Melanocortin Receptor 4 (MC4R) Signaling System in Nile Tilapia
Abstract
1. Introduction
2. Results
2.1. Cloning of Tilapia MC4R, MRAP2b, POMCa1, POMCb, AgRP, and AgRP2 Coding Regions
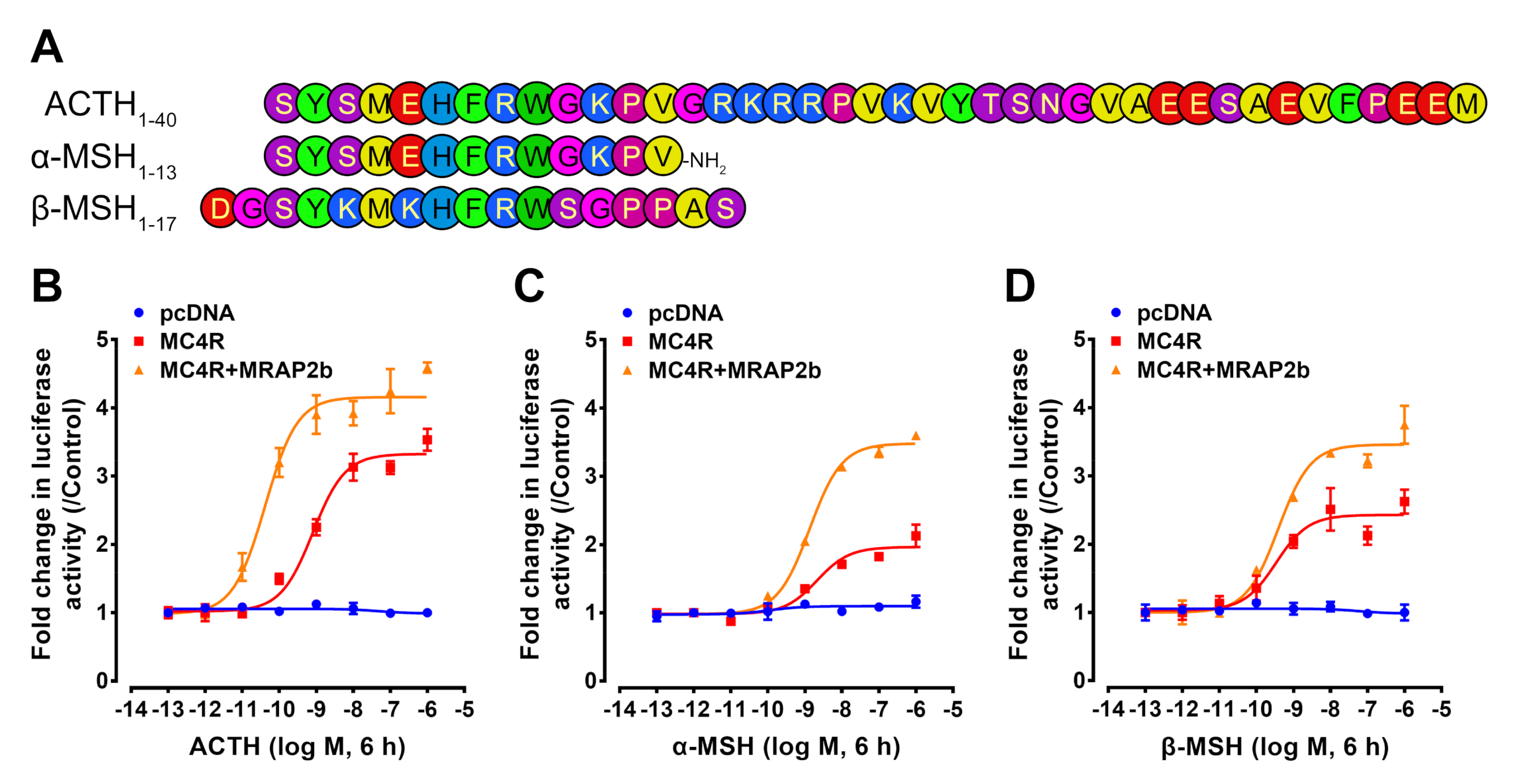
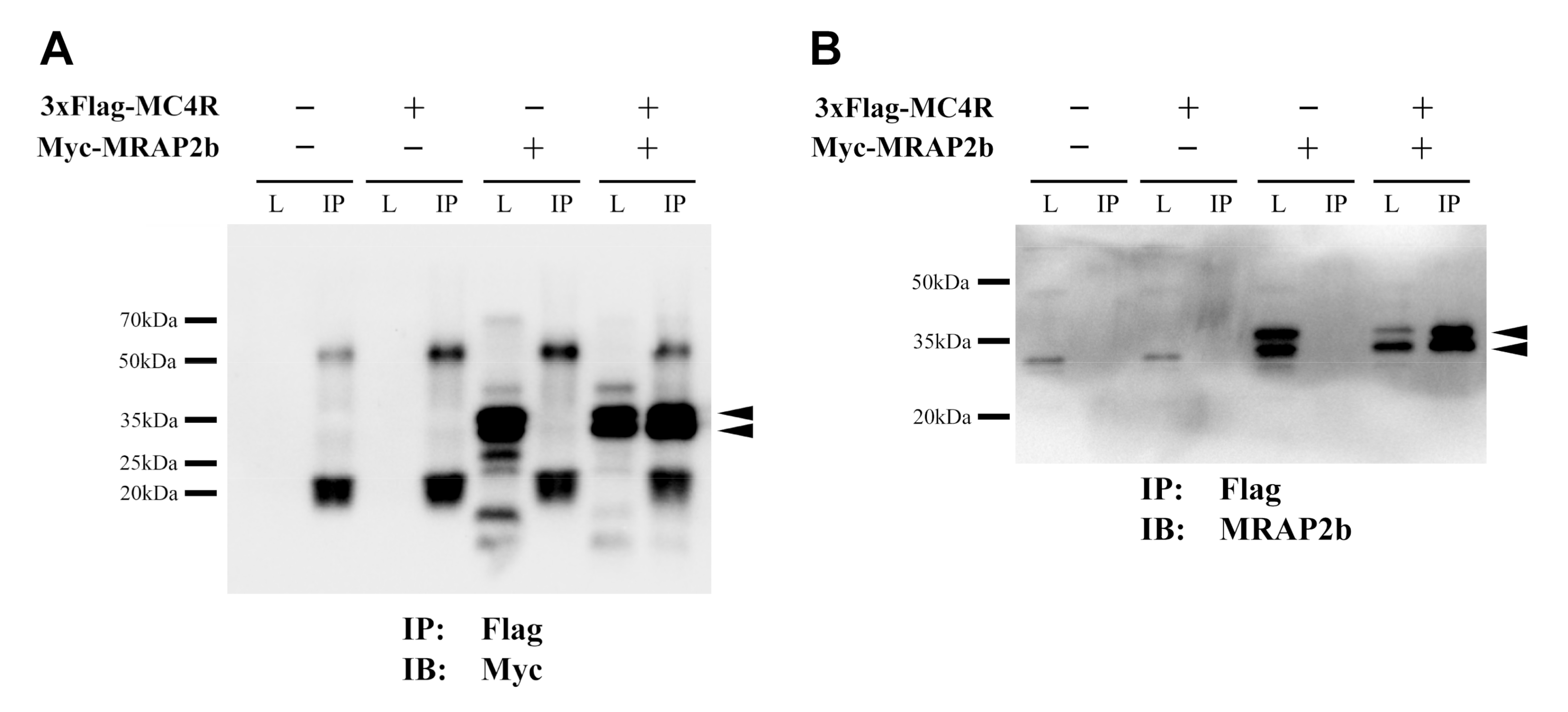
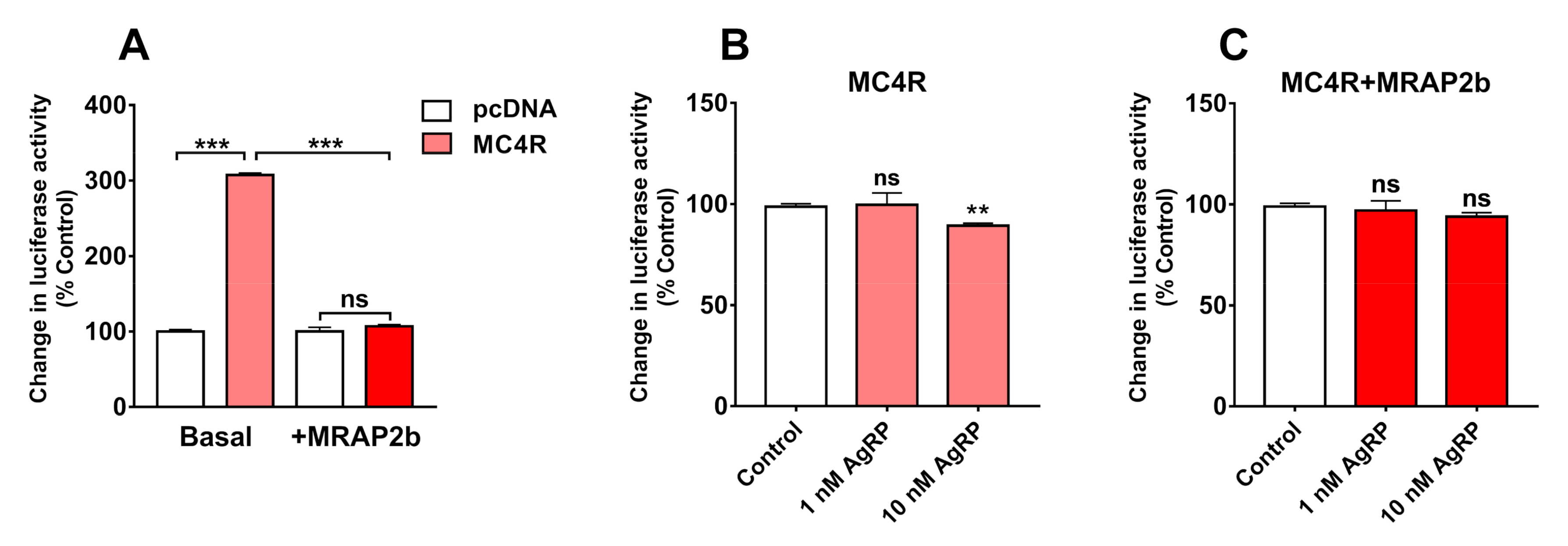
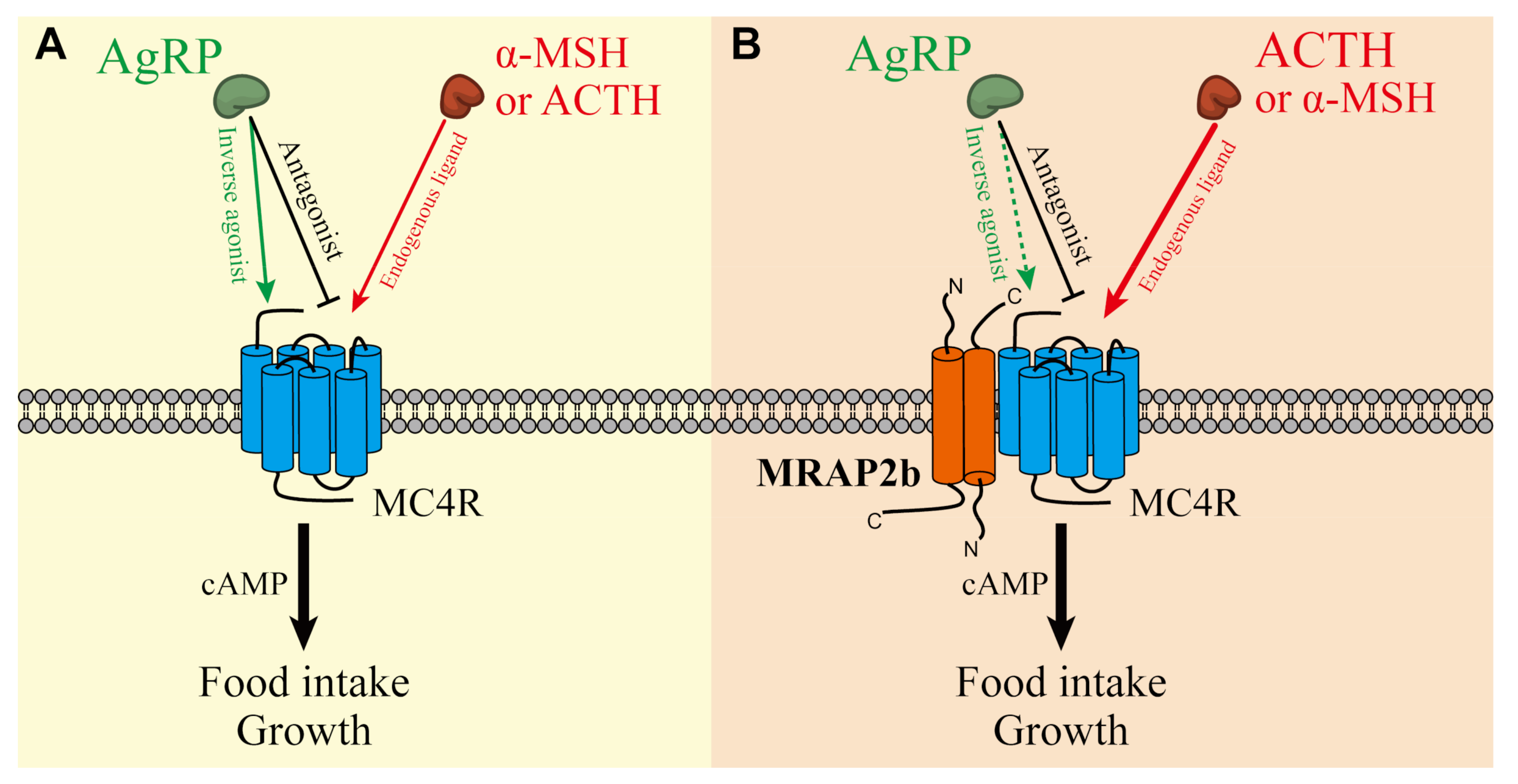
2.2. Functionality of Tilapia MC4R in the Absence or Presence of MRAP2b
2.3. AgRP Can Act as an Inverse Agonist of Tilapia MC4R
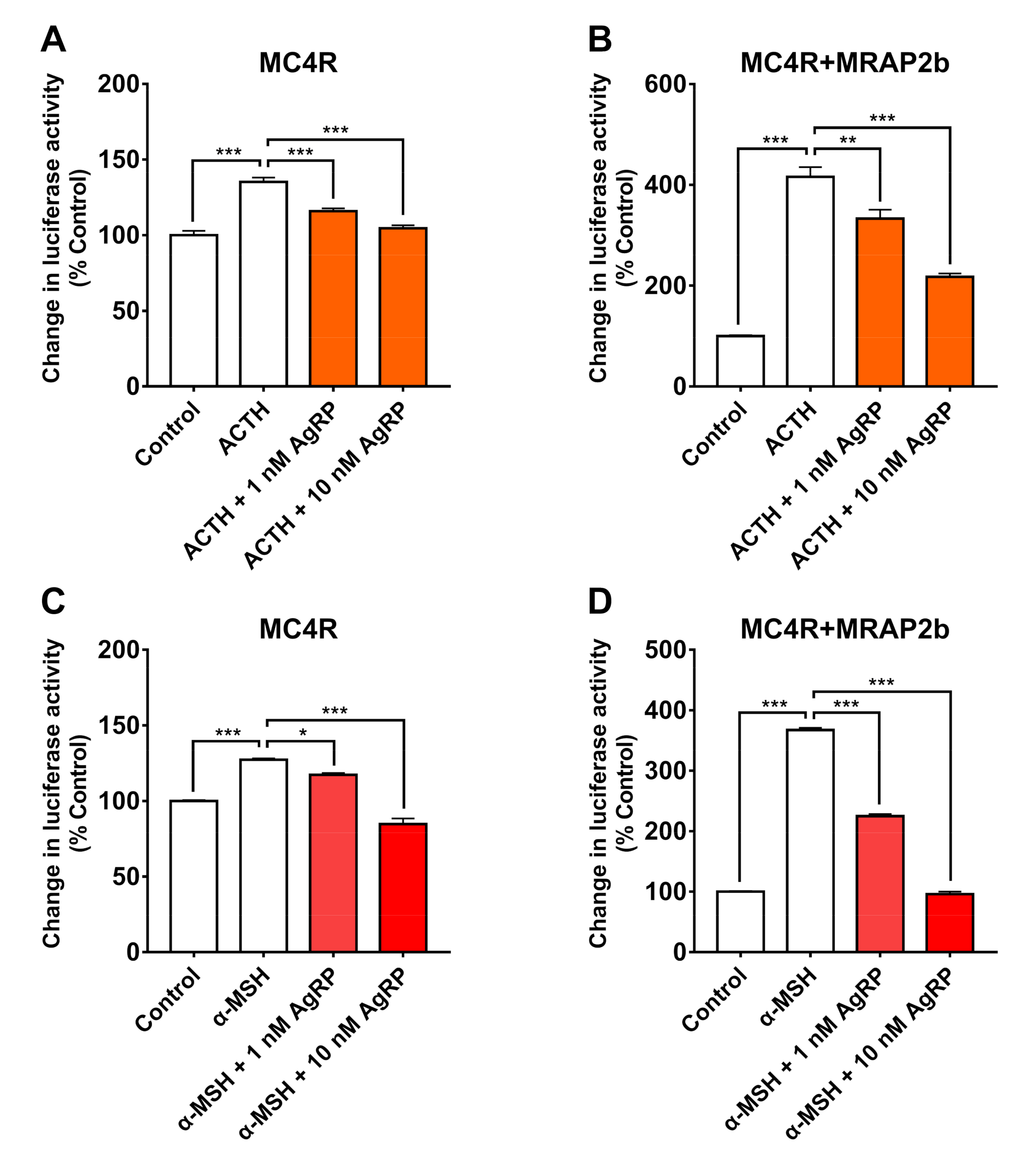
2.4. AgRP Can Antagonize ACTH/α-MSH Actions on MC4R
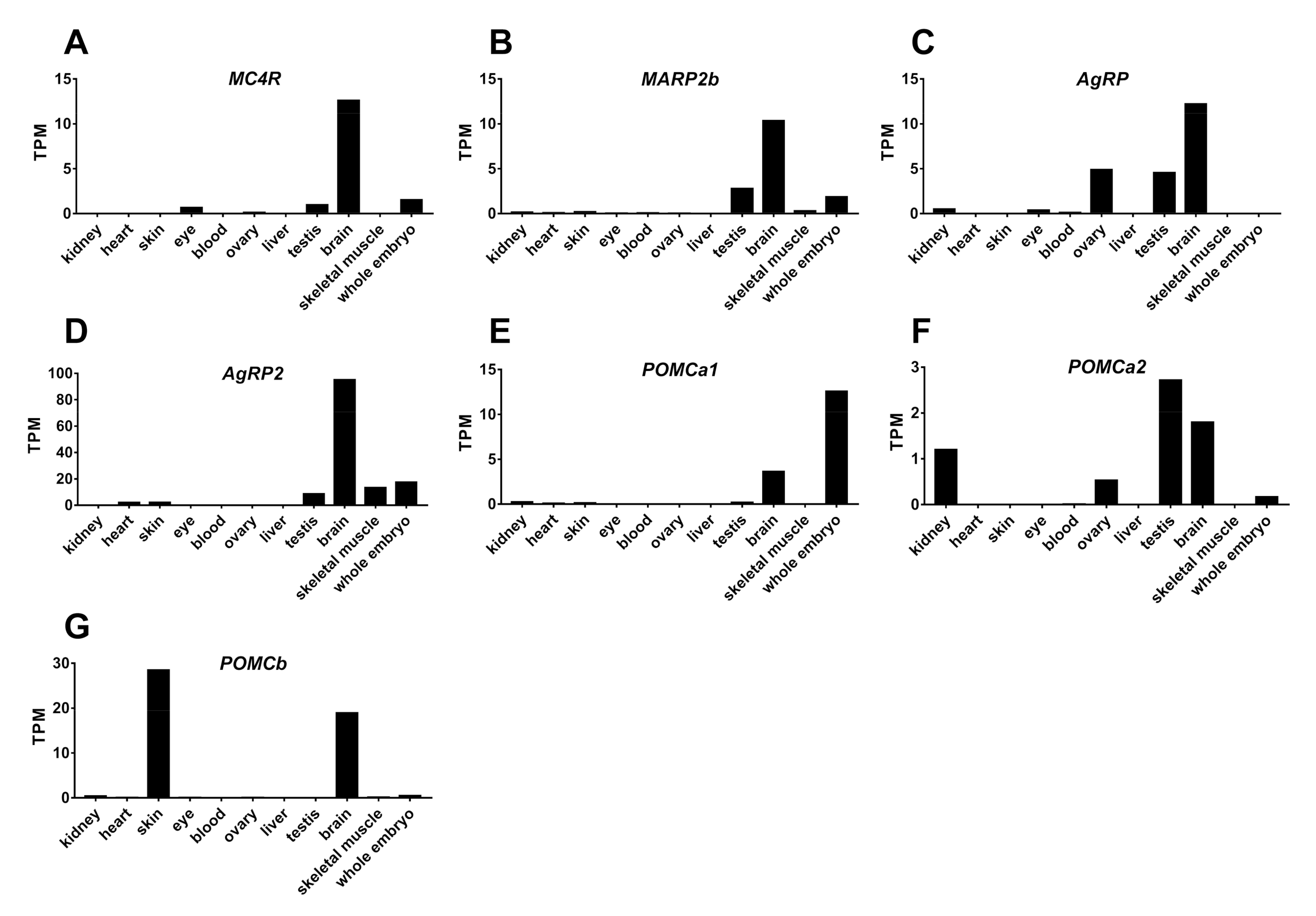
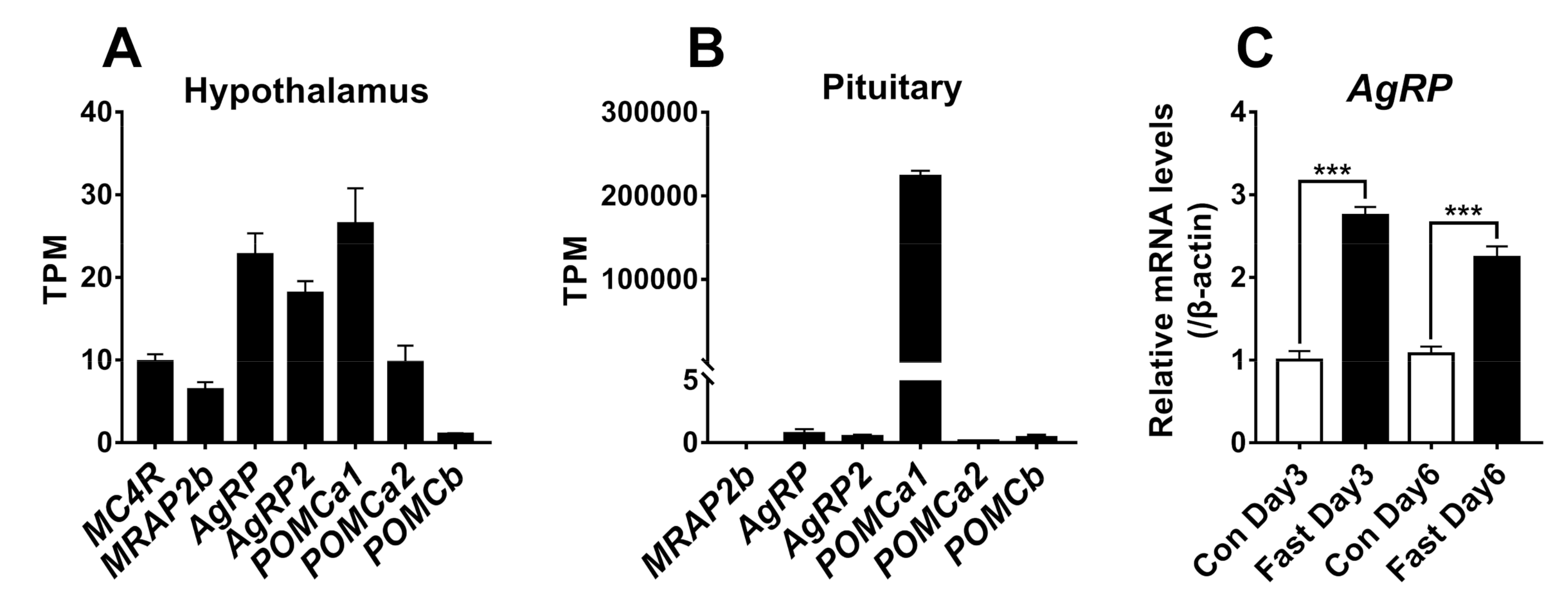
2.5. Tissue Expression of MC4R, MRAP2b, POMCs, and AgRPs in Tilapia
2.6. Fasting Induces Hypothalamic AgRP Expression in Growing Tilapia
3. Discussion
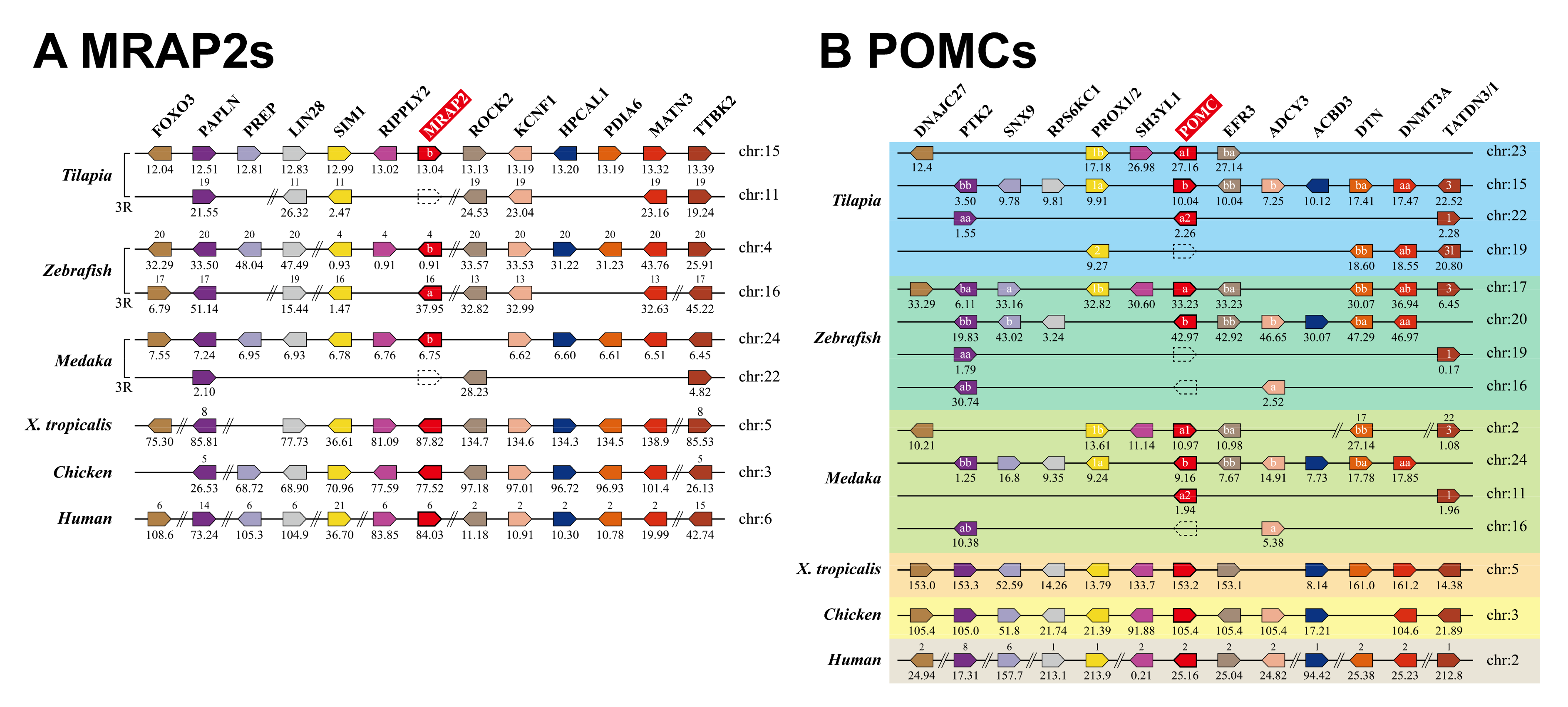
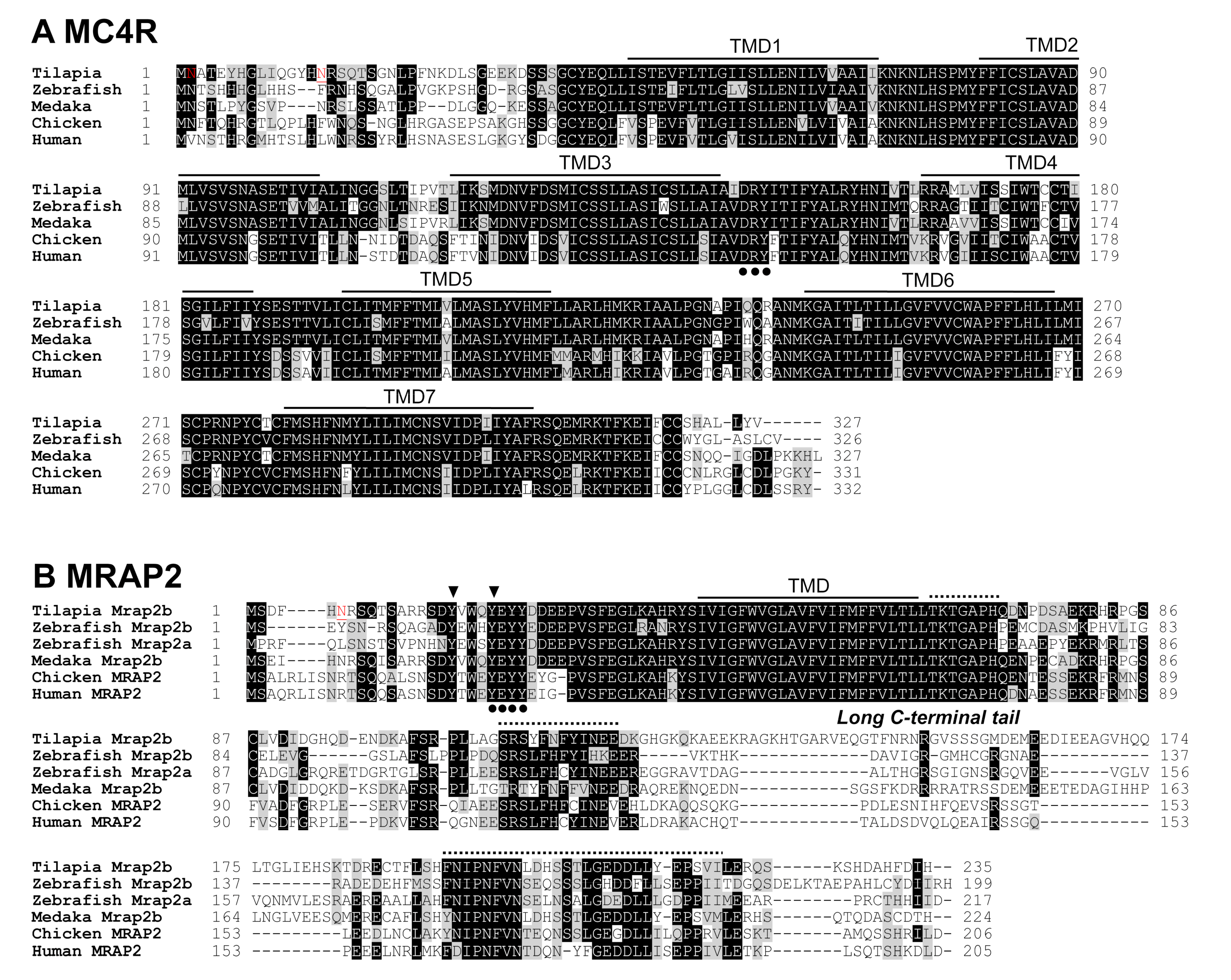
3.1. Identification of MC4R Signaling System in Nile Tilapia
3.2. The Functionality of the Tilapia MC4R Signaling System
3.3. Tissue Expression of MC4R Signaling System in Tilapia
4. Materials and Methods
4.1. Chemicals, Primers, Peptides, and Antibodies
4.2. Animals, Tissues, and Ethical Statement
4.3. Total RNA Extraction and Reverse Transcription (RT)
4.4. Cloning of Tilapia MC4R, MRAP2, POMCs, and AgRPs
4.5. RNA-Seq Analysis of MC4R, MRAP2b, POMCs, and AgRPs in Tilapia Tissues
4.6. Quantitative Real-Time RT-PCR Assay (qRT-PCR)
4.7. Functional Analysis of tiMC4R and tiMRAP2b
4.8. Co-Immunoprecipitation (Co-IP) Assay
4.9. Detection of the Inverse Agonistic and Antagonistic Actions of AgRP
4.10. Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cone, R.D. Studies on the physiological functions of the melanocortin system. Endocr. Rev. 2006, 27, 736–749. [Google Scholar] [CrossRef] [PubMed]
- Asai, M.; Ramachandrappa, S.; Joachim, M.; Shen, Y.; Zhang, R.; Nuthalapati, N.; Ramanathan, V.; Strochlic, D.E.; Ferket, P.; Linhart, K.; et al. Loss of function of the melanocortin 2 receptor accessory protein 2 is associated with mammalian obesity. Science 2013, 341, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Josep Agulleiro, M.; Cortes, R.; Fernandez-Duran, B.; Navarro, S.; Guillot, R.; Meimaridou, E.; Clark, A.J.; Cerda-Reverter, J.M. Melanocortin 4 receptor becomes an ACTH receptor by coexpression of melanocortin receptor accessory protein 2. Mol. Endocrinol. 2013, 27, 1934–1945. [Google Scholar] [CrossRef]
- Sebag, J.A.; Zhang, C.; Hinkle, P.M.; Bradshaw, A.M.; Cone, R.D. Developmental control of the melanocortin-4 receptor by MRAP2 proteins in zebrafish. Science 2013, 341, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, X.; Zhou, Y.; Cui, L.; Li, J.; Wu, C.; Wan, Y.; Li, J.; Wang, Y. The interaction of MC3R and MC4R with MRAP2, ACTH, alpha-MSH and AgRP in chickens. J. Endocrinol. 2017, 234, 155–174. [Google Scholar] [CrossRef]
- Liu, R.; Kinoshita, M.; Adolfi, M.C.; Schartl, M. Analysis of the Role of the Mc4r System in Development, Growth, and Puberty of Medaka. Front. Endocrinol. 2019, 10, 213. [Google Scholar] [CrossRef]
- Dores, R.M.; Baron, A.J. Evolution of POMC: Origin, phylogeny, posttranslational processing, and the melanocortins. Ann. N. Y. Acad. Sci. 2011, 1220, 34–48. [Google Scholar] [CrossRef]
- Kleinau, G.; Heyder, N.A.; Tao, Y.-X.; Scheerer, P. Structural Complexity and Plasticity of Signaling Regulation at the Melanocortin-4 Receptor. Int. J. Mol. Sci. 2020, 21, 5728. [Google Scholar] [CrossRef]
- Sebag, J.A.; Hinkle, P.M. Regions of melanocortin 2 (MC2) receptor accessory protein necessary for dual topology and MC2 receptor trafficking and signaling. J. Biol. Chem. 2009, 284, 610–618. [Google Scholar] [CrossRef]
- Huszar, D.; Lynch, C.A.; Fairchild-Huntress, V.; Dunmore, J.H.; Fang, Q.; Berkemeier, L.R.; Gu, W.; Kesterson, R.A.; Boston, B.A.; Cone, R.D.; et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 1997, 88, 131–141. [Google Scholar] [CrossRef]
- Ollmann, M.M.; Wilson, B.D.; Yang, Y.K.; Kerns, J.A.; Chen, Y.; Gantz, I.; Barsh, G.S. Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science 1997, 278, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.; Shutter, J.R.; Sarmiento, U.; Sarosi, I.; Stark, K.L. Overexpression of Agrt leads to obesity in transgenic mice. Nat. Genet. 1997, 17, 273–274. [Google Scholar] [CrossRef] [PubMed]
- Vastermark, A.; Krishnan, A.; Houle, M.E.; Fredriksson, R.; Cerda-Reverter, J.M.; Schioth, H.B. Identification of distant Agouti-like sequences and re-evaluation of the evolutionary history of the Agouti-related peptide (AgRP). PLoS ONE 2012, 7, e40982. [Google Scholar] [CrossRef] [PubMed]
- Dores, R.M. Hypothesis and Theory: Revisiting Views on the Co-evolution of the Melanocortin Receptors and the Accessory Proteins, MRAP1 and MRAP2. Front. Endocrinol. 2016, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.Z.; Chen, R.; Zhang, Y.; Tao, Y.X. Orange-spotted grouper melanocortin-4 receptor: Modulation of signaling by MRAP2. Gen. Comp. Endocrinol. 2019, 284, 113234. [Google Scholar] [CrossRef] [PubMed]
- Volkoff, H. The Neuroendocrine Regulation of Food Intake in Fish: A Review of Current Knowledge. Front. Neurosci. 2016, 10, 540. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Cone, R.D. Creation of a genetic model of obesity in a teleost. FASEB J. 2007, 21, 2042–2049. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Song, Y.; Wang, Y.; Zhang, T.; Duan, M.; Li, Y.; Liao, L.; Zhu, Z.; Hu, W. Increased food intake in growth hormone-transgenic common carp (Cyprinus carpio L.) may be mediated by upregulating Agouti-related protein (AgRP). Gen. Comp. Endocrinol. 2013, 192, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Cerda-Reverter, J.M.; Schioth, H.B.; Peter, R.E. The central melanocortin system regulates food intake in goldfish. Regul. Pept. 2003, 115, 101–113. [Google Scholar] [CrossRef]
- White, S.L.; Volkoff, H.; Devlin, R.H. Regulation of feeding behavior and food intake by appetite-regulating peptides in wild-type and growth hormone-transgenic coho salmon. Horm. Behav. 2016, 84, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, J.; Wu, C.; Hu, Z.; An, L.; Wan, Y.; Fang, C.; Zhang, X.; Li, J.; Wang, Y. The Asp298Asn polymorphism of melanocortin-4 receptor (MC4R) in pigs: Evidence for its potential effects on MC4R constitutive activity and cell surface expression. Anim. Genet. 2020, 51, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Amano, M.; Itoh, T.; Yasuda, A.; Yamanome, T.; Amemiya, Y.; Sasaki, K.; Sakai, M.; Yamamori, K.; Kawauchi, H. Nucleotide sequence and expression of three subtypes of proopiomelanocortin mRNA in barfin flounder. Gen. Comp. Endocrinol. 2005, 141, 291–303. [Google Scholar] [CrossRef]
- Harris, R.M.; Dijkstra, P.D.; Hofmann, H.A. Complex structural and regulatory evolution of the pro-opiomelanocortin gene family. Gen. Comp. Endocrinol. 2014, 195, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Golling, G.; Thacker, T.L.; Cone, R.D. Agouti-related protein (AGRP) is conserved and regulated by metabolic state in the zebrafish, Danio rerio. Endocrine 2003, 22, 257–265. [Google Scholar] [CrossRef]
- Zhang, C.; Song, Y.; Thompson, D.A.; Madonna, M.A.; Millhauser, G.L.; Toro, S.; Varga, Z.; Westerfield, M.; Gamse, J.; Chen, W.; et al. Pineal-specific agouti protein regulates teleost background adaptation. Proc. Natl. Acad. Sci. USA 2010, 107, 20164–20171. [Google Scholar] [CrossRef] [PubMed]
- Webb, T.R.; Clark, A.J. Minireview: The melanocortin 2 receptor accessory proteins. Mol. Endocrinol. 2010, 24, 475–484. [Google Scholar] [CrossRef]
- Navarro, S.; Soletto, L.; Puchol, S.; Rotllant, J.; Soengas, J.L.; Cerda-Reverter, J.M. 60 YEARS OF POMC: POMC: An evolutionary perspective. J. Mol. Endocrinol. 2016, 56, T113–T118. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Wang, C.Y.; Kwok, A.H.; Leung, F.C. Identification of the endogenous ligands for chicken growth hormone-releasing hormone (GHRH) receptor: Evidence for a separate gene encoding GHRH in submammalian vertebrates. Endocrinology 2007, 148, 2405–2416. [Google Scholar] [CrossRef]
- Nijenhuis, W.A.; Oosterom, J.; Adan, R.A. AgRP(83-132) acts as an inverse agonist on the human-melanocortin-4 receptor. Mol. Endocrinol. 2001, 15, 164–171. [Google Scholar] [CrossRef][Green Version]
- Cerda-Reverter, J.M.; Ringholm, A.; Schioth, H.B.; Peter, R.E. Molecular cloning, pharmacological characterization, and brain mapping of the melanocortin 4 receptor in the goldfish: Involvement in the control of food intake. Endocrinology 2003, 144, 2336–2349. [Google Scholar] [CrossRef]
- Cerda-Reverter, J.M.; Peter, R.E. Endogenous melanocortin antagonist in fish: Structure, brain mapping, and regulation by fasting of the goldfish agouti-related protein gene. Endocrinology 2003, 144, 4552–4561. [Google Scholar] [CrossRef] [PubMed]
- Metz, J.R.; Peters, J.J.; Flik, G. Molecular biology and physiology of the melanocortin system in fish: A review. Gen. Comp. Endocrinol. 2006, 148, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Danielson, P.; Sollars, C.; Alrubaian, J.; Balm, P.; Dores, R.M. Cloning of a neoteleost (Oreochromis mossambicus) pro-opiomelanocortin (POMC) cDNA reveals a deletion of the gamma-melanotropin region and most of the joining peptide region: Implications for POMC processing. Peptides 1999, 20, 1391–1399. [Google Scholar] [CrossRef]
- Cardoso, J.C.; Laiz-Carrion, R.; Louro, B.; Silva, N.; Canario, A.V.; Mancera, J.M.; Power, D.M. Divergence of duplicate POMC genes in gilthead sea bream Sparus auratus. Gen. Comp. Endocrinol. 2011, 173, 396–404. [Google Scholar] [CrossRef]
- Kang, D.Y.; Kim, H.C. Functional relevance of three proopiomelanocortin (POMC) genes in darkening camouflage, blind-side hypermelanosis, and appetite of Paralichthys olivaceus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2015, 179, 44–56. [Google Scholar] [CrossRef]
- Dores, R.M.; Sollars, C.; Danielson, P.; Lee, J.; Alrubaian, J.; Joss, J.M. Cloning of a proopiomelanocortin cDNA from the pituitary of the Australian lungfish, Neoceratodus forsteri: Analyzing trends in the organization of this prohormone precursor. Gen. Comp. Endocrinol. 1999, 116, 433–444. [Google Scholar] [CrossRef]
- Lee, J.; Lecaude, S.; Danielson, P.; Sollars, C.; Alrubaian, J.; Propper, C.R.; Lihrmann, I.; Vaudry, H.; Dores, R.M. Cloning of proopiomelanocortin from the brain of the african lungfish, Protopterus annectens, and the brain of the western spadefoot toad, Spea multiplicatus. Neuroendocrinology 1999, 70, 43–54. [Google Scholar] [CrossRef]
- Danielson, P.B.; Alrubaian, J.; Muller, M.; Redding, J.M.; Dores, R.M. Duplication of the POMC gene in the paddlefish (Polyodon spathula): Analysis of gamma-MSH, ACTH, and beta-endorphin regions of ray-finned fish POMC. Gen. Comp. Endocrinol. 1999, 116, 164–177. [Google Scholar] [CrossRef]
- Alrubaian, J.; Danielson, P.; Fitzpatrick, M.; Schreck, C.; Dores, R.M. Cloning of a second proopiomelanocortin cDNA from the pituitary of the sturgeon, Acipenser transmontanus. Peptides 1999, 20, 431–436. [Google Scholar] [CrossRef]
- Bu, G.; Cui, L.; Lv, C.; Lin, D.; Huang, L.; Li, Z.; Li, J.; Zeng, X.; Wang, Y. Opioid Peptides and Their Receptors in Chickens: Structure, Functionality, and Tissue Distribution. Peptides 2020, 128, 170307. [Google Scholar] [CrossRef]
- Wang, M.; Chen, Y.; Zhu, M.; Xu, B.; Guo, W.; Lyu, Y.; Zhang, C. Pharmacological modulation of melanocortin-4 receptor by melanocortin receptor accessory protein 2 in Nile tilapia. Gen. Comp. Endocrinol. 2019, 282, 113219. [Google Scholar] [CrossRef]
- Tao, M.; Ji, R.-L.; Huang, L.; Fan, S.-Y.; Liu, T.; Liu, S.-J.; Tao, Y.-X. Regulation of melanocortin-4 receptor pharmacology by two isoforms of melanocortin receptor accessory protein 2 in topmouth culter (Culter alburnus). Front. Endocrinol. 2020, 11, 538. [Google Scholar]
- Soletto, L.; Hernandez-Balfago, S.; Rocha, A.; Scheerer, P.; Kleinau, G.; Cerda-Reverter, J.M. Melanocortin Receptor Accessory Protein 2-Induced Adrenocorticotropic Hormone Response of Human Melanocortin 4 Receptor. J. Endocr. Soc. 2019, 3, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Agulleiro, M.J.; Cortes, R.; Leal, E.; Rios, D.; Sanchez, E.; Cerda-Reverter, J.M. Characterization, tissue distribution and regulation by fasting of the agouti family of peptides in the sea bass (Dicentrarchus labrax). Gen. Comp. Endocrinol. 2014, 205, 251–259. [Google Scholar] [CrossRef]
- Schjolden, J.; Schioth, H.B.; Larhammar, D.; Winberg, S.; Larson, E.T. Melanocortin peptides affect the motivation to feed in rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 2009, 160, 134–138. [Google Scholar] [CrossRef]
- Takahashi, A.; Amano, M.; Amiya, N.; Yamanome, T.; Yamamori, K.; Kawauchi, H. Expression of three proopiomelanocortin subtype genes and mass spectrometric identification of POMC-derived peptides in pars distalis and pars intermedia of barfin flounder pituitary. Gen. Comp. Endocrinol. 2006, 145, 280–286. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Chiba, H.; Amiya, N.; Yamanome, T.; Mizusawa, K.; Amano, M.; Takahashi, A. Transcription elements and functional expression of proopiomelanocortin genes in the pituitary gland of the barfin flounder. Gen. Comp. Endocrinol. 2008, 158, 259–267. [Google Scholar] [CrossRef]
- Takahashi, A.; Kobayashi, Y.; Amano, M.; Yamanome, T. Structural and functional diversity of proopiomelanocortin in fish with special reference to barfin flounder. Peptides 2009, 30, 1374–1382. [Google Scholar] [CrossRef]
- Masini, M.A.; Sturla, M.; Pestarino, M.; Facchinetti, F.; Gallinelli, A.; Uva, B.M. Proopiomelanocortin (POMC) mRNA and POMC-derived peptides immunolocalization in the skin of Protopterus annectens, an African lungfish. Peptides 1999, 20, 87–91. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Tsuchiya, K.; Yamanome, T.; Schioth, H.B.; Kawauchi, H.; Takahashi, A. Food deprivation increases the expression of melanocortin-4 receptor in the liver of barfin flounder, Verasper moseri. Gen. Comp. Endocrinol. 2008, 155, 280–287. [Google Scholar] [CrossRef]
- Sanchez, E.; Rubio, V.C.; Thompson, D.; Metz, J.; Flik, G.; Millhauser, G.L.; Cerda-Reverter, J.M. Phosphodiesterase inhibitor-dependent inverse agonism of agouti-related protein on melanocortin 4 receptor in sea bass (Dicentrarchus labrax). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 296, R1293–R1306. [Google Scholar] [CrossRef] [PubMed]
- O’Shaughnessy, P.J.; Fleming, L.M.; Jackson, G.; Hochgeschwender, U.; Reed, P.; Baker, P.J. Adrenocorticotropic hormone directly stimulates testosterone production by the fetal and neonatal mouse testis. Endocrinology 2003, 144, 3279–3284. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Luo, Q.; Zhang, J.; Mo, C.; Wang, Y.; Li, J. Endothelins (EDN1, EDN2, EDN3) and their receptors (EDNRA, EDNRB, EDNRB2) in chickens: Functional analysis and tissue distribution. Gen. Comp. Endocrinol. 2019, 283, 113231. [Google Scholar] [CrossRef] [PubMed]
- Brawand, D.; Wagner, C.E.; Li, Y.I.; Malinsky, M.; Keller, I.; Fan, S.; Simakov, O.; Ng, A.Y.; Lim, Z.W.; Bezault, E. The genomic substrate for adaptive radiation in African cichlid fish. Nature 2014, 513, 375–381. [Google Scholar]
- Mo, C.; Huang, L.; Cui, L.; Lv, C.; Lin, D.; Song, L.; Zhu, G.; Li, J.; Wang, Y. Characterization of NMB, GRP and their receptors (BRS3, NMBR and GRPR) in chickens. J. Mol. Endocrinol. 2017, 59, 61–79. [Google Scholar] [CrossRef]


| Peptide | Gene | Amino Acid Sequence |
|---|---|---|
| ACTHs | POMCa1 | SYSMEHFRWGKPVGRKRRPVKVYTSNGVAEESAEVFPEEM |
| POMCa2 | SYSMEHFRWGKPVGRKRRPIKIYTTNGLEEESAELFPGEM | |
| POMCb | SYSMEHFRWGKPSGRKRRPVKVFASSLEGGSSSEGRFPFQ | |
| α-MSHs | POMCa1 | SYSMEHFRWGKPV |
| POMCa2 | SYSMEHFRWGKPV | |
| POMCb | SYSMEHFRWGKPS | |
| β-MSHs | POMCa1 | DGSYKMKHFRWSGPPAS |
| POMCa2 | DEMYKMKHFRWGGLPAS | |
| POMCb | DRTYKMSHFRWGSPPAS | |
| ε-MSH | POMCb | SYRMEHFRWGKPA |
| β-endorphins | POMCa1 | YGGFMKSWDERSQRPLLTLFKNVINKEGQQQK |
| POMCa2 | YGGFMKSWDERSQRPLITLFKNVINKEAQEEKREQ | |
| aPOMCb | NGGFMKPWEEKPQGQLAKFFRNILVKDVKRIMG |
| Peptides | EC50 (nM) | |
|---|---|---|
| MC4R | MC4R + MRAP2b | |
| ACTH | 0.246 ± 0.048 | 0.032 ± 0.005 |
| α-MSH | 0.955 ± 0.262 | 0.786 ± 0.162 |
| β-MSH | 0.421 ± 0.190 | 0.572 ± 0.122 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Deng, Y.; Zhang, Z.; Cao, B.; Li, J.; Sun, C.; Hu, Z.; Zhang, J.; Li, J.; Wang, Y. Melanocortin Receptor 4 (MC4R) Signaling System in Nile Tilapia. Int. J. Mol. Sci. 2020, 21, 7036. https://doi.org/10.3390/ijms21197036
Liu T, Deng Y, Zhang Z, Cao B, Li J, Sun C, Hu Z, Zhang J, Li J, Wang Y. Melanocortin Receptor 4 (MC4R) Signaling System in Nile Tilapia. International Journal of Molecular Sciences. 2020; 21(19):7036. https://doi.org/10.3390/ijms21197036
Chicago/Turabian StyleLiu, Tianqiang, Yue Deng, Zheng Zhang, Baolong Cao, Jing Li, Caiyun Sun, Zhixing Hu, Jiannan Zhang, Juan Li, and Yajun Wang. 2020. "Melanocortin Receptor 4 (MC4R) Signaling System in Nile Tilapia" International Journal of Molecular Sciences 21, no. 19: 7036. https://doi.org/10.3390/ijms21197036
APA StyleLiu, T., Deng, Y., Zhang, Z., Cao, B., Li, J., Sun, C., Hu, Z., Zhang, J., Li, J., & Wang, Y. (2020). Melanocortin Receptor 4 (MC4R) Signaling System in Nile Tilapia. International Journal of Molecular Sciences, 21(19), 7036. https://doi.org/10.3390/ijms21197036







