3D Bone Morphology Alters Gene Expression, Motility, and Drug Responses in Bone Metastatic Tumor Cells
Abstract
1. Introduction
2. Results
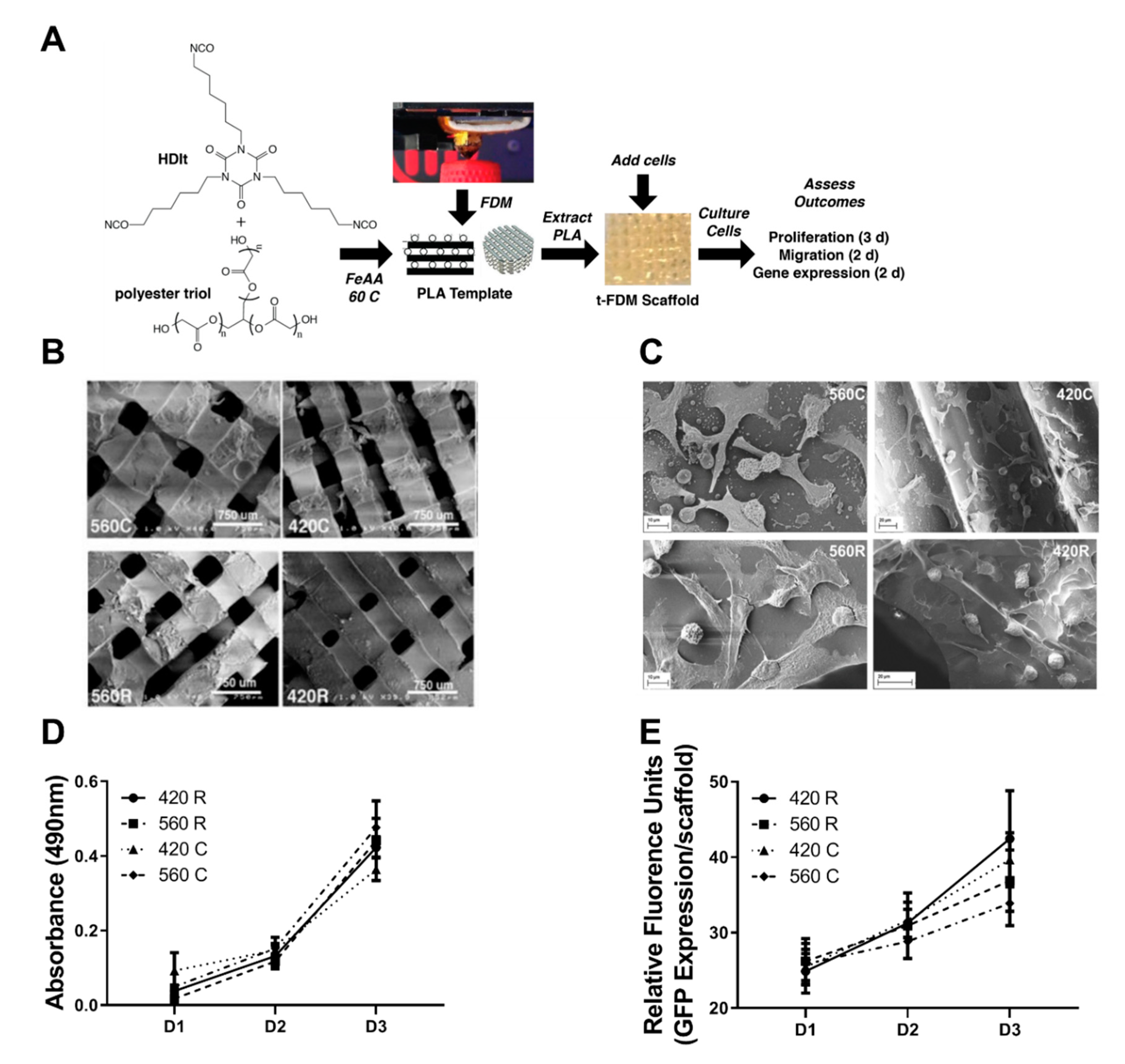
2.1. 3D Scaffolds Recapitulate the Mechanical and Morphometric Properties of Trabecular Bone
2.2. 3D Scaffolds Support Proliferation of Tumor Cells
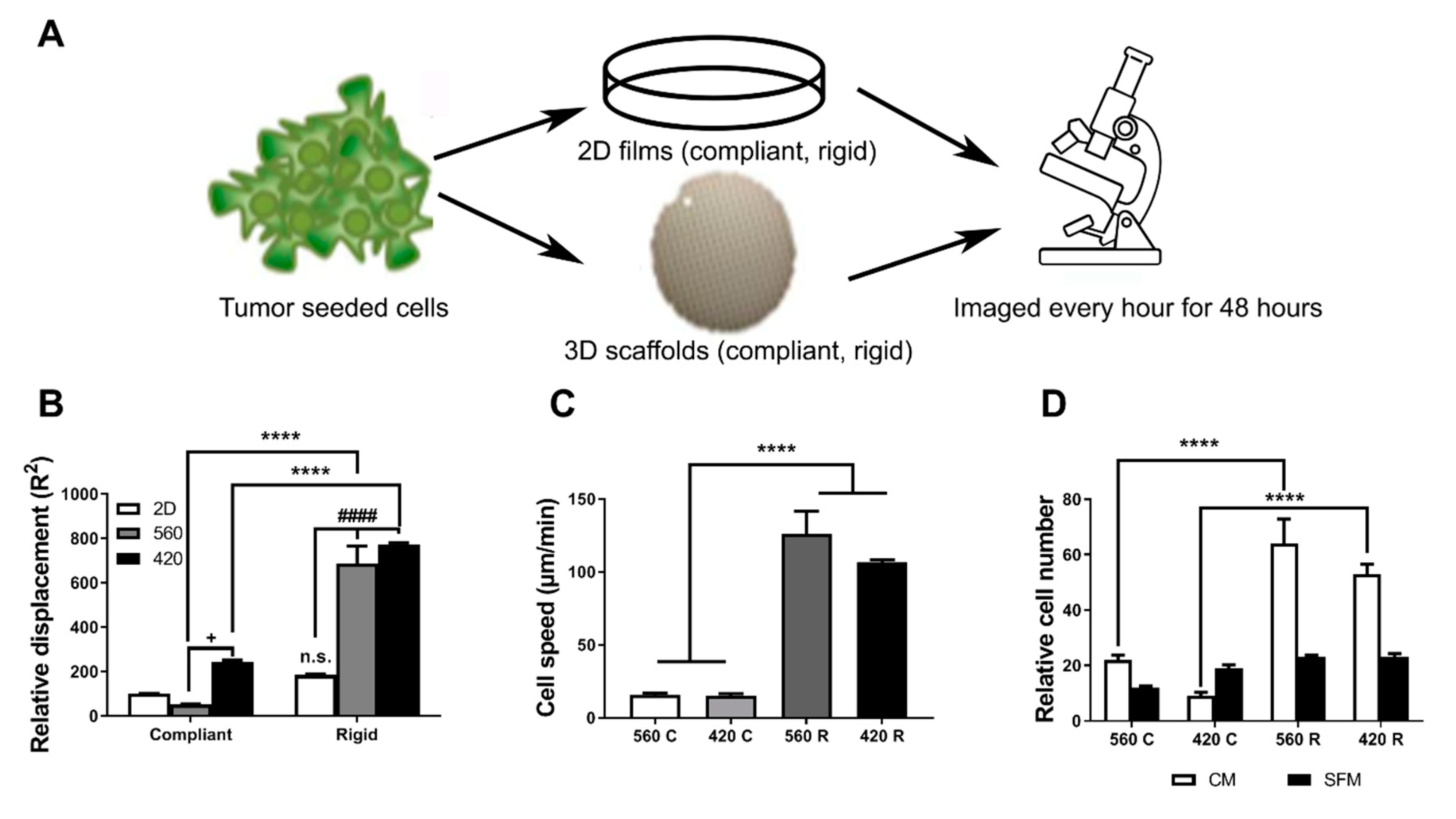
2.3. Substrate Modulus and Pore Size Regulate the Motility of Tumor Cells
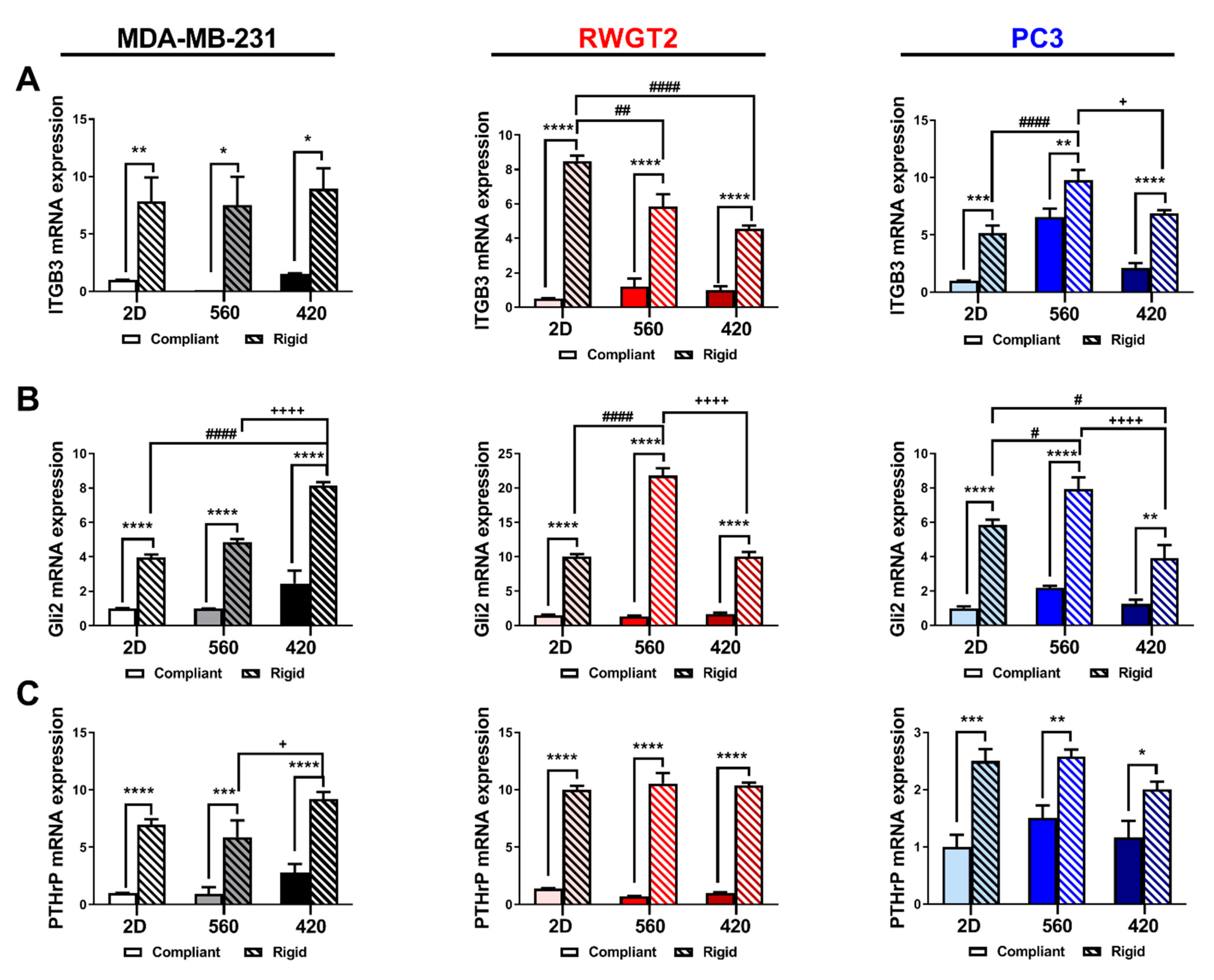
2.4. 3D Scaffolds Influence the Expression of Bone-Metastatic Genes In Vitro
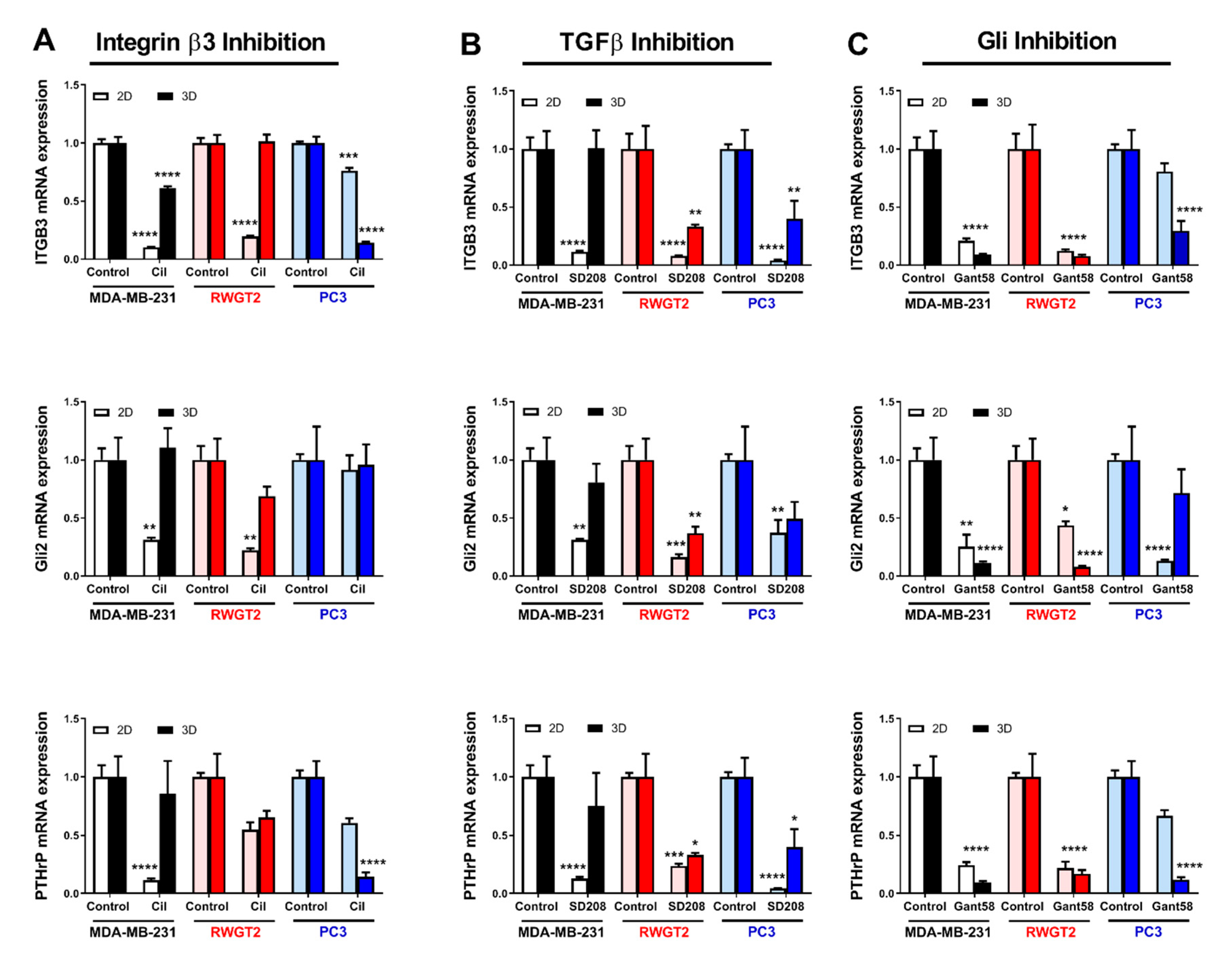
2.5. 3D Scaffolds Influence the Response of Tumor Cells to Therapeutics
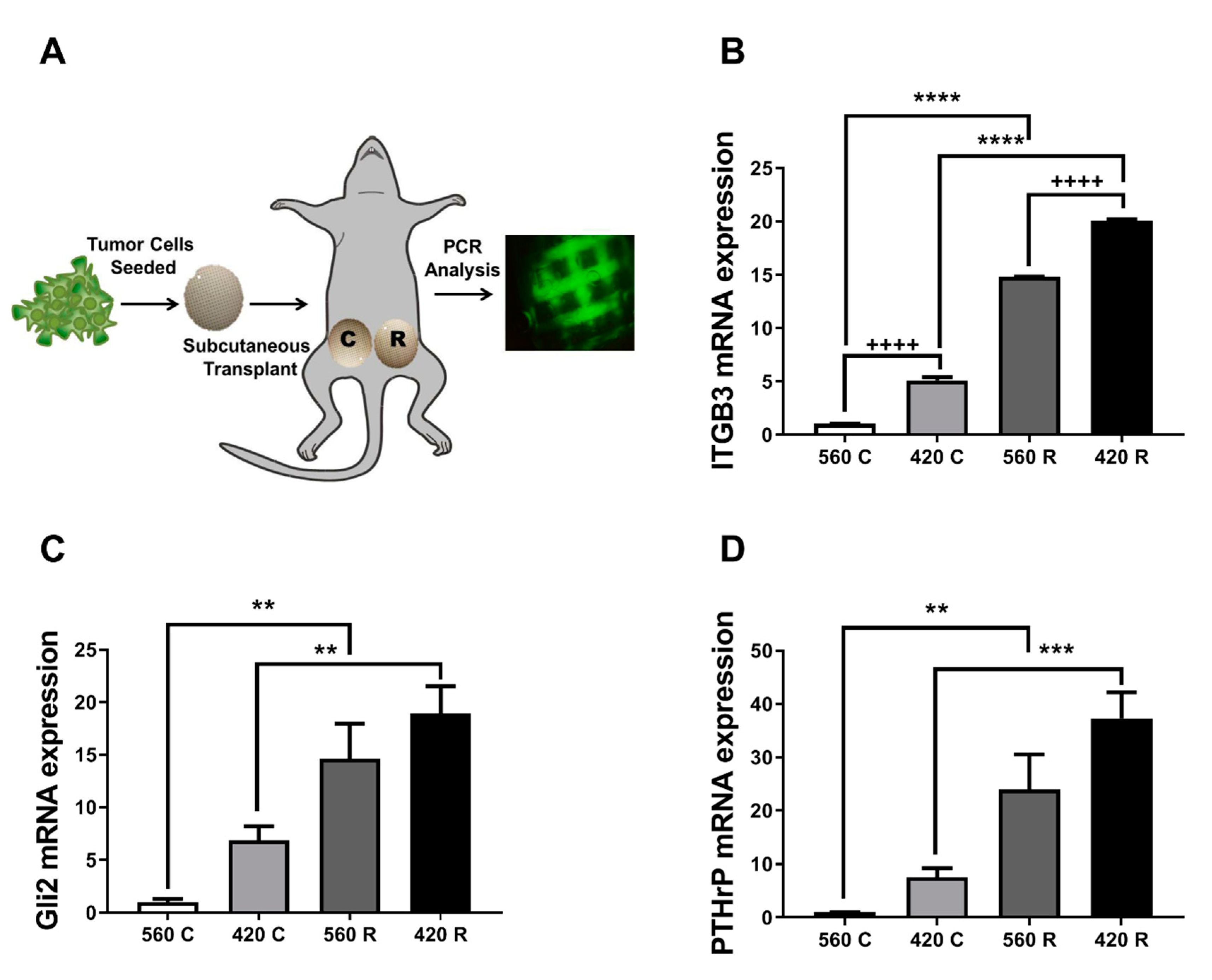
2.6. Substrate Modulus and Pore Size Mediate Expression of Bone-Metastatic Genes in a Breast Cancer Xenograft Model
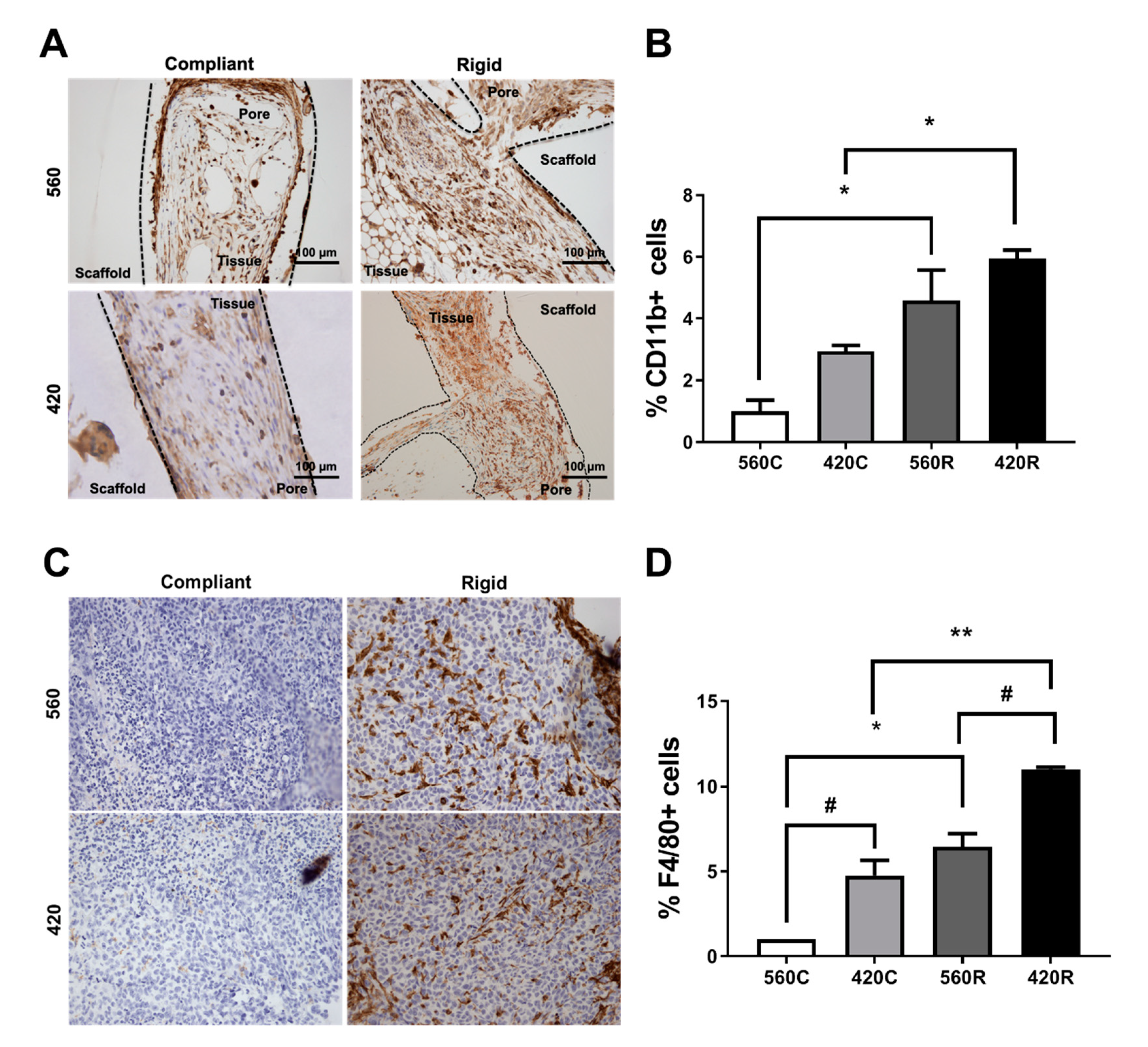
2.7. Substrate Modulus Influences Immune Cell Infiltration
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Synthesis of 2D Poly(Ester Urethane) (PEUR) Films
4.3. Fabrication and Characterization of 3D Scaffolds by templated-Fused Deposition Modeling (t-FDM)
4.4. Cell Culture
4.5. Cell Viability, Proliferation, and Metabolic Activity
4.6. Cell Migration Assays
4.7. Scanning Electron Microscopy
4.8. Drug Treatments
4.9. Quantitative RT-PCR
4.10. In Vivo Studies
4.11. Histomorphometry
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johnson, R.W.; Schipani, E.; Giaccia, A.J. HIF targets in bone remodeling and metastatic disease. Pharmacol. Ther. 2015, 150, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Saad, F.; Lipton, A.; Cook, R.; Chen, Y.-M.; Smith, M.; Coleman, R. Pathologic fractures correlate with reduced survival in patients with malignant bone disease. Cancer 2007, 110, 1860–1867. [Google Scholar] [CrossRef] [PubMed]
- Schrader, J.; Gordon-Walker, T.T.; Aucott, R.L.; Van Deemter, M.; Quaas, A.; Walsh, S.; Benten, D.; Forbes, S.J.; Wells, R.G.; Iredale, J.P. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology 2011, 53, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Tilghman, R.W.; Blais, E.M.; Cowan, C.R.; Sherman, N.E.; Grigera, P.R.; Jeffery, E.D.; Fox, J.W.; Blackman, B.R.; Tschumperlin, D.J.; A Papin, J.; et al. Matrix Rigidity Regulates Cancer Cell Growth by Modulating Cellular Metabolism and Protein Synthesis. PLoS ONE 2012, 7, e37231. [Google Scholar] [CrossRef] [PubMed]
- Page, J.M.; Merkel, A.R.; Ruppender, N.S.; Guo, R.; Dadwal, U.C.; Cannonier, S.A.; Basu, S.; Guelcher, S.A.; Sterling, J.A. Matrix rigidity regulates the transition of tumor cells to a bone-destructive phenotype through integrin β3 and TGF-β receptor type II. Biomaterials 2015, 64, 33–44. [Google Scholar] [CrossRef]
- Ruppender, N.S.; Merkel, A.R.; Martin, T.J.; Mundy, G.R.; Sterling, J.A.; Guelcher, S.A. Matrix Rigidity Induces Osteolytic Gene Expression of Metastatic Breast Cancer Cells. PLoS ONE 2010, 5, e15451. [Google Scholar] [CrossRef]
- Bianchi, M.; Edreira, E.R.U.; Wolke, J.G.C.; Birgani, Z.T.; Habibović, P.; Jansen, J.A.; Tampieri, A.; Marcacci, M.; Leeuwenburgh, S.; Beucken, J.J.J.P.V.D. Substrate geometry directs the in vitro mineralization of calcium phosphate ceramics. Acta Biomater. 2014, 10, 661–669. [Google Scholar] [CrossRef]
- Bidan, C.M.; Kommareddy, K.P.; Rumpler, M.; Kollmannsberger, P.; Bréchet, Y.J.M.; Fratzl, P.; Dunlop, J.W.C. How Linear Tension Converts to Curvature: Geometric Control of Bone Tissue Growth. PLoS ONE 2012, 7, e36336. [Google Scholar] [CrossRef]
- Bidan, C.M.; Kommareddy, K.; Rumpler, M.; Kollmannsberger, P.; Fratzl, P.; Dunlop, J.W.C. Geometry as a Factor for Tissue Growth: Towards Shape Optimization of Tissue Engineering Scaffolds. Adv. Healthc. Mater. 2012, 2, 186–194. [Google Scholar] [CrossRef]
- Gamsjäger, E.; Bidan, C.; Fischer, F.; Fratzl, P.; Dunlop, J.W.C. Modelling the role of surface stress on the kinetics of tissue growth in confined geometries. Acta Biomater. 2013, 9, 5531–5543. [Google Scholar] [CrossRef]
- Vanderburgh, J.; Sterling, J.A.; Guelcher, S.A. 3D Printing of Tissue Engineered Constructs for In Vitro Modeling of Disease Progression and Drug Screening. Ann. Biomed. Eng. 2016, 45, 164–179. [Google Scholar] [CrossRef]
- Cowin, S.C.; Cardoso, L. Blood and interstitial flow in the hierarchical pore space architecture of bone tissue. J. Biomech. 2014, 48, 842–854. [Google Scholar] [CrossRef] [PubMed]
- Fritton, S.P.; Weinbaum, S. Fluid and Solute Transport in Bone: Flow-Induced Mechanotransduction. Annu. Rev. Fluid Mech. 2009, 41, 347–374. [Google Scholar] [CrossRef] [PubMed]
- Bixel, M.G.; Kusumbe, A.P.; Ramasamy, S.; Sivaraj, K.K.; Butz, S.; Vestweber, D.; Adams, R.H. Flow Dynamics and HSPC Homing in Bone Marrow Microvessels. Cell Rep. 2017, 18, 1804–1816. [Google Scholar] [CrossRef] [PubMed]
- Harmata, A.J.; Uppuganti, S.; Granke, M.; Guelcher, S.A.; Nyman, J.S. Compressive fatigue and fracture toughness behavior of injectable, settable bone cements. J. Mech. Behav. Biomed. Mater. 2015, 51, 345–355. [Google Scholar] [CrossRef]
- Lynch, M.E.; Fischbach, C. Biomechanical forces in the skeleton and their relevance to bone metastasis: Biology and engineering considerations. Adv. Drug Deliv. Rev. 2014, 119–134. [Google Scholar] [CrossRef]
- Fischbach, C.; Chen, R.; Matsumoto, T.; Schmelzle, T.; Brugge, J.S.; Polverini, P.J.; Mooney, D.J. Engineering tumors with 3D scaffolds. Nat. Methods 2007, 4, 855–860. [Google Scholar] [CrossRef]
- Schuessler, T.K.; Chan, X.Y.; Chen, H.J.; Ji, K.; Park, K.M.; Roshan-Ghias, A.; Sethi, P.; Thakur, A.; Tian, X.; Villasante, A.; et al. Biomimetic tissue-engineered systems for advancing cancer research: NCI Strategic Workshop report. Cancer Res. 2014, 74, 5359–5363. [Google Scholar] [CrossRef]
- Infanger, D.W.; Lynch, M.E.; Fischbach, C. Engineered Culture Models for Studies of Tumor-Microenvironment Interactions. Annu. Rev. Biomed. Eng. 2013, 15, 29–53. [Google Scholar] [CrossRef]
- Aveic, S.; Davtalab, R.; Vogt, M.; Weber, M.; Buttler, P.; Tonini, G.P.; Fischer, H. Calcium phosphate scaffolds with defined interconnecting channel structure provide a mimetic 3D niche for bone marrow metastasized tumor cell growth. Acta Biomater. 2019, 88, 527–539. [Google Scholar] [CrossRef]
- Pathi, S.P.; Kowalczewski, C.; Tadipatri, R.; Fischbach, C. A Novel 3-D Mineralized Tumor Model to Study Breast Cancer Bone Metastasis. PLoS ONE 2010, 5, e8849. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Guo, J.; Tierney, E.G.; Curtin, C.M.; Malhotra, M.; Darcy, R.; O’Brien, F.J.; O’Driscoll, C.M. The use of collagen-based scaffolds to simulate prostate cancer bone metastases with potential for evaluating delivery of nanoparticulate gene therapeutics. Biomaterials 2015, 66, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.S.; Vomund, A.N.; Phillips, C.L.; Grandbois, M. Structural changes in human type I collagen fibrils investigated by force spectroscopy. Exp. Cell Res. 2004, 299, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhao, X. A 3D model of ovarian cancer cell lines on peptide nanofiber scaffold to explore the cell–scaffold interaction and chemotherapeutic resistance of anticancer drugs. Int. J. Nanomed. 2011, 6, 303–310. [Google Scholar] [CrossRef]
- Fong, E.L.S.; Lamhamedi-Cherradi, S.-E.; Burdett, E.; Ramamoorthy, V.; Lazar, A.J.; Kasper, F.K.; Farach-Carson, M.C.; Vishwamitra, D.; Demicco, E.G.; Menegaz, B.A.; et al. Modeling Ewing sarcoma tumors in vitro with 3D scaffolds. Proc. Natl. Acad. Sci. USA 2013, 110, 6500–6505. [Google Scholar] [CrossRef]
- McCoy, R.J.; O’Brien, F.J. Influence of Shear Stress in Perfusion Bioreactor Cultures for the Development of Three-Dimensional Bone Tissue Constructs: A Review. Tissue Eng. Part B: Rev. 2010, 16, 587–601. [Google Scholar] [CrossRef]
- Guo, R.; Lu, S.; Merkel, A.R.; Sterling, J.A.; Guelcher, S.A. Substrate Modulus Regulates Osteogenic Differentiation of Rat Mesenchymal Stem Cells through Integrin β1 and BMP Receptor Type IA. J. Mater. Chem. B 2016, 4, 3584–3593. [Google Scholar] [CrossRef]
- Guo, R.; Lu, S.; Page, J.M.; Merkel, A.R.; Basu, S.; Sterling, J.A.; Guelcher, S.A. Fabrication of 3D Scaffolds with Precisely Controlled Substrate Modulus and Pore Size by Templated-Fused Deposition Modeling to Direct Osteogenic Differentiation. Adv. Healthc. Mater. 2015, 4, 1826–1832. [Google Scholar] [CrossRef]
- Gupta, M.K.; Meyer, T.A.; Nelson, C.E.; Duvall, C.L. Poly(PS-b-DMA) micelles for reactive oxygen species triggered drug release. J. Control. Release 2012, 162, 591–598. [Google Scholar] [CrossRef]
- Guo, R.; Merkel, A.; Sterling, J.; Davidson, J.; Guelcher, S.A. Substrate modulus of 3D-printed scaffolds regulates the regenerative response in subcutaneous implants through the macrophage phenotype and Wnt signaling. Biomaterials 2015, 73, 85–95. [Google Scholar] [CrossRef]
- Cyster, L.; Grant, D.M.; Howdle, S.; Rose, F.R.A.J.; Irvine, D.; Freeman, D.; Scotchford, C.A.; Shakesheff, K.M. The influence of dispersant concentration on the pore morphology of hydroxyapatite ceramics for bone tissue engineering. Biomaterials 2005, 26, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Karageorgiou, V.; Kaplan, D. Porosity of 3D biomaterial scaffolds and osteogenesis. Biomaterials 2005, 26, 5474–5491. [Google Scholar] [CrossRef] [PubMed]
- Penk, A.; Forster, Y.; Scheidt, H.A.; Nimptsch, A.; Hacker, M.; Schulz, M.B.; Ahnert, P.; Schiller, J.; Rammelt, S.; Huster, D. The pore size of PLGA bone implants determines the de novo formation of bone tissue in tibial head defects in rats. Magn. Reson. Med. 2012, 70, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Santoro, M.; Perale, G. Polymeric scaffolds as stem cell carriers in bone repair. J. Tissue Eng. Regen. Med. 2013, 9, 1093–1119. [Google Scholar] [CrossRef]
- Guise, T.A. Breast cancer bone metastases: It’s all about the neighborhood. Cell 2013, 154, 957–959. [Google Scholar] [CrossRef]
- Zhang, X.H.-F.; Jin, X.; Malladi, S.; Zou, Y.; Wen, Y.H.; Brogi, E.; Smid, M.; Foekens, J.A.; Massagué, J. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell 2013, 154, 1060–1073. [Google Scholar] [CrossRef]
- Yin, J.J.; Selander, K.; Chirgwin, J.M.; Dallas, M.; Grubbs, B.G.; Wieser, R.; Massagué, J.; Mundy, G.R.; A Guise, T. TGF-β signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J. Clin. Investig. 1999, 103, 197–206. [Google Scholar] [CrossRef]
- Buenrostro, D.; Kwakwa, K.A.; Putnam, N.E.; Merkel, A.R.; Johnson, J.R.; Cassat, J.; Sterling, J.A. Early TGF-β inhibition in mice reduces the incidence of breast cancer induced bone disease in a myeloid dependent manner. Bone 2018, 113, 77–88. [Google Scholar] [CrossRef]
- Azarin, S.M.; Yi, J.; Gower, R.M.; Aguado, B.A.; Sullivan, M.E.; Goodman, A.G.; Jiang, E.J.; Rao, S.S.; Ren, Y.; Tucker, S.L.; et al. In vivo capture and label-free detection of early metastatic cells. Nat. Commun. 2015, 6, 8094. [Google Scholar] [CrossRef]
- Roca, H.; McCauley, L.K. Inflammation and skeletal metastasis. BoneKEy Rep. 2015, 4, 706. [Google Scholar] [CrossRef]
- Yang, L.; Huang, J.; Ren, X.; Gorska, A.E.; Chytil, A.; Aakre, M.; Carbone, D.P.; Matrisian, L.M.; Richmond, A.; Lin, P.C.; et al. Abrogation of TGFβ Signaling in Mammary Carcinomas Recruits Gr-1+CD11b+ Myeloid Cells that Promote Metastasis. Cancer Cell 2008, 13, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Mastro, A.M.; Vogler, E.A. A Three-Dimensional Osteogenic Tissue Model for the Study of Metastatic Tumor Cell Interactions with Bone. Cancer Res. 2009, 69, 4097–4100. [Google Scholar] [CrossRef] [PubMed]
- Moreau, J.E.; Anderson, K.; Mauney, J.R.; Nguyen, T.; Kaplan, D.L.; Rosenblatt, M. Tissue-Engineered Bone Serves as a Target for Metastasis of Human Breast Cancer in a Mouse Model. Cancer Res. 2007, 67, 10304–10308. [Google Scholar] [CrossRef] [PubMed]
- Sieh, S.; Taubenberger, A.; Lehman, M.L.; Clements, J.; Nelson, C.C.; Hutmacher, D.W. Paracrine interactions between LNCaP prostate cancer cells and bioengineered bone in 3D in vitro culture reflect molecular changes during bone metastasis. Bone 2014, 63, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Sterling, J.A.; Edwards, J.R.; Martin, T.J.; Mundy, G.R. Advances in the biology of bone metastasis: How the skeleton affects tumor behavior. Bone 2011, 48, 6–15. [Google Scholar] [CrossRef]
- Weilbaecher, K.N.; Guise, T.A.; McCauley, L.K. Cancer to bone: A fatal attraction. Nat. Rev. Cancer 2011, 11, 411–425. [Google Scholar] [CrossRef]
- E Lynch, M.; Brooks, D.; Mohanan, S.; Lee, M.J.; Polamraju, P.; Dent, K.; Bonassar, L.J.; Van Der Meulen, M.C.H.; Fischbach, C. In vivo tibial compression decreases osteolysis and tumor formation in a human metastatic breast cancer model. J. Bone Miner. Res. 2013, 28, 2357–2367. [Google Scholar] [CrossRef]
- Rumpler, M.; Woesz, A.; Dunlop, J.W.C.; Van Dongen, J.T.; Fratzl, P. The effect of geometry on three-dimensional tissue growth. J. R. Soc. Interface 2008, 5, 1173–1180. [Google Scholar] [CrossRef]
- Sanz-Herrera, J.; Moreo, P.; García-Aznar, J.M.; Doblaré, M. On the effect of substrate curvature on cell mechanics. Biomaterials 2009, 30, 6674–6686. [Google Scholar] [CrossRef]
- Pickl, M.; Ries, C.H. Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene 2008, 28, 461–468. [Google Scholar] [CrossRef]
- Mohammad, K.S.; Javelaud, D.; Fournier, P.G.; Niewolna, M.; McKenna, C.R.; Peng, X.H.; Duong, V.; Dunn, L.K.; Mauviel, A.; Guise, T.A. TGF-beta-RI kinase inhibitor SD-208 reduces the development and progression of melanoma bone metastases. Cancer Res. 2010, 71, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Nyman, J.S.; Merkel, A.R.; Uppuganti, S.; Nayak, B.; Rowland, B.; Makowski, A.J.; Oyajobi, B.O.; Sterling, J.A. Combined treatment with a transforming growth factor beta inhibitor (1D11) and bortezomib improves bone architecture in a mouse model of myeloma-induced bone disease. Bone 2016, 91, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Nyman, J.S.; Alvarez, J.; Chakrabarti, A.; Ayres, A.; Sterling, J.; Edwards, J.; Rana, T.; Johnson, R.; Perrien, D.S.; et al. Anti-Transforming Growth Factor ß Antibody Treatment Rescues Bone Loss and Prevents Breast Cancer Metastasis to Bone. PLoS ONE 2011, 6, e27090. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.R.; Nyman, J.S.; Lwin, S.T.; Moore, M.M.; Esparza, J.; O’Quinn, E.C.; Hart, A.J.; Biswas, S.; A Patil, C.; Lonning, S.; et al. Inhibition of TGF-β signaling by 1D11 antibody treatment increases bone mass and quality in vivo. J. Bone Miner. Res. 2010, 25, 2419–2426. [Google Scholar] [CrossRef]
- Zhao, Y.; Bachelier, R.; Treilleux, I.; Pujuguet, P.; Peyruchaud, O.; Baron, R.; Clément-Lacroix, P.; Clezardin, P. Tumor v 3 Integrin Is a Therapeutic Target for Breast Cancer Bone Metastases. Cancer Res. 2007, 67, 5821–5830. [Google Scholar] [CrossRef]
- Nakamura, I.; Pilkington, M.F.; Lakkakorpi, P.T.; Lipfert, L.; Sims, S.M.; Dixon, S.J.; A Rodan, G.; Duong, L.T. Role of alpha(v)beta(3) integrin in osteoclast migration and formation of the sealing zone. J. Cell Sci. 1999, 112, 3985–3993. [Google Scholar]
- Vanderburgh, J.P.; Kwakwa, K.A.; Werfel, T.A.; Merkel, A.R.; Gupta, M.K.; Johnson, R.W.; Guelcher, S.A.; Duvall, C.L.; Rhoades, J.A. Systemic delivery of a Gli inhibitor via polymeric nanocarriers inhibits tumor-induced bone disease. J. Control. Release 2019, 311-312, 257–272. [Google Scholar] [CrossRef]
- Vanderburgh, J.; Hill, J.L.; Gupta, M.K.; Kwakwa, K.A.; Wang, S.K.; Moyer, K.; Bedingfield, S.K.; Merkel, A.R.; D’Arcy, R.; Guelcher, S.A.; et al. Tuning Ligand Density To Optimize Pharmacokinetics of Targeted Nanoparticles for Dual Protection against Tumor-Induced Bone Destruction. ACS Nano 2020, 14, 311–327. [Google Scholar] [CrossRef]
- Sterling, J.A.; Oyajobi, B.O.; Grubbs, B.; Padalecki, S.S.; Munoz, S.A.; Gupta, A.; Story, B.; Zhao, M.; Mundy, G.R. The Hedgehog Signaling Molecule Gli2 Induces Parathyroid Hormone-Related Peptide Expression and Osteolysis in Metastatic Human Breast Cancer Cells. Cancer Res. 2006, 66, 7548–7553. [Google Scholar] [CrossRef]
- Guise, T.A.; Yoneda, T.; Yates, A.J.; Mundy, G.R. The combined effect of tumor-produced parathyroid hormone-related protein and transforming growth factor-alpha enhance hypercalcemia in vivo and bone resorption in vitro. J. Clin. Endocrinol. Metab. 1993, 77, 40–45. [Google Scholar]
- Johnson, R.W.; Nguyen, M.P.; Padalecki, S.S.; Grubbs, B.G.; Merkel, A.R.; Oyajobi, B.O.; Matrisian, L.M.; Mundy, G.R.; Sterling, J.A. TGF-beta promotion of Gli2-induced expression of parathyroid hormone-related protein, an important osteolytic factor in bone metastasis, is independent of canonical Hedgehog signaling. Cancer Res. 2010, 71, 822–831. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.P.; Merkel, A.R.; Masood-Campbell, S.K.; Elefteriou, F.; Sterling, J.A. Models of Bone Metastasis. J. Vis. Exp. 2012, 10, e4260. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]

| Treatment Group | Label | Pore Size (μm) | Bulk Substrate Modulus Ks (MPa) |
|---|---|---|---|
| 2D compliant film | 2DC | N/A | 5 ± 0.4 |
| 2D rigid film | 2DR | N/A | 266 ± 27 |
| 3D compliant scaffolds (small pores) | 420C | 423 ± 34 | 5 ± 0.4 |
| 3D compliant scaffolds (large pores) | 560C | 557 ± 44 | 5 ± 0.4 |
| 3D rigid scaffolds (small pores) | 420R | 423 ± 34 | 266 ± 27 |
| 3D rigid scaffolds (large pores) | 560R | 557 ± 44 | 266 ± 27 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dadwal, U.C.; Merkel, A.R.; Page, J.M.; Kwakwa, K.A.; Kessler, M.; Rhoades, J.A. 3D Bone Morphology Alters Gene Expression, Motility, and Drug Responses in Bone Metastatic Tumor Cells. Int. J. Mol. Sci. 2020, 21, 6913. https://doi.org/10.3390/ijms21186913
Dadwal UC, Merkel AR, Page JM, Kwakwa KA, Kessler M, Rhoades JA. 3D Bone Morphology Alters Gene Expression, Motility, and Drug Responses in Bone Metastatic Tumor Cells. International Journal of Molecular Sciences. 2020; 21(18):6913. https://doi.org/10.3390/ijms21186913
Chicago/Turabian StyleDadwal, Ushashi C., Alyssa R. Merkel, Jonathan M. Page, Kristin A. Kwakwa, Michael Kessler, and Julie A. Rhoades. 2020. "3D Bone Morphology Alters Gene Expression, Motility, and Drug Responses in Bone Metastatic Tumor Cells" International Journal of Molecular Sciences 21, no. 18: 6913. https://doi.org/10.3390/ijms21186913
APA StyleDadwal, U. C., Merkel, A. R., Page, J. M., Kwakwa, K. A., Kessler, M., & Rhoades, J. A. (2020). 3D Bone Morphology Alters Gene Expression, Motility, and Drug Responses in Bone Metastatic Tumor Cells. International Journal of Molecular Sciences, 21(18), 6913. https://doi.org/10.3390/ijms21186913







