Functional Characterization of a Dendrobium officinale Geraniol Synthase DoGES1 Involved in Floral Scent Formation
Abstract
:1. Introduction
2. Results
2.1. Identification of Candidate GES Genes from the D. officinale Genome
2.2. Phylogenetic Analysis of DoGES Proteins in the D. officinale Genome
2.3. Molecular Cloning and Analysis of DoGES1 from D. officinale Flowers
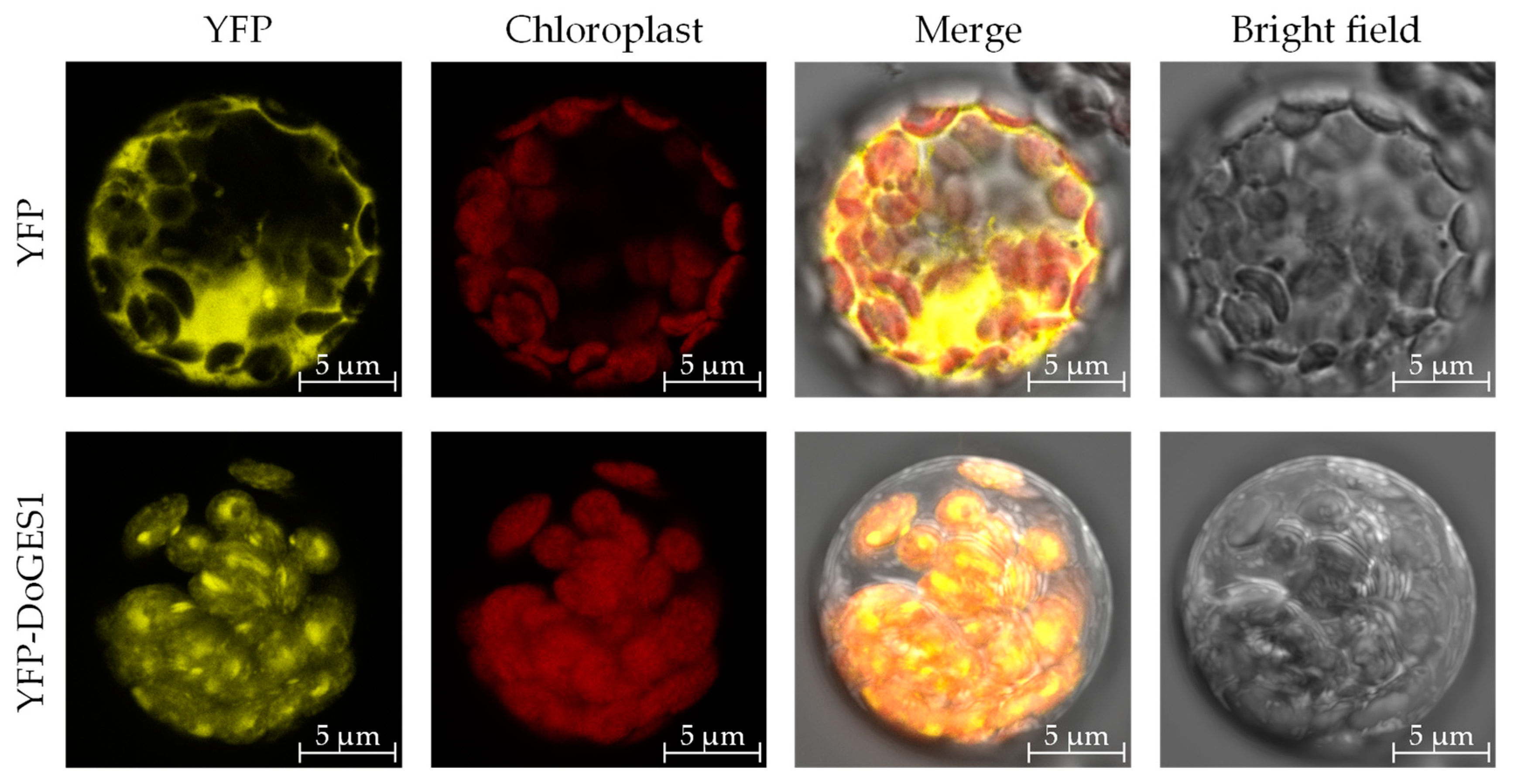
2.4. Subcellular Localization of DoGES1
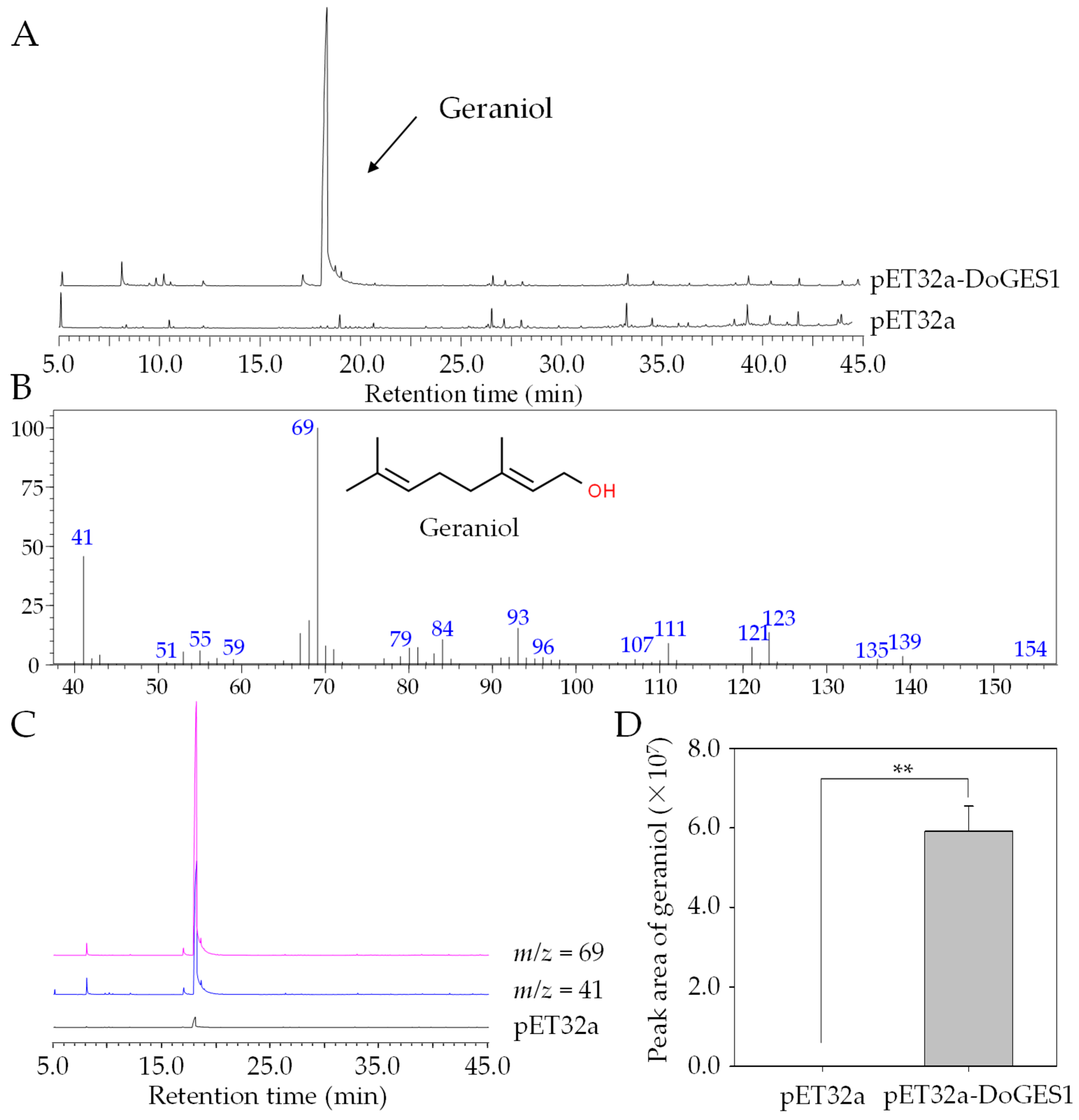
2.5. Functional Characterization of Enzyme Encoded by DoGES1 in Escherichia coli
2.6. Ectopic Expression of DoGES1 in N. benthamiana
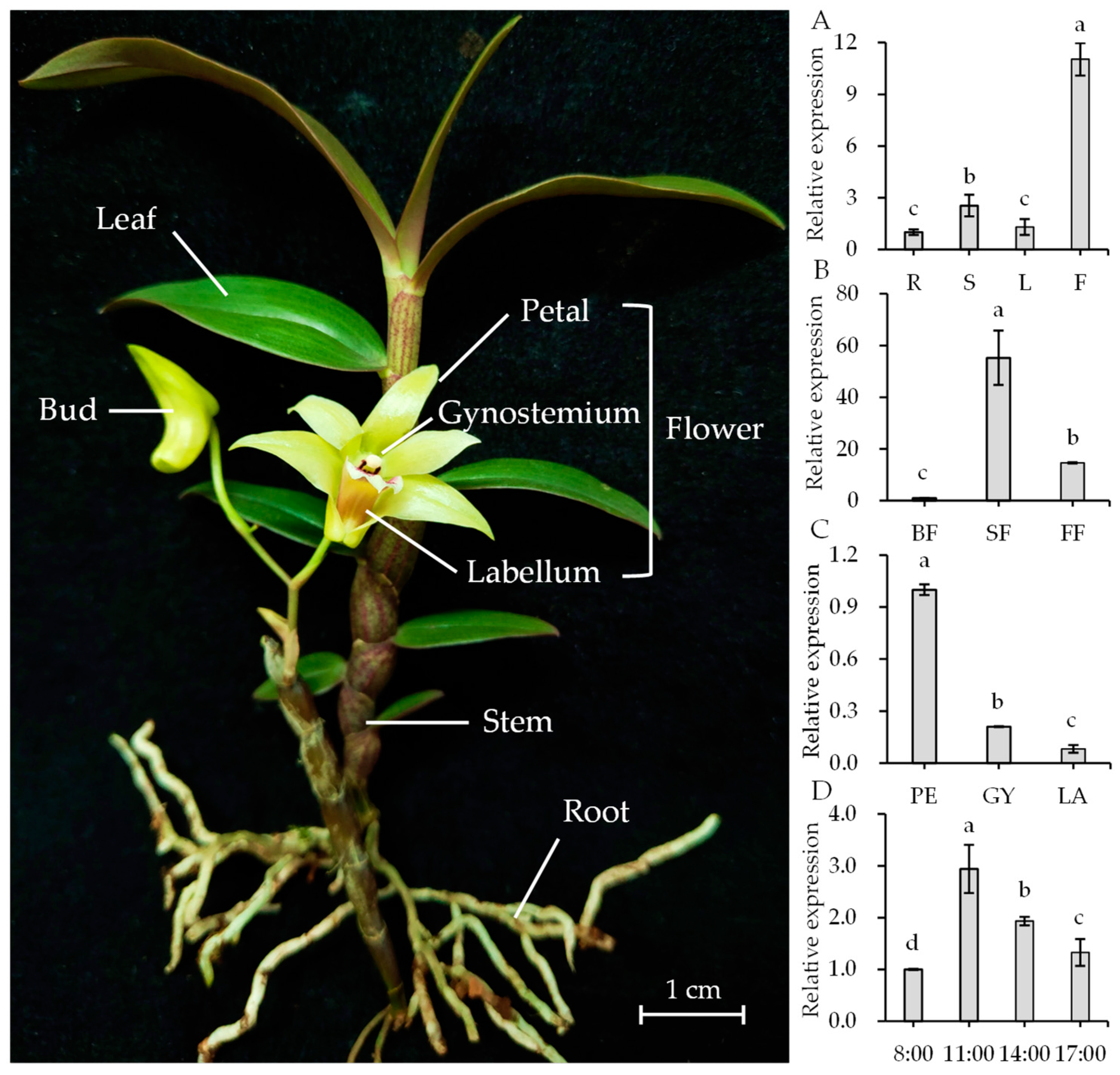
2.7. Temporal-Spatial Expression Patterns Analysis of DoGES1 in D. officinale
2.8. Activation of DoGES1 Gene Expression in Response to Methyl Jasmonate
3. Discussion
4. Materials and Methods
4.1. Plant Materials and MeJA Treatment
4.2. Molecular Cloning of the DoGES1 Gene and Bioinformatics Analysis
4.3. Prokaryotic Expression and Purification of DoGES1 Protein
4.4. In Vitro Enzyme Assay of DoGES1
4.5. Transient Expression of DoGES1 in N. benthamiana
4.6. Quantification of Volatile Monoterpenes Using GC-MS
4.7. Subcellular Location of DoGES1 Protein
4.8. Real-Time Quantitative PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DMAPP | Dimethylallyl diphosphate |
| DXR | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase |
| DXS | 1-Deoxy-d-xylulose 5-phosphate synthase |
| GC–MS | Gas chromatography–mass spectrometry |
| GES | Geraniol synthase |
| GPP | Geranyl pyrophosphate |
| GPPS | GPP synthase |
| HDS | 4-Hydroxy-3-methylbut-2-en-1-yl diphosphate synthase |
| IPP | Isopentenyl diphosphate |
| IPTG | Isopropyl-β-d-thiogalactopyranoside |
| MeJA | Methyl jasmonate |
| MEP | Methylerythritol phosphate |
| MVA | Mevalonic acid |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| YFP | Yellow fluorescent protein |
References
- Goff, S.A.; Klee, H.J. Plant volatile compounds: Sensory cues for health and nutritional value? Science 2006, 311, 815–819. [Google Scholar] [CrossRef]
- Holopainen, J.K.; Gershenzon, J. Multiple stress factors and the emission of plant VOCs. Trends Plant Sci. 2010, 15, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, H.; Schuurink, R.C.; Bleeker, P.M.; Schiestl, F. The role of volatiles in plant communication. Plant J. 2019, 100, 892–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramya, M.; Jang, S.; An, H.R.; Lee, S.Y.; Park, P.M.; Park, P.H. Volatile organic compounds from orchids: From synthesis and function to gene regulation. Int. J. Mol. Sci. 2020, 21, 1160. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, Y.Y.; Pan, Z.J.; Hsu, C.C.; Yang, Y.P.; Hsu, Y.C.; Chuang, Y.C.; Shih, H.H.; Chen, W.H.; Tsai, W.C.; Chen, H.H. Research on orchid biology and biotechnology. Plant Cell Physiol. 2011, 52, 1467–1486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iijima, Y.; Gang, D.R.; Fridman, E.; Lewinsohn, E.; Pichersky, E. Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol. 2004, 134, 370–379. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Zhao, C.; Zhang, G.; Teixeira da Silva, J.A.; Duan, J. Genome-wide identification and expression profile of TPS gene family in Dendrobium officinale and the role of DoTPS10 in linalool biosynthesis. Int. J. Mol. Sci. 2020, 21, 5419. [Google Scholar] [CrossRef] [PubMed]
- Chacón, M.G.; Marriott, A.; Kendrick, E.G.; Styles, M.Q.; Leak, D.J. Esterification of geraniol as a strategy for increasing product titre and specificity in engineered Escherichia coli. Microb. Cell Factories 2019, 18, 105. [Google Scholar] [CrossRef]
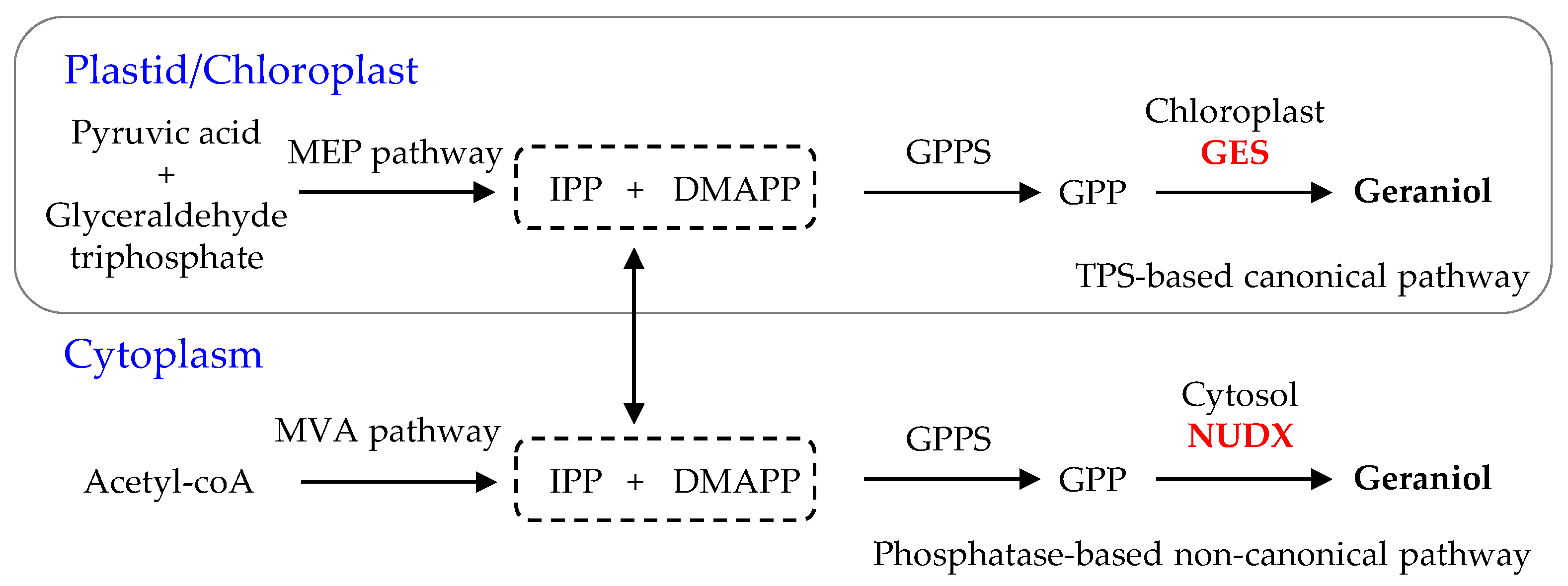
- Sun, P.; Schuurink, R.C.; Caissard, J.C.; Hugueney, P.; Baudino, S. My way: Noncanonical biosynthesis pathway for plant volatiles. Trends Plant Sci. 2016, 21, 884–894. [Google Scholar] [CrossRef]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef]
- Magnard, J.L.; Roccia, A.; Caissard, J.C.; Vergne, P.; Sun, P.; Hecquet, R.; Dubois, A.; Hibrand-Saint Oyant, L.; Jullien, F.; Nicolè, F.; et al. Plant volatiles. Biosynthesis of monoterpene scent compounds in roses. Science 2015, 349, 81–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Xu, Y.; Shen, S.; Yin, X.; Klee, H.; Zhang, B.; Chen, K.; Hancock, R. Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J. Exp. Bot. 2017, 68, 4929–4938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, T.; Shadrack, K.; Yang, S.; Xue, X.; Li, S.; Wang, N.; Wang, Q.; Wang, L.; Gao, X.; Cronk, Q. Functional characterization of terpene synthases accounting for the volatilized-terpene heterogeneity in Lathyrus odoratus cultivar flowers. Plant Cell Physiol. 2020, 61, pcaa100. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, Y.Y.; Jeng, M.F.; Tsai, W.C.; Chuang, Y.C.; Li, C.Y.; Wu, T.S.; Kuoh, C.S.; Chen, W.H.; Chen, H.H. A novel homodimeric geranyl diphosphate synthase from the orchid Phalaenopsis bellina lacking a DD(X)2-4D motif. Plant J. 2008, 55, 719–733. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.C.; Hung, Y.C.; Tsai, W.C.; Chen, W.H.; Chen, H.H. PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids. J. Exp. Bot. 2018, 69, 4363–4377. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Wang, X.; Liu, H.; Tian, Y.; Lian, J.; Yang, R.; Hao, S.; Wang, X.; Yang, S.; Li, Q.; et al. The genome of Dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb. Mol. Plant 2015, 8, 922–934. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Xu, Q.; Bian, C.; Tsai, W.C.; Yeh, C.M.; Liu, K.; Yoshida, K.; Zhang, L.; Chang, S.; Chen, F.; et al. The Dendrobium catenatum Lindl genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.Y.; Jin, J.; Sarojam, R.; Ramachandran, S.A. Comprehensive survey on the terpene synthase gene family provides new insight into its evolutionary patterns. Genome Biol. Evol. 2019, 11, 2078–2098. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Bohlmann, J.; Meyer-Gauen, G.; Croteau, R. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 1998, 95, 4126–4133. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Liu, B.; Li, M.; Gao, X.; Fang, Q.; Liu, C.; Ding, H.; Wang, L.; Gao, X. Identification and characterization of terpene synthase genes accounting for volatile terpene emissions in flowers of Freesia × hybrida. J. Exp. Bot. 2018, 69, 4249–4265. [Google Scholar] [CrossRef] [Green Version]
- Kaundal, R.; Saini, R.; Zhao, P.X. Combining machine learning and homology-based approaches to accurately predict subcellular localization in Arabidopsis. Plant Physiol. 2010, 154, 36–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, X.; Xiao, X.; Chou, K.C. PLoc-mPlant: Predict subcellular localization of multi-location plant proteins by incorporating the optimal GO information into general PseAAC. Mol. Biosyst. 2017, 13, 1722–1727. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Guo, Y.; Shi, X.; Yang, Y.; Chen, J.; Zhang, Q.; Sun, M. Overexpression of LiTPS2 from a cultivar of lily (Lilium ‘Siberia′) enhances the monoterpenoids content in tobacco flowers. Plant Physiol. Biochem. 2020, 151, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.M.; Gershenzon, J.; Bohlmann, J. Induction of volatile terpene biosynthesis and diurnal emission by methyl jasmonate in foliage of Norway spruce. Plant Physiol. 2003, 132, 1586–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Viljoen, A.M. Geraniol—A review of a commercially important fragrance material. S. Afr. J. Bot. 2010, 76, 643–651. [Google Scholar] [CrossRef] [Green Version]
- Vranová, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef]
- Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef]
- Ye, P.; Liang, S.; Wang, X.; Duan, L.; Jiang-Yan, F.; Yang, J.; Zhan, R.; Ma, D. Transcriptome analysis and targeted metabolic profiling for pathway elucidation and identification of a geraniol synthase involved in iridoid biosynthesis from Gardenia jasminoides. Ind. Crop Prod. 2019, 132, 48–58. [Google Scholar] [CrossRef]
- Hamachi, A.; Nisihara, M.; Saito, S.; Rim, H.; Takahashi, H.; Islam, M.; Uemura, T.; Ohnishi, T.; Ozawa, R.; Maffei, M.E.; et al. Overexpression of geraniol synthase induces heat stress susceptibility in Nicotiana tabacum. Planta 2019, 249, 235–249. [Google Scholar] [CrossRef]
- Liu, J.; Guan, Z.; Liu, H.; Qi, L.; Zhang, D.; Zou, T.; Yin, P. Structural insights into the substrate recognition mechanism of Arabidopsis GPP-bound NUDX1 for noncanonical monoterpene biosynthesis. Mol. Plant 2018, 11, 218–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, F.; Pichersky, E. More is better: The diversity of terpene metabolism in plants. Curr. Opin. Plant Biol. 2020, 55, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Miettinen, K.; Goedbloed, M.; Verstappen, F.W.; Voster, A.; Jongsma, M.A.; Memelink, J.; van der Krol, S.; Bouwmeester, H.J. Characterization of two geraniol synthases from Valeriana officinalis and Lippia dulcis: Similar activity but difference in subcellular localization. Metab. Eng. 2013, 20, 198–211. [Google Scholar] [CrossRef]
- Yue, Y.; Yu, R.; Fan, Y. Characterization of two monoterpene synthases involved in floral scent formation in Hedychium coronarium. Planta 2014, 240, 745–762. [Google Scholar] [CrossRef] [PubMed]
- Nagegowda, D.A.; Gutensohn, M.; Wilkerson, C.G.; Dudareva, N. Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J. 2008, 55, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, H.; Wang, X.; Li, K.; Dong, M.; Li, Y.; Zhu, Q.; Shang, F. Mechanism of floral scent production in Osmanthus fragrans and the production and regulation of its key floral constituents, β-ionone and linalool. Hortic. Res. 2019, 6, 106. [Google Scholar] [CrossRef] [Green Version]
- Flach, A.; Dondon, R.C.; Singer, R.B.; Koehler, S.; Amaral Mdo, C.; Marsaioli, A.J. The chemistry of pollination in selected Brazilian Maxillariinae orchids: Floral rewards and fragrance. J. Chem. Ecol. 2004, 30, 1045–1056. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Lee, M.C.; Chang, Y.L.; Chen, W.H.; Chen, H.H. Diurnal regulation of the floral scent emission by light and circadian rhythm in the Phalaenopsis orchids. Bot. Stud. 2017, 58, 50–58. [Google Scholar] [CrossRef]
- Zhou, H.; Shamala, L.F.; Yi, X.; Yan, Z.; Wei, S. Analysis of terpene synthase family genes in Camellia sinensis with an emphasis on abiotic stress conditions. Sci. Rep. 2020, 10, 933. [Google Scholar] [CrossRef]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Yu, Z.; Zhang, G.; Teixeira da Silva, J.A.; Yang, Z.; Duan, J. The β-1,3-galactosetransferase gene DoGALT2 is essential for stigmatic mucilage production in Dendrobium officinale. Plant Sci. 2019, 287, 110179. [Google Scholar] [CrossRef]
- Yu, Z.; He, C.; Teixeira da Silva, J.A.; Luo, J.; Yang, Z.; Duan, J. The GDP-mannose transporter gene (DoGMT) from Dendrobium officinale is critical for mannan biosynthesis in plant growth and development. Plant Sci. 2018, 277, 43–54. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, C.; Yu, Z.; Silva, J.A.T.d.; He, C.; Wang, H.; Si, C.; Zhang, M.; Zeng, D.; Duan, J. Functional Characterization of a Dendrobium officinale Geraniol Synthase DoGES1 Involved in Floral Scent Formation. Int. J. Mol. Sci. 2020, 21, 7005. https://doi.org/10.3390/ijms21197005
Zhao C, Yu Z, Silva JATd, He C, Wang H, Si C, Zhang M, Zeng D, Duan J. Functional Characterization of a Dendrobium officinale Geraniol Synthase DoGES1 Involved in Floral Scent Formation. International Journal of Molecular Sciences. 2020; 21(19):7005. https://doi.org/10.3390/ijms21197005
Chicago/Turabian StyleZhao, Conghui, Zhenming Yu, Jaime A. Teixeira da Silva, Chunmei He, Haobin Wang, Can Si, Mingze Zhang, Danqi Zeng, and Jun Duan. 2020. "Functional Characterization of a Dendrobium officinale Geraniol Synthase DoGES1 Involved in Floral Scent Formation" International Journal of Molecular Sciences 21, no. 19: 7005. https://doi.org/10.3390/ijms21197005
APA StyleZhao, C., Yu, Z., Silva, J. A. T. d., He, C., Wang, H., Si, C., Zhang, M., Zeng, D., & Duan, J. (2020). Functional Characterization of a Dendrobium officinale Geraniol Synthase DoGES1 Involved in Floral Scent Formation. International Journal of Molecular Sciences, 21(19), 7005. https://doi.org/10.3390/ijms21197005




