, C.F. Notch-1 immunoexpression is increased in Alzheimer’s and Pick’s disease. J. Neurol. Sci. 2006, 244, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.F.; van Dijk, R.; Sluijs, J.A.; Nair, S.M.; Racchi, M.; Levelt, C.N.; van Leeuwen, F.W.; Ho, E.M. Activation of the Notch pathway in Down syndrome: Cross-talk of Notch and APP. FASEB J. 2005, 19, 1451–1458. [Google Scholar] [PubMed]
- Wang, S.; Barres, B.A. Up a notch: Instructing gliogenesis minireview. Neuron 2000, 27, 197–200. [Google Scholar] [CrossRef] [Green Version]
- Kurabayashi, N.; Sanada, K. Increased dosage of DYRK1A and DSCR1 delays neuronal differentiation in neocortical progenitor cells. Genes Dev. 2013, 27, 2708–2721. [Google Scholar] [CrossRef] [Green Version]
- Abelaira, H.M.; Reus, G.Z.; Neotti, M.V.; Quevedo, J. The role of mTOR in depression and antidepressant responses. Life Sci. 2014, 101, 10–14. [Google Scholar] [CrossRef]
- Kassai, H.; Sugaya, Y.; Noda, S.; Nakao, K.; Maeda, T.; Kano, M.; Aiba, A. Selective activation of mTORC1 signaling recapitulates microcephaly, tuberous sclerosis, and neurodegenerative diseases. Cell Rep. 2014, 7, 1626–1639. [Google Scholar] [CrossRef] [Green Version]
- Chao, M.V. Neurotrophins and their receptors: A convergence point for many signalling pathways. Nat. Rev. Neurosci. 2003, 4, 299. [Google Scholar] [CrossRef]
- Zweifel, L.S.; Kuruvilla, R.; Ginty, D.D. Functions and mechanisms of retrograde neurotrophin signaling. Nat. Rev. Neurosci. 2005, 6, 615–625. [Google Scholar] [CrossRef]
- Chao, M.V.; Rajagopal, R.; Lee, F.S. Neurotrophin signalling in health and disease. Clin. Sci. 2006, 110, 167–173. [Google Scholar] [CrossRef]
- Tapia-Arancibia, L.; Aliaga, E.; Silhol, M.; Arancibia, S. New insights into brain BDNF function in normal aging and Alzheimer disease. Brain Res. Rev. 2008, 59, 201–220. [Google Scholar] [CrossRef] [PubMed]
- Roper, R.J.; Baxter, L.L.; Saran, N.G.; Klinedinst, D.K.; Beachy, P.A.; Reeves, R.H. Defective cerebellar response to mitogenic Hedgehog signaling in Down syndrome mice. Proc. Natl. Acad. Sci. USA 2006, 103, 1452–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trazzi, S.; Mitrugno, V.M.; Valli, E.; Fuchs, C.; Rizzi, S.; Guidi, S.; Perini, G.; Bartesaghi, R.; Ciani, E. APP-dependent up-regulation of Ptch1 underlies proliferation impairment of neural precursors in Down syndrome. Hum. Mol. Genet. 2011, 20, 1560–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trazzi, S.; Fuchs, C.; Valli, E.; Perini, G.; Bartesaghi, R.; Ciani, E. The amyloid precursor protein (APP) triplicated gene impairs neuronal precursor differentiation and neurite development through two different domains in the Ts65Dn mouse model for Down syndrome. J. Biol. Chem. 2013, 288, 20817–20829. [Google Scholar] [CrossRef] [Green Version]
- Nalivaeva, N.N.; Turner, A.J. The amyloid precursor protein: A biochemical enigma in brain development, function and disease. FEBS Lett. 2013, 587, 2046–2054. [Google Scholar] [CrossRef] [Green Version]
- Giacomini, A.; Stagni, F.; Trazzi, S.; Guidi, S.; Emili, M.; Brigham, E.; Ciani, E.; Bartesaghi, R. Inhibition of APP gamma-secretase restores sonic hedgehog signaling and neurogenesis in the Ts65Dn mouse model of Down syndrome. Neurobiol. Dis. 2015, 82, 385–396. [Google Scholar] [CrossRef] [Green Version]
- London, J.; Rouch, C.; Bui, L.C.; Assayag, E.; Souchet, B.; Daubigne, F.; Medjaoui, H.; Luquet, S.; Magnan, C.; Delabar, J.M.; et al. Overexpression of the DYRK1A Gene (Dual-Specificity Tyrosine Phosphorylation- Regulated Kinase 1A) Induces Alterations of the Serotoninergic and Dopaminergic Processing in Murine Brain Tissues. Mol. Neurobiol. 2018, 55, 3822–3831. [Google Scholar] [CrossRef]
- Yuen, E.Y.; Qin, L.; Wei, J.; Liu, W.; Liu, A.; Yan, Z. Synergistic regulation of glutamatergic transmission by serotonin and norepinephrine reuptake inhibitors in prefrontal cortical neurons. J. Biol. Chem. 2014, 289, 25177–25185. [Google Scholar] [CrossRef] [Green Version]
- Xing, B.; Li, Y.C.; Gao, W.J. Norepinephrine versus dopamine and their interaction in modulating synaptic function in the prefrontal cortex. Brain Res. 2016, 1641, 217–233. [Google Scholar] [CrossRef] [Green Version]
- Keating, D.J.; Dubach, D.; Zanin, M.P.; Yu, Y.; Martin, K.; Zhao, Y.-F.; Chen, C.; Porta, S.; Arbonés, M.L.; Mittaz, L.; et al. DSCR1/RCAN1 regulates vesicle exocytosis and fusion pore kinetics: Implications for Down syndrome and Alzheimer’s disease. Hum. Mol. Genet. 2008, 1, 1020–1030. [Google Scholar] [CrossRef] [Green Version]
- Zanin, M.P.; Mackenzie, K.D.; Peiris, H.; Pritchard, M.A.; Keating, D.J. RCAN1 regulates vesicle recycling and quantal release kinetics via effects on calcineurin activity. J. Neurochem. 2013, 124, 290–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peiris, H.; Keating, D. The neural and endocrine roles of RCAN1 in health and disease. Clin. Exp. Pharm. Physiol 2018, 45, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Arbonés, M.L.; Thomazeau, A.; Nakano-Kobayashi, A.; Hagiwara, M.; Delabar, J.M. DYRK1A and cognition: A lifelong relationship. Pharmacol. Ther. 2019, 194, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Aranda, S.; Alvarez, M.; Turro, S.; Laguna, A.; de la Luna, S. Sprouty2-mediated inhibition of fibroblast growth factor signaling is modulated by the protein kinase DYRK1A. Mol. Cell. Biol. 2008, 28, 5899–5911. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Sung, J.Y.; Park, J.; Song, W.J.; Chang, S.; Chung, K.C. Dyrk1A negatively regulates the actin cytoskeleton through threonine phosphorylation of N-WASP. J. Cell Sci. 2012, 125, 67–80. [Google Scholar] [CrossRef] [Green Version]
- Lepagnol-Bestel, A.M.; Zvara, A.; Maussion, G.; Quignon, F.; Ngimbous, B.; Ramoz, N.; Imbeaud, S.; Loe-Mie, Y.; Benihoud, K.; Agier, N.; et al. DYRK1A interacts with the REST/NRSF-SWI/SNF chromatin remodelling complex to deregulate gene clusters involved in the neuronal phenotypic traits of Down syndrome. Hum. Mol. Genet. 2009, 18, 1405–1414. [Google Scholar] [CrossRef] [Green Version]
- Martinez de Lagran, M.; Benavides-Piccione, R.; Ballesteros-Yanez, I.; Calvo, M.; Morales, M.; Fillat, C.; Defelipe, J.; Ramakers, G.J.A.; Dierssen, M. Dyrk1A influences neuronal morphogenesis through regulation of cytoskeletal dynamics in mammalian cortical neurons. Cereb. Cortex 2012, 22, 2867–2877. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Park, J.; Song, W.J.; Chang, S. Overexpression of Dyrk1A causes the defects in synaptic vesicle endocytosis. Neurosignals 2010, 18, 164–172. [Google Scholar] [CrossRef]
- Kwon, H.B.; Kozorovitskiy, Y.; Oh, W.J.; Peixoto, R.T.; Akhtar, N.; Saulnier, J.L.; Gu, C.; Sabatini, B.L. Neuroligin-1-dependent competition regulates cortical synaptogenesis and synapse number. Nat. Neurosci. 2012, 15, 1667–1674. [Google Scholar] [CrossRef] [Green Version]
- Toiber, D.; Azkona, G.; Ben-Ari, S.; Toran, N.; Soreq, H.; Dierssen, M. Engineering DYRK1A overdosage yields Down syndrome-characteristic cortical splicing aberrations. Neurobiol. Dis. 2010, 40, 348–359. [Google Scholar] [CrossRef]
- Fan, F.; Funk, L.; Lou, X. Dynamin 1- and 3-Mediated Endocytosis Is Essential for the Development of a Large Central Synapse In Vivo. J. Neurosci. 2016, 36, 6097–6115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Cerro, S.; Martinez, P.; Vidal, V.; Corrales, A.; Florez, J.; Vidal, R.; Rueda, N.; Arbonés, M.L.; Martínez-Cué, C. Overexpression of Dyrk1A is implicated in several cognitive, electrophysiological and neuromorphological alterations found in a mouse model of Down syndrome. PLoS ONE 2014, 9, e106572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Rai, A.; Hur, E.M.; Smilansky, Z.; Chang, K.T.; Min, K.T. DSCR1 is required for both axonal growth cone extension and steering. J. Cell Biol. 2016, 213, 451–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.; Xiong, H.; Liu, T.; Zhang, L.; Godzik, A.; Zhuohua, Z. Aggregate formation and synaptic abnormality induced by DSCR1. J. Neurochem. 2004, 88, 1485–1496. [Google Scholar] [CrossRef] [PubMed]
- Martin, K.R.; Corlett, A.; Dubach, D.; Mustafa, T.; Coleman, H.A.; Parkington, H.C.; Mersen, T.D.; Bourne, J.A.; Porta, S.; Arbonés, M.L.; et al. Over-expression of RCAN1 causes Down syndrome-like hippocampal deficits that alter learning and memory. Hum. Mol. Genet. 2012, 21, 3025–3041. [Google Scholar] [CrossRef]
- Sugiura, R.; Sio, S.O.; Shuntoh, H.; Kuno, T. Molecular genetic analysis of the calcineurin signaling pathways. Cell. Mol. Life Sci. 2001, 58, 278–288. [Google Scholar] [CrossRef]
- Marks, B.; McMahon, H.T. Calcium triggers calcineurin-dependent synaptic vesicle recycling in mammalian nerve terminals. Curr. Biol. 1998, 8, 740–749. [Google Scholar] [CrossRef] [Green Version]
- Cousin, M.A.; Tan, T.C.; Robinson, P.J. Protein phosphorylation is required for endocytosis in nerve terminals: Potential role for the dephosphins dynamin I and synaptojanin, but not AP180 or amphiphysin. J. Neurochem. 2001, 76, 105–116. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Sukeena, J.M.; Simmons, A.B.; Hansen, E.J.; Nuhn, R.E.; Samuels, I.S.; Fuerst., P.G. DSCAM promotes refinement in the mouse retina through cell death and restriction of exploring dendrites. J. Neurosci. 2015, 35, 5640–5654. [Google Scholar] [CrossRef]
- Alves-Sampaio, A.; Troca-Marín, J.A.; Montesinos, M.L. NMDA mediated regulation of DSCAM dendritic local translation is lost in a mouse model of Down’s syndrome. J. Neurosci. 2009, 30, 13537–13548. [Google Scholar] [CrossRef]
- Sache, S.M.; Lievens, S.; Ribeiro, L.; Dascenco, D.; Massachaele, D.; Horré, K.; Misbaer, A.; Vanderroost, N.; De Smet, A.S.; Salta, E.; et al. Nuclear import of the DSCAM-cytoplasmatic domain drives signaling capable of inhibiting synapse formation. EMBO J. 2019, 38, e99669. [Google Scholar]
- O’Bryan, J.P. Intersecting pathways in cell biology. Sci. Signal. 2010, 3, re10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guipponi, M.; Scott, H.S.; Hattori, M.; Ishii, K.; Sakaki, Y.; Antonarakis, S.E. Genomic structure, sequence, and refined mapping of the human intersectin gene (itsn), which encompasses 250 kb on chromosome 21q22.1→q22.2. Cytogenet. Cell. Genet. 1998, 83, 218–220. [Google Scholar] [CrossRef]
- Pucharcos, C.; Fuentes, J.J.; Casas, C.; de la Luna, S.; Alcantara, S.; Arbones, M.L.; Soriano, E.; Estivill, X.; Pritchard., M. Alu-splice cloning of human intersectin (itsn), a putative multivalent binding protein expressed in proliferating and differentiating neurons and overexpressed in Down syndrome. Eur. J. Hum. Genet. 1999, 7, 704–712. [Google Scholar] [CrossRef]
- Wilmot, B.; McWeeney, S.K.; Nixon, R.R.; Montine, T.J.; Laut, J.; Harrington, C.A.; Kaye, J.A.; Kramer, P.L. Translational gene mapping of cognitive decline. Neurobiol. Aging 2008, 29, 524–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nixon, R.A. Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases. Neurobiol. Aging 2005, 26, 373–382. [Google Scholar] [CrossRef]
- Nishimura, T.; Yamaguch, T.; Tokunaga, A.; Hara, A.; Hamaguchi, T.; Kato, K.; Iwamatsu, A.; Okano, H.; Kaibuchi, K. Role of numb in dendritic spine development with a Cdc42 GEF intersectin and EphB2. Mol. Biol. Cell 2009, 17, 1273–1285. [Google Scholar] [CrossRef]
- Gunner, G.; Cheadle, L.; Johnson, K.M.; Ayata, P.; Badimon, A.; Mondo, E.; Nagy, M.A.; Liu, L.; Beiller, S.M.; Kim, K.-W.; et al. Sensory lesioning induces microglial synapse elimination via ADAM10 and fractalkine signaling. Nat. Neurosci. 2019, 22, 1075–1088. [Google Scholar] [CrossRef]
- Jiang, T.; Zhang, Y.D.; Gao, Q.; Zhou, J.S.; Zhu, X.C.; Lu, H.; Shi, J.-Q.; Tan, L.; Chen, Q.; Yu, J.-T. TREM1 facilitates microglial phagocytosis of amyloid beta. Acta Neuropathol. 2016, 132, 667–683. [Google Scholar] [CrossRef]
- Edwards, F.A. A unifying hypothesis for Alzheimer’s disease: From plaques to neurodegeneration. Trends Neurosci. 2019, 42, 310–322. [Google Scholar] [CrossRef]
- Magdesian, M.H.; Carvalho, M.M.; Mendes, F.A.; Saraiva, L.M.; Juliano, M.A.; Juliano, L.; Garcia-Abreu, J.; Ferreira, S.T. Amyloid-beta binds to the extracellular cysteine-richdomain of Frizzled and inhibits Wnt/beta-catenin signaling. J. Biol. Chem. 2008, 283, 9359–9368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araujo, B.H.S.; Kaid, C.; De Souza, J.S.; Gomes da Silva, S.; Goulart, E.; Caires, L.C.J.; Musso, C.M.; Torres, L.B.; Ferrasa, A.; Herai, R.; et al. Down syndrome iPSC-derived astrocytes impair neuronal synaptogenesis and the mTOR pathway in vitro. Mol. Neurobiol. 2018, 55, 5962–5975. [Google Scholar] [CrossRef]
- Weston, M.C.; Chen, H.; Swann, J.W. Multiple roles for mammalian target of rapamycin signaling in both glutamatergic and GABAergic synaptic transmission. J. Neurosci. 2012, 32, 11441–11452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Zhou, F.; ten Dijke, P. Signaling interplay between transforming growth factor-beta receptor and PI3K/AKT pathways in cancer. Trends Biochem. Sci. 2013, 38, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Liu, H.; Yan, R.; Hu, M. PI3K/Akt and ERK/MAPK Signaling Promote Different Aspects of Neuron Survival and Axonal Regrowth Following Rat Facial Nerve Axotomy. Neurochem. Res. 2017, 42, 3515–3524. [Google Scholar] [CrossRef] [PubMed]
- Chidambaram, H.; Chinnathambi, S. G-protein coupled receptors and tau-different roles in Alzheimer’s disease. Neuroscience 2020, 438, 198–214. [Google Scholar] [CrossRef] [PubMed]
- Duka, V.; Lee, J.-H.; Credle, J.; Wills, J.; Oaks, A.; Smolinsky, C.; Shah, K.; Mash, D.C.; Masliah, E.; Sidhu, A. Identification of the sites of tau hyperphosphorylation and activation of tau kinases in synucleinopathies and Alzheimer’s diseases. PLoS ONE 2013, 8, e75025. [Google Scholar] [CrossRef]
- Zhu, C.; Xu, B.; Sun, X.; Zhu, Q.; Sui, Y. Targeting CCR3 to reduce amyloid-b production, Tau hyperphosphorylation, and synaptic loss in a mouse model of Alzheimer’s disease. Mol. Neurobiol. 2017, 54, 7964–7978. [Google Scholar] [CrossRef]
- Thathiah, A.; De Strooper, B. The role of G protein-coupled receptors in the pathology of Alzheimer’s disease. Nat. Rev. Neurosci. 2011, 12, 73. [Google Scholar] [CrossRef]
- Niswender, C.M.; Conn, P.J. Metabotropic glutamate receptors: Physiology, pharmacology, and disease. Annu. Rev. Pharm. Toxicol. 2010, 50, 295–322. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-S.; Silva, A.J. The molecular and cellular biology of enhanced cognition. Nat. Rev. Neurosci. 2009, 10, 126. [Google Scholar] [CrossRef] [PubMed]
- Rueda, N.; Flórez, J.; Dierssen, M.; Martínez-Cué, C. Translational validity and implications of pharmacotherapies in preclinical models of Down syndrome. Prog. Brain Res. 2020, 251, 245–268. [Google Scholar] [PubMed]
- Panza, F.; Lozupone, M.; Seripa, D.; Imbimbo, B.P. Amyloid-β immunotherapy for alzheimer disease: Is it now a long shot? Ann. Neurol. 2019, 85, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Zmijewski, P.A.; Gao, L.Y.; Saxena, A.R.; Chavannes, N.K.; Hushmendy, S.F.; Bhoiwala, D.L.; Crawford, D.R. Fish oil improves gene targets of Down syndrome in C57BL and BALB/c mice. Nutr. Res. 2015, 35, 440–448. [Google Scholar] [CrossRef]
- Martínez-Martínez, S.; Redondo, J.M. Inhibitors of the calcineurin/NFAT pathway. Curr. Med. Chem. 2004, 11, 997–1007. [Google Scholar] [CrossRef]
- Wang, C.; Yu, J.T.; Miao, D.; Wu, Z.C.; Tan, M.S.; Tan, L. Targeting the mTOR signaling network for Alzheimer’s disease therapy. Mol. Neurobiol. 2014, 49, 120–135. [Google Scholar] [CrossRef]
- Amidfar, M.; de Oliveira, J.; Kucharska, E.; Budni, J.; Kim, Y.K. The role of CREB and BDNF in neurobiology and treatment of Alzheimer’s disease. Life Sci. 2020, 257, 118020. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, L.; Yang, C.; Li, Z.; Rong, S. Procyanidins and Alzheimer’s Disease. Mol. Neurobiol. 2019, 56, 5556–5567. [Google Scholar] [CrossRef]
- Congdon, E.E.; Sigurdsson, E.M. Tau-targeting therapies for Alzheimer disease. Nat. Rev. Neurol. 2018, 14, 399–415. [Google Scholar] [CrossRef]
- Allen, S.J.; Watson, J.J.; Shoemark, D.K.; Barua, N.U.; Patel, N.K. GDNF, NGF and BDNF as therapeutic options for neurodegeneration. Pharm. Ther. 2013, 138, 155–175. [Google Scholar] [CrossRef]
- Chu, L.W. Alzheimer’s disease: Early diagnosis and treatment. Hong Kong Med. J. 2012, 18, 228–237. [Google Scholar] [PubMed]
- Macdonald, R.; Barnes, K.; Hastings, C.; Mortiboys, H. Mitochondrial abnormalities in Parkinson’s disease and Alzheimer’s disease: Can mitochondria be targeted therapeutically? Biochem. Soc. Trans. 2018, 46, 891–909. [Google Scholar] [CrossRef] [PubMed]
- Boccardi, V.; Murasecco, I.; Mecocci, P. Diabetes drugs in the fight against Alzheimer’s disease. Ageing Res. Rev. 2019, 54, 100936. [Google Scholar] [CrossRef] [PubMed]
- Frautschy, S.A.; Cole, G.M. Why pleiotropic interventions are needed for Alzheimer’s disease. Mol. Neurobiol. 2010, 41, 392–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Colonna, M. Microglia in Alzheimer’s disease: A target for immunotherapy. J. Leukoc. Biol. 2019, 106, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.L.; Tong, G.; Ballard, C. Treatment Combinations for Alzheimer’s Disease: Current and Future Pharmacotherapy Options. J. Alzheimers Dis. 2019, 67, 779–794. [Google Scholar] [CrossRef] [Green Version]
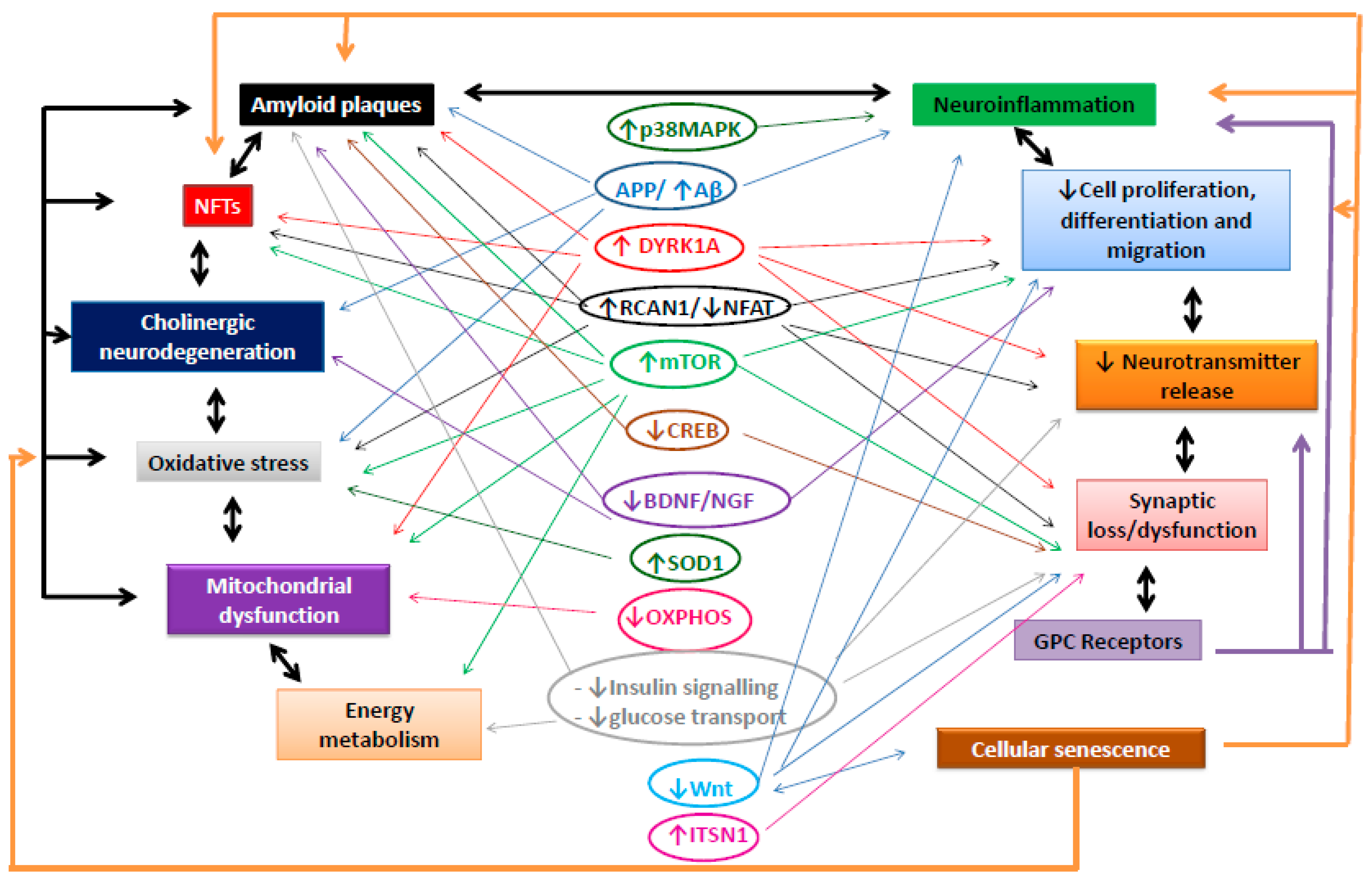
| Neuropathological Characteristic | Signalling Pathway | Up- or Downregulation | Pathophysiological Role in AD |
|---|---|---|---|
| Amyloid plaques | APP [31] | ↑ in DS and AD | Generation of Aβ oligomers |
| DYR1A [37,38,39,40,47] | ↑ in DS and AD | Aβ degradation, APP phosphorylation | |
| RCAN1/NFAT [42,43,45,46,47,48] | ↑ RCAN/↓ NFAT in DS and AD | Mediation of Aβ-induced neuronal death, disruption of Ca2+ homeostasis | |
| PIK3/Akm/mTOR [51,52,55,56,57,64,65,66,67,68,74,75] | ↑ in DS and AD | Contribution to Aβ generation and aggregation, inhibition of autophagy, reduction of Aβ clearance | |
| CREB [80,81,82,83,84,85,86] | ↓ in DS and AD | Induction of synaptic loss by Aβ | |
| BDNF/NGF [88,92,93,98,99,100] | ↓ in DS and AD | Accumulation of APP C-terminal fragments and aggregation of Aβ | |
| Neurofibrillary tangles | DYRK1A [117,118,119,120,121,122,123,124,125] | ↑ in DS and AD | Modifications in tau splicing and enhancement of tau phosphorylation |
| RCAN1/NFAT [126,127,128,129,130,131] | ↑ RCAN/↓ NFAT in DS and AD | Prevention of tau degradation and enhancement of tau phosphorylation | |
| CDK5 [133,134,135,136] | ↑ in DS | Enhancement of tau phosphorylation | |
| PP2A [137,138,139,140,141] | ↓ in DS | Enhancement of tau phosphorylation | |
| mTOR/SIRT1 [56,143,144,145] | ↑ mTOR/↓ SIRT in DS and AD | Enhancement of tau phosphorylation, promotion of tau accumulation | |
| Cholinergic system [156,157,158,159,160,161,162,163,164,165,166,167,168,169] | ↓ in DS and AD | Tau pathology in cholinergic neurons that aggravates neurodegeneration | |
| Cholinergic neurodegeneration | NGF/proNGF/TrkA/p75NTR [170,171,172,173] | ↓ in DS and AD | Reduction in survival of cholinergic neurons |
| Aβ [172,173,174] | ↑ in DS and AD | Facilitation of cholinergic neurodegeneration | |
| Oxidative stress | SOD1 [182] | ↑ in DS | Induction of Redox imbalance |
| RCAN1/NFAT [46,183,184,185,186,187] | ↑ RCAN/↓ NFAT in DS and AD | Alterations in mitochondrial function and increase in ROS production | |
| APP/Aβ [188,191] | ↑ in DS and AD | Enhancement of lipid, DNA, and RNA oxidation | |
| Glutamatergic system [189,190] | ↑ in AD | Promotion of OS-induced excitotoxicity | |
| Cholinergic system [191] | ↓ in DS and AD | Aβ-induced enhancement of OS in cholinergic neurons | |
| mTOR [192,193,194,196,198] | ↑ in DS and AD | OS disruption of mTOR function and mTOR enhancement of oxidative damage | |
| Mitochondrial dysfunction | Enhanced oxidative stress [191,200,201] | ↑ in DS and AD | Enhancement of ROS-mediated disruption of mitochondrial integrity and function |
| OXPHOS [203,204,205,206,209,210,211,212,213,214,215,216,217,218,219,220,221,222] | ↓ in DS and AD | Enhancement of Aβ production, alterations in cell membranes and synapses, reduction in mitochondrial inner membrane potential, reduction in energy production, and lower mitochondrial function | |
| Raptor/mTOR [197,223,224] | ↑ in DS and AD | Alterations in mitochondrial activity and metabolism | |
| Energy metabolism | Insulin signaling [241,242,243] | ↓ in DS and AD | Alterations in energy metabolism, impairment of neuronal activity, plasticity and survival, and facilitation of Aβ aggregation |
| Glucose transport and metabolism [245,246,247,248,249,250] | ↓ in DS and AD | Reduction in energy for synaptic transmission and neurotransmitter biosynthesis, alterations in autophagy | |
| PI3-K/Akt/mTOR [65,67,253–256 | ↑ in DS and AD | Dysregulation of energy balance, induction of insulin resistance, altered autophagy | |
| Cellular senescence | Release of proinflammatory cytokines [269,270] | ↑ in DS and AD | Induction of cellular senescence and enhancement by senescence |
| Oxidative stress and mitochondrial dysfunction [15,262] | ↑ in DS and AD | Induction of cellular senescence and enhancement by senescence | |
| Proteostasis (Aβ and tau) [267,268] | ↑ in DS and AD | Induction of cellular senescence and enhancement by senescence, induction of cellular senescence and enhancement by senescence | |
| USP16-Wnt [271,272,273] | ↑ UPS16 in DS/ ↓ Wnt in DS and AD | Induction of senescence through DNA damage, downregulation of the Wnt pathway reducing stem cell renewal | |
| Immune response/inflammation | p38MAPK [281,282,283,284,285] | ↑ in DS and AD | Increase in release of cytokines |
| Aβ/APP [287,290] | ↑ in DS and AD | Increase in release of cytokines which further aggravates Aβ pathology | |
| HPA [292,293] | ↑ in AD | Cytokines produce excessive activation of the HPA, which aggravates the energy deficits and enhances OS | |
| Wnt [304] | ↓ in DS and AD | Altered microglia activation, enhancement of neuroinflammation, tau hyperphosphorylation, and synaptic loss | |
| Cell proliferation/differentiation and migration | DYRK1A [305,306,307] | ↑ in DS and AD | Induction of cell cycle exit, premature differentiation or precursors resulting in a reduced number of adult neurons |
| DYRK1A/DREAM [308,309] | - | Inhibition of cell proliferation due to cell cycle arrest | |
| DYRK1A/Cyclin D1 [310,311] | Inhibition of proliferation and promotion of premature differentiation, prevention the entry into the S phase of the cycle | ||
| DYRK1A/Notch [312,313,314] | Inhibition of notch signaling that controls neurogenesis, induction of a shift from neurogenic to glionenic fate of progenitors | ||
| DYRK1A/NFAT [316] | Delay of neurogenesis by the synergic effect with RCAN1 | ||
| mTOR [317,318] | ↑ in DS and AD | Apoptotic death of NPCs | |
| BDNF [92,322] | ↓ in DS and AD | Impairment of cell proliferation and differentiation | |
| Shh [323,324,325] | ↓ in DS | Impairment of proliferation of NPCs | |
| APP [327] | ↑ in DS and AD | Alterations in cell cycle regulation, neural precursor maturation | |
| Neurotransmitter release | DYRK1A [328,330] | ↑ in DS and AD | Reductions in neurotransmitter synthesis and release |
| RCAN1 [331,332,333] | ↑ in DS and AD | Reductions in neurotransmitter synthesis and release | |
| Synapses | DYRK1A [334,335,336,337,338,339,340,341,342,343] | ↑ in DS and AD | Impairment in dendritic growth and complexity; dendritic spine formation; reduction of synaptic components necessary for synapse formation, maintenance, and functioning |
| RCAN1/NFAT [344,345,346,349] | ↑ RCAN/↓ NFAT in DS and AD | Modification of the localization of synaptic proteins, decreased phosphorylation of proteins necessary for synaptic plasticity | |
| DSCAM [351] | ↑ in DS and AD | Inhibition of dendritic branching and synapse formation | |
| ITSN [353,355,356,357,358] | ↑ in DS and AD | Enlargement of the early endosomal compartment, altered endocytic trafficking, leading to a reduced number and recycling of synaptic vesicles | |
| Wnt [359,362] | ↓ in DS and AD | Alterations in synapse number and function | |
| PI3K/AKT/mTOR [365,366] | ↑ in DS and AD | Loss of synapses partly mediated by enhanced cytokine release, impairment of synaptic development | |
| Receptors | Muscarinic ACh receptors [158] | ↓ in DS and AD | Impairment of cholinergic transmission: the loss of these receptors is mediated by tau phosphorylation |
| CXR2 and CC3 chemokine receptors [369,370] | ↑ in AD | Enhancement of tau phosphorylation ad cytokine release | |
| mGluR2 receptors [371] | ↑ in AD | Enhancement of tau phosphorylation | |
| Other GPCRs [367] | - | Alteration of neurotransmission by different mechanisms including tau phosphorylation, increased cytokine release, and aggravation of amyloid pathology |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Cué, C.; Rueda, N. Signalling Pathways Implicated in Alzheimer′s Disease Neurodegeneration in Individuals with and without Down Syndrome. Int. J. Mol. Sci. 2020, 21, 6906. https://doi.org/10.3390/ijms21186906
Martínez-Cué C, Rueda N. Signalling Pathways Implicated in Alzheimer′s Disease Neurodegeneration in Individuals with and without Down Syndrome. International Journal of Molecular Sciences. 2020; 21(18):6906. https://doi.org/10.3390/ijms21186906
Chicago/Turabian StyleMartínez-Cué, Carmen, and Noemí Rueda. 2020. "Signalling Pathways Implicated in Alzheimer′s Disease Neurodegeneration in Individuals with and without Down Syndrome" International Journal of Molecular Sciences 21, no. 18: 6906. https://doi.org/10.3390/ijms21186906
APA StyleMartínez-Cué, C., & Rueda, N. (2020). Signalling Pathways Implicated in Alzheimer′s Disease Neurodegeneration in Individuals with and without Down Syndrome. International Journal of Molecular Sciences, 21(18), 6906. https://doi.org/10.3390/ijms21186906





