OsExo70B1 Positively Regulates Disease Resistance to Magnaporthe oryzae in Rice
Abstract
:1. Introduction
2. Results
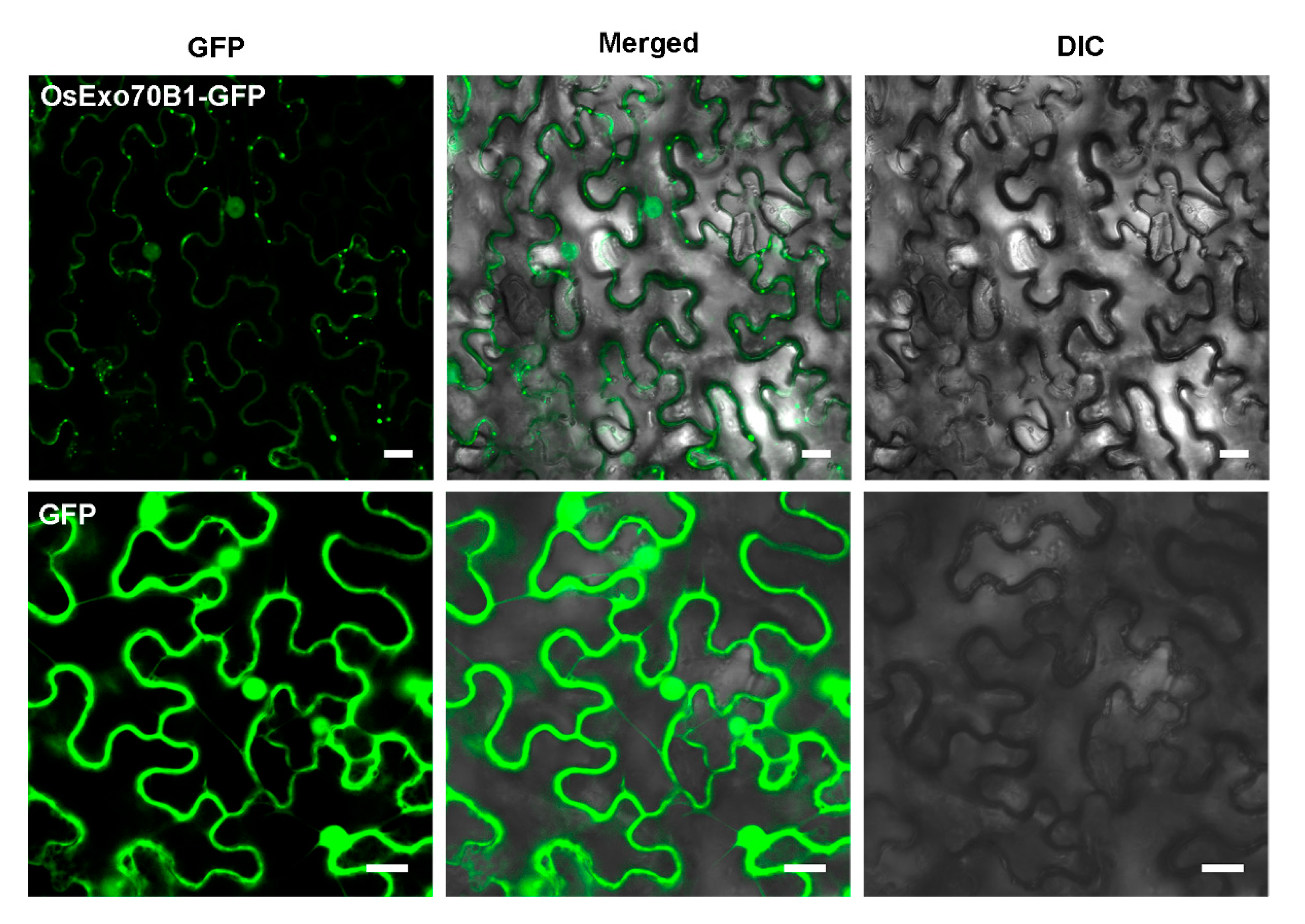
2.1. Characterization of OsExo70B1
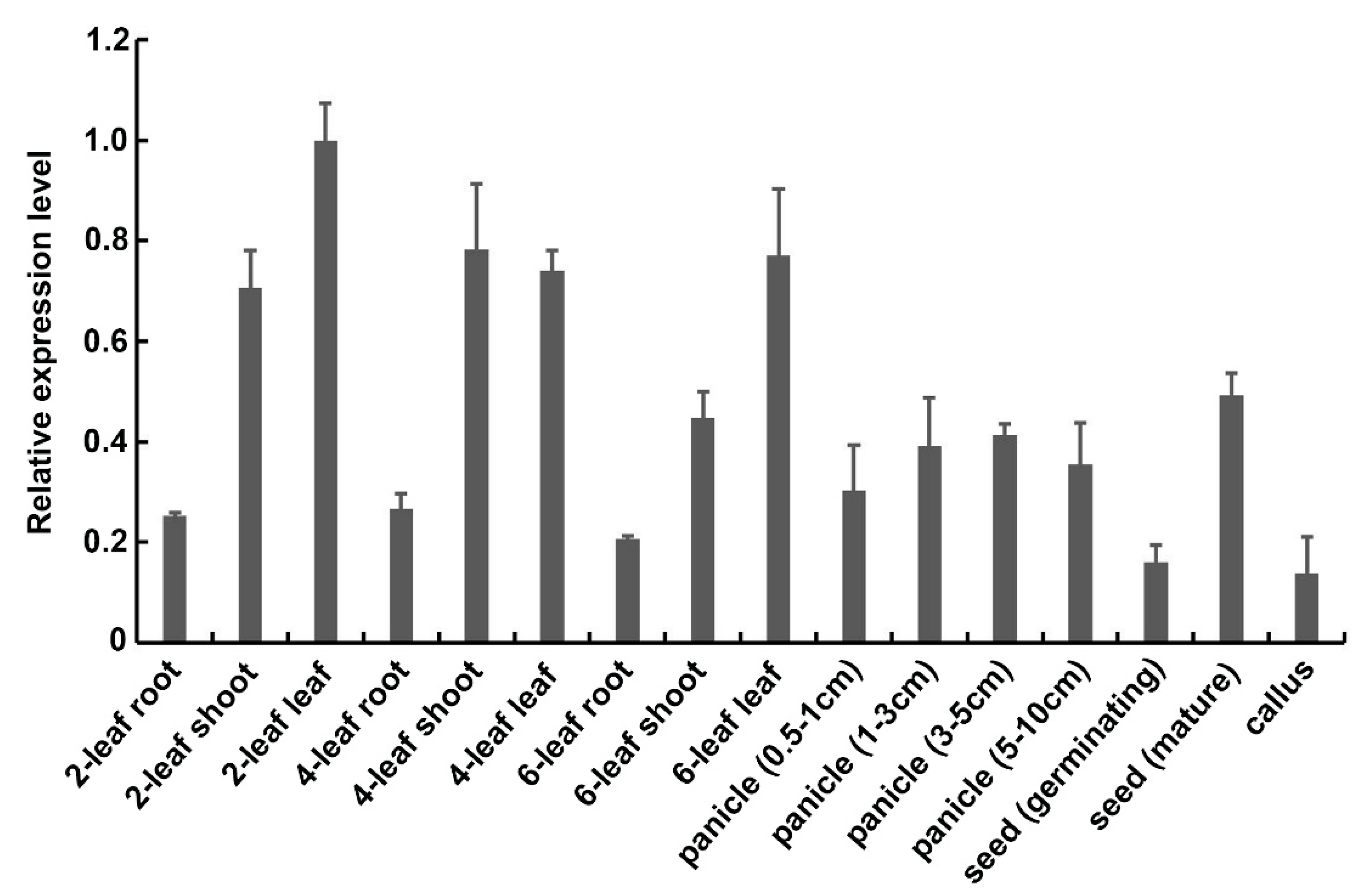
2.2. Transcription of OsExo70B1 Is Induced by PAMPs and M. oryza
2.3. Knocking out OsExo70B1 Compromises Plant Resistance to M. oryzae
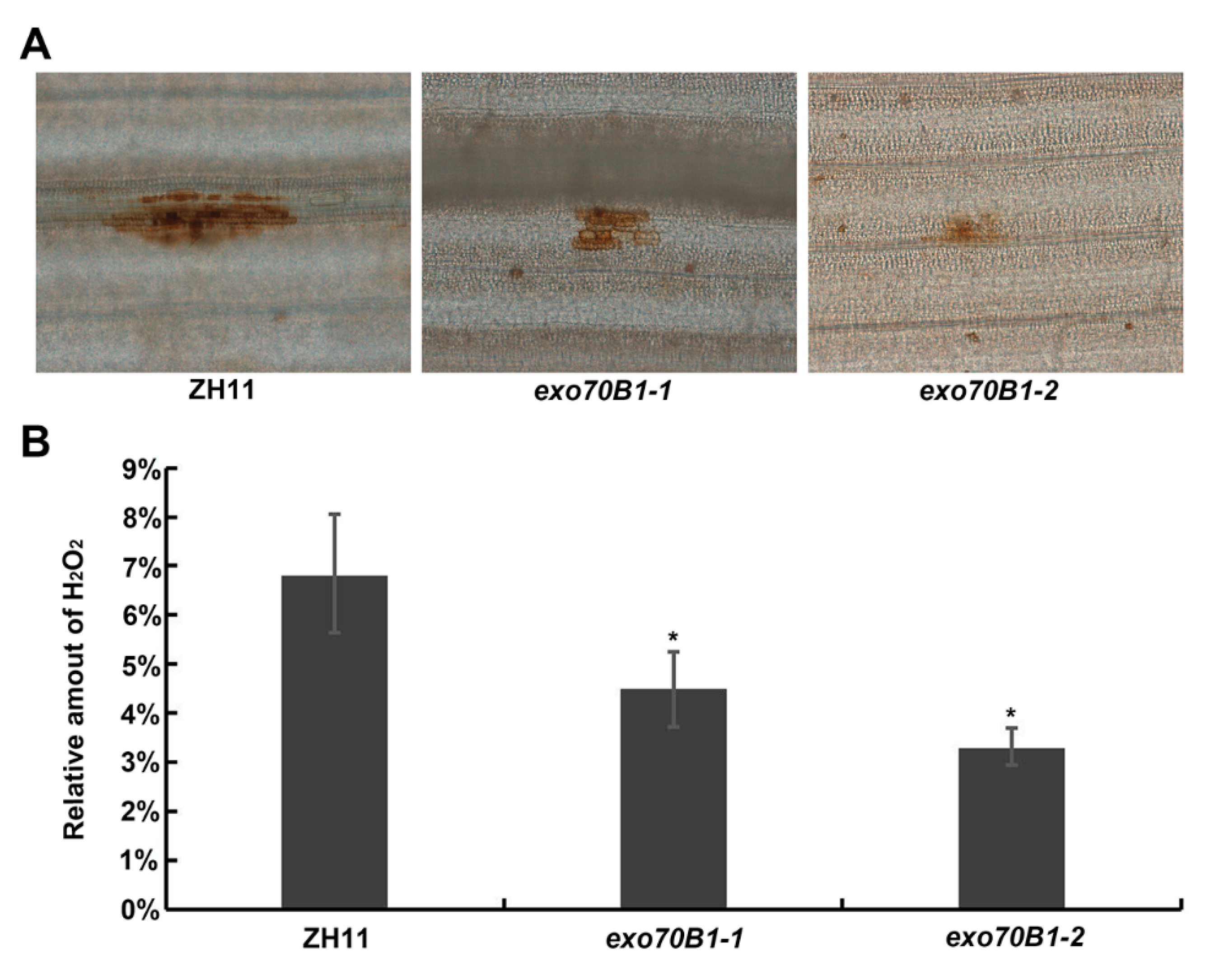
2.4. Accumulation of H2O2 Is Lower and the Infection Progress Is Faster in exo70B1-1 Mutant Compared to the Wild Type
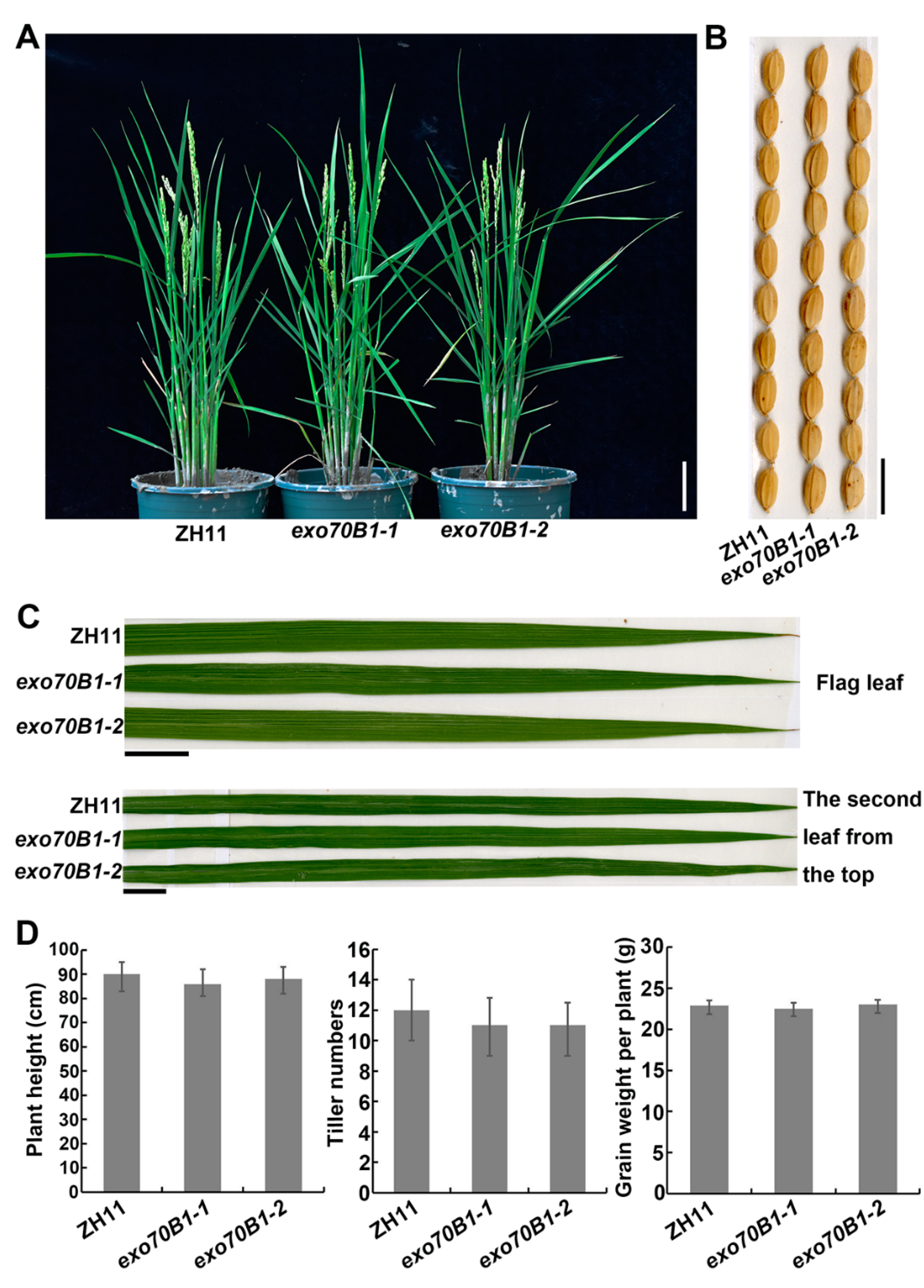
2.5. Knocking out OsExo70B1 Does Not Alter Plant Architecture or Grain Yield
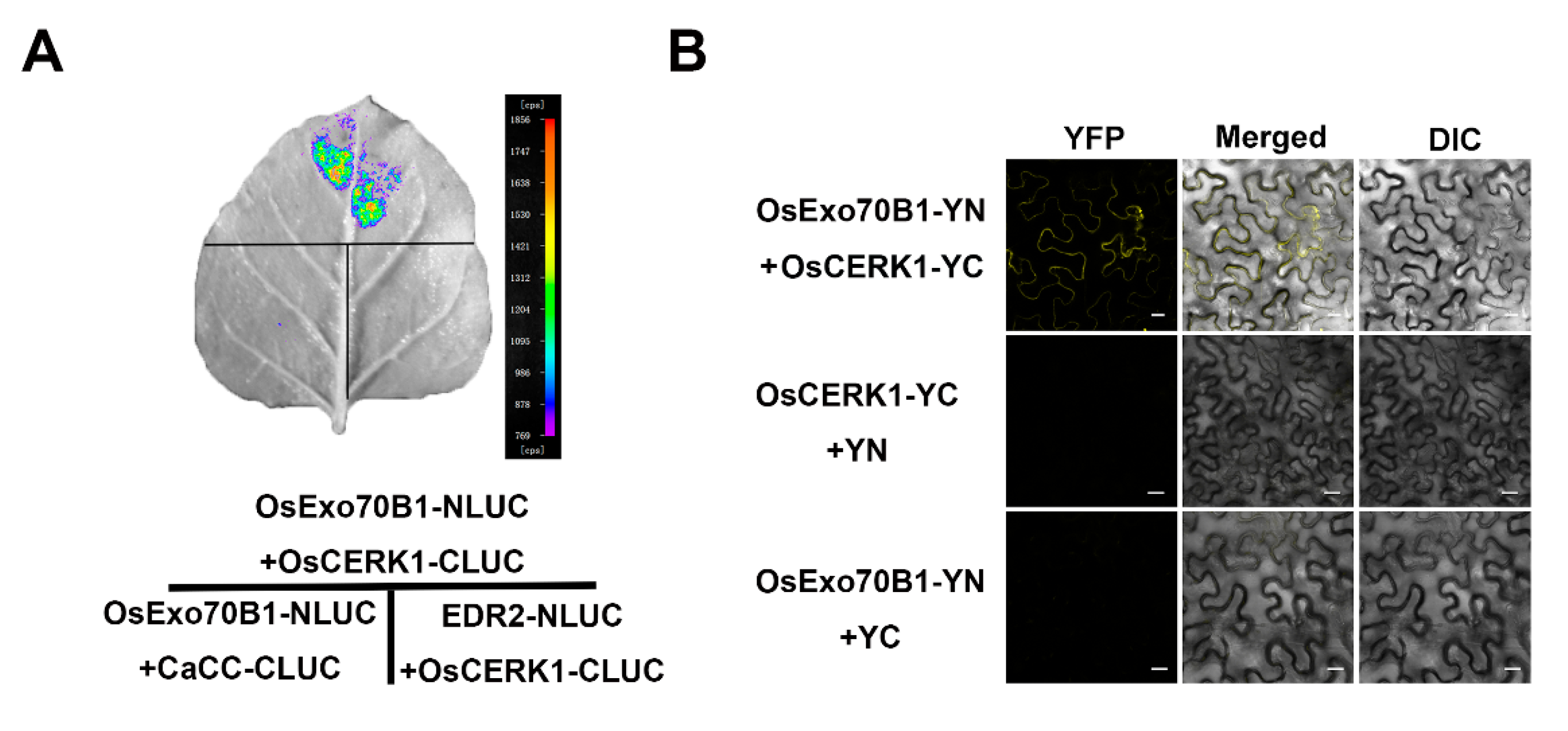
2.6. OsExo70B1 Associates with OsCERK1
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Subcell Localization Analysis
4.3. Construction of Complementary Transgenic Plant of exo70B1-1
4.4. Gene Expression Analysis
4.5. Chitin and flg22 Treatments
4.6. Rice Blast Fungus Incubation and Rice Sheath Penetration Assay
4.7. DAB Staining
4.8. Split-Luciferase Complementation Assay
4.9. Bimolecular Fluorescence Complementation (BiFC) Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Gene Accession Numbers
| Sequence data from this article can be found in Rice Genome Annotation Project databases under the following accession numbers: | |
| OsExo70B1 | (LOC_Os01g61180) |
| OsCERK1 | (LOC_Os08g425880) |
| Ubiquitin | (LOC_Os03g13170) |
| Actin | (LOC_Os03g50885) |
| PR5 | (LOC_Os03g46070) |
| PR10 | (LOC_Os12g36880) |
| Pot2 | (EMBL accession Z33638) |
References
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Feng, B.; Zhou, J.M.; Tang, D. Plant immune signaling: Advancing on two frontiers. J. Integr. Plant Biol. 2020, 62, 2–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.M.; Liu, P.Q.; Xu, Y.J.; Xiao, S. Protein trafficking during plant innate immunity. J. Integr. Plant Biol. 2016, 58, 284–298. [Google Scholar] [CrossRef]
- Gu, Y.; Zavaliev, R.; Dong, X. Membrane Trafficking in Plant Immunity. Mol. Plant 2017, 10, 1026–1034. [Google Scholar] [CrossRef]
- Zarsky, V.; Cvrckova, F.; Potocky, M.; Hala, M. Exocytosis and cell polarity in plants—Exocyst and recycling domains. New Phytol. 2009, 183, 255–272. [Google Scholar] [CrossRef]
- Zarsky, V.; Kulich, I.; Fendrych, M.; Pecenkova, T. Exocyst complexes multiple functions in plant cells secretory pathways. Curr. Opin. Plant Biol. 2013, 16, 726–733. [Google Scholar] [CrossRef]
- Pecenkova, T.; Sabol, P.; Kulich, I.; Ortmannova, J.; Zarsky, V. Constitutive Negative Regulation of R Proteins in Arabidopsis also via Autophagy Related Pathway? Front. Plant Sci. 2016, 7, 260. [Google Scholar] [CrossRef] [Green Version]
- Pecenkova, T.; Markovic, V.; Sabol, P.; Kulich, I.; Zarsky, V. Exocyst and autophagy-related membrane trafficking in plants. J. Exp. Bot. 2017, 69, 47–57. [Google Scholar] [CrossRef] [Green Version]
- Saeed, B.; Brillada, C.; Trujillo, M. Dissecting the plant exocyst. Curr. Opin. Plant Biol. 2019, 52, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Mei, K.; Guo, W. The exocyst complex. Curr. Biol. 2018, 28, R922–R925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.; Wang, Y.; Yao, X.; Xu, Z.; An, L.; Yin, M. The role of Exo70 in vascular smooth muscle cell migration. Cell Mol. Biol. Lett. 2016, 21, 20. [Google Scholar] [CrossRef] [Green Version]
- Cvrckova, F.; Zarsky, V. Old AIMs of the exocyst: Evidence for an ancestral association of exocyst subunits with autophagy-associated Atg8 proteins. Plant Signal. Behav. 2013, 8, e27099. [Google Scholar] [CrossRef] [Green Version]
- Elias, M.; Drdova, E.; Ziak, D.; Bavlnka, B.; Hala, M.; Cvrckova, F.; Soukupova, H.; Zarsky, V. The exocyst complex in plants. Cell Biol. Int. 2003, 27, 199–201. [Google Scholar] [CrossRef]
- Chong, Y.T.; Gidda, S.K.; Sanford, C.; Parkinson, J.; Mullen, R.T.; Goring, D.R. Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol. 2010, 185, 401–419. [Google Scholar] [CrossRef]
- Cvrckova, F.; Grunt, M.; Bezvoda, R.; Hala, M.; Kulich, I.; Rawat, A.; Zarsky, V. Evolution of the land plant exocyst complexes. Front. Plant Sci. 2012, 3, 159. [Google Scholar] [CrossRef] [Green Version]
- Tu, B.; Hu, L.; Chen, W.; Li, T.; Hu, B.; Zheng, L.; Lv, Z.; You, S.; Wang, Y.; Ma, B.; et al. Disruption of OsEXO70A1 Causes Irregular Vascular Bundles and Perturbs Mineral Nutrient Assimilation in Rice. Sci. Rep. 2015, 5, 18609. [Google Scholar] [CrossRef]
- Du, Y.; Overdijk, E.J.R.; Berg, J.A.; Govers, F.; Bouwmeester, K. Solanaceous exocyst subunits are involved in immunity to diverse plant pathogens. J. Exp. Bot. 2018, 69, 655–666. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Zhang, X.; Wan, W.; Zhang, H.; Liu, J.; Li, M.; Wang, H.; Xiao, J.; Wang, X. Identification and Characterization of the EXO70 Gene Family in Polyploid Wheat and Related Species. Int. J. Mol. Sci. 2018, 20, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stegmann, M.; Anderson, R.G.; Ichimura, K.; Pecenkova, T.; Reuter, P.; Zarsky, V.; McDowell, J.M.; Shirasu, K.; Trujillo, M. The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 2012, 24, 4703–4716. [Google Scholar] [CrossRef] [Green Version]
- Drdova, E.J.; Synek, L.; Pecenkova, T.; Hala, M.; Kulich, I.; Fowler, J.E.; Murphy, A.S.; Zarsky, V. The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. Plant J. 2013, 73, 709–719. [Google Scholar] [CrossRef]
- Zhao, T.; Rui, L.; Li, J.; Nishimura, M.T.; Vogel, J.P.; Liu, N.; Liu, S.; Zhao, Y.; Dangl, J.L.; Tang, D. A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet. 2015, 11, e1004945. [Google Scholar] [CrossRef]
- Seo, D.H.; Ahn, M.Y.; Park, K.Y.; Kim, E.Y.; Kim, W.T. The N-Terminal UND Motif of the Arabidopsis U-Box E3 Ligase PUB18 Is Critical for the Negative Regulation of ABA-Mediated Stomatal Movement and Determines Its Ubiquitination Specificity for Exocyst Subunit Exo70B1. Plant Cell 2016, 28, 2952–2973. [Google Scholar] [CrossRef] [Green Version]
- Synek, L.; Vukasinovic, N.; Kulich, I.; Hala, M.; Aldorfova, K.; Fendrych, M.; Zarsky, V. EXO70C2 Is a Key Regulatory Factor for Optimal Tip Growth of Pollen. Plant Physiol. 2017, 174, 223–240. [Google Scholar] [CrossRef] [Green Version]
- Vukasinovic, N.; Oda, Y.; Pejchar, P.; Synek, L.; Pecenkova, T.; Rawat, A.; Sekeres, J.; Potocky, M.; Zarsky, V. Microtubule-dependent targeting of the exocyst complex is necessary for xylem development in Arabidopsis. New Phytol. 2017, 213, 1052–1067. [Google Scholar] [CrossRef]
- Kulich, I.; Vojtikova, Z.; Sabol, P.; Ortmannova, J.; Nedela, V.; Tihlarikova, E.; Zarsky, V. Exocyst Subunit EXO70H4 Has a Specific Role in Callose Synthase Secretion and Silica Accumulation. Plant Physiol. 2018, 176, 2040–2051. [Google Scholar] [CrossRef] [Green Version]
- Kulich, I.; Vojtikova, Z.; Glanc, M.; Ortmannova, J.; Rasmann, S.; Zarsky, V. Cell wall maturation of Arabidopsis trichomes is dependent on exocyst subunit EXO70H4 and involves callose deposition. Plant Physiol. 2015, 168, 120–131. [Google Scholar] [CrossRef] [Green Version]
- Pecenkova, T.; Hala, M.; Kulich, I.; Kocourkova, D.; Drdova, E.; Fendrych, M.; Toupalova, H.; Zarsky, V. The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant-pathogen interaction. J. Exp. Bot. 2011, 62, 2107–2116. [Google Scholar] [CrossRef] [Green Version]
- Kulich, I.; Pecenkova, T.; Sekeres, J.; Smetana, O.; Fendrych, M.; Foissner, I.; Hoftberger, M.; Zarsky, V. Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic 2013, 14, 1155–1165. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Hake, K.; Wang, W.; Zhao, T.; Romeis, T.; Tang, D. CALCIUM-DEPENDENT PROTEIN KINASE5 Associates with the Truncated NLR Protein TIR-NBS2 to Contribute to exo70B1-Mediated Immunity. Plant Cell 2017, 29, 746–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Chen, J.; Wang, M.; Ren, Y.; Wang, S.; Lei, C.; Cheng, Z.; Sodmergen. Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J. Exp. Bot. 2018, 69, 1051–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Mpina, M.H.; Birch, P.R.; Bouwmeester, K.; Govers, F. Phytophthora infestans RXLR Effector AVR1 Interacts with Exocyst Component Sec5 to Manipulate Plant Immunity. Plant Physiol. 2015, 169, 1975–1990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Liu, N.; Gao, C.; Rui, L.; Tang, D. The Pseudomonas Syringae Effector AvrPtoB Associates With and Ubiquitinates Arabidopsis Exocyst Subunit EXO70B1. Front. Plant Sci. 2019, 10, 1027. [Google Scholar] [CrossRef] [Green Version]
- Fujisaki, K.; Abe, Y.; Ito, A.; Saitoh, H.; Yoshida, K.; Kanzaki, H.; Kanzaki, E.; Utsushi, H.; Yamashita, T.; Kamoun, S.; et al. Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity. Plant J. 2015, 83, 875–887. [Google Scholar] [CrossRef] [Green Version]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef] [Green Version]
- Carotenuto, G.; Chabaud, M.; Miyata, K.; Capozzi, M.; Takeda, N.; Kaku, H.; Shibuya, N.; Nakagawa, T.; Barker, D.G.; Genre, A. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol. 2017, 214, 1440–1446. [Google Scholar] [CrossRef] [Green Version]
- Sabol, P.; Kulich, I.; Zarsky, V. RIN4 recruits the exocyst subunit EXO70B1 to the plasma membrane. J. Exp. Bot. 2017, 68, 3253–3265. [Google Scholar] [CrossRef]
- Wang, J.; Ding, Y.; Wang, J.; Hillmer, S.; Miao, Y.; Lo, S.W.; Wang, X.; Robinson, D.G.; Jiang, L. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 2010, 22, 4009–4030. [Google Scholar] [CrossRef] [Green Version]
- Stegmann, M.; Anderson, R.G.; Westphal, L.; Rosahl, S.; McDowell, J.M.; Trujillo, M. The exocyst subunit Exo70B1 is involved in the immune response of Arabidopsis thaliana to different pathogens and cell death. Plant Signal Behav. 2013, 8, e27421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.H.; Chen, S.; Shirsekar, G.; Zhou, B.; Khang, C.H.; Songkumarn, P.; Afzal, A.J.; Ning, Y.; Wang, R.; Bellizzi, M.; et al. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 2012, 24, 4748–4762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimono, M.; Koga, H.; Akagi, A.; Hayashi, N.; Goto, S.; Sawada, M.; Kurihara, T.; Matsushita, A.; Sugano, S.; Jiang, C.J.; et al. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 2012, 13, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Kim, S.G.; Tsuda, K.; Gupta, R.; Park, S.Y.; Kim, S.T.; Kang, K.Y. Magnaporthe oryzae-Secreted Protein MSP1 Induces Cell Death and Elicits Defense Responses in Rice. Mol. Plant Microbe. Interact. 2016, 29, 299–312. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Cao, X.L.; Zhu, Y.; Yang, X.M.; Zhang, K.N.; Xiao, Z.Y.; Wang, H.; Zhao, J.H.; Zhang, L.L.; Li, G.B.; et al. Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol. 2019, 222, 1507–1522. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Liu, N.; Gao, C.; Cai, H.; Romeis, T.; Tang, D. The Arabidopsis exocyst subunits EXO70B1 and EXO70B2 regulate FLS2 homeostasis at the plasma membrane. New Phytol. 2020, 227, 529–544. [Google Scholar] [CrossRef]
- Nandety, R.S.; Caplan, J.L.; Cavanaugh, K.; Perroud, B.; Wroblewski, T.; Michelmore, R.W.; Meyers, B.C. The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 2013, 162, 1459–1472. [Google Scholar] [CrossRef] [Green Version]
- McHale, L.; Tan, X.; Koehl, P.; Michelmore, R.W. Plant NBS-LRR proteins: Adaptable guards. Genome Biol. 2006, 7, 212. [Google Scholar] [CrossRef] [Green Version]
- Hong, D.; Jeon, B.W.; Kim, S.Y.; Hwang, J.U.; Lee, Y. The ROP2-RIC7 pathway negatively regulates light-induced stomatal opening by inhibiting exocyst subunit Exo70B1 in Arabidopsis. New Phytol. 2016, 209, 624–635. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Ye, X.; Guo, R.; Huang, J.; Wang, W.; Tang, J.; Tan, L.; Zhu, J.K.; Chu, C.; Qian, Y. Genome-wide Targeted Mutagenesis in Rice Using the CRISPR/Cas9 System. Mol. Plant 2017, 10, 1242–1245. [Google Scholar] [CrossRef] [Green Version]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Shi, H.; Zhou, L.; Peng, C.; Liu, D.; Zhou, X.; Wu, W.; Yin, J.; Qin, H.; Ma, W.; et al. OsBSK1-2, an Orthologous of AtBSK1, Is Involved in Rice Immunity. Front. Plant Sci. 2017, 8, 908. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Li, W.; Cao, J.; Meng, F.; Yu, Y.; Huang, J.; Jiang, L.; Liu, M.; Zhang, Z.; Chen, X.; et al. Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 2017, 89, 338–353. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.B.; Xu, R.; Liu, C.; Shen, N.; Han, L.B.; Tang, D. Magnaporthe oryzae fimbrin organizes actin networks in the hyphal tip during polar growth and pathogenesis. PLoS Pathog. 2020, 16, e1008437. [Google Scholar] [CrossRef] [Green Version]
- Kachroo, P.; Leong, S.A.; Chattoo, B.B. Pot2, an inverted repeat transposon from the rice blast fungus Magnaporthe grisea. Mol. Gen. Genet. 1994, 245, 339–348. [Google Scholar] [CrossRef]
- Chen, H.; Zou, Y.; Shang, Y.; Lin, H.; Wang, Y.; Cai, R.; Tang, X.; Zhou, J.M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Wu, G.; Shi, H.; Tang, D. Receptor-like kinase 902 associates with and phosphorylates brassinosteroid-signaling kinase1 to regulate plant immunity. Mol. Plant 2019, 12, 59–70. [Google Scholar] [CrossRef] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, H.; Fang, J.; Liang, J.; Diao, Z.; Wang, W.; Yang, D.; Li, S.; Tang, D. OsExo70B1 Positively Regulates Disease Resistance to Magnaporthe oryzae in Rice. Int. J. Mol. Sci. 2020, 21, 7049. https://doi.org/10.3390/ijms21197049
Hou H, Fang J, Liang J, Diao Z, Wang W, Yang D, Li S, Tang D. OsExo70B1 Positively Regulates Disease Resistance to Magnaporthe oryzae in Rice. International Journal of Molecular Sciences. 2020; 21(19):7049. https://doi.org/10.3390/ijms21197049
Chicago/Turabian StyleHou, Hongna, Jianbo Fang, Jiahui Liang, Zhijuan Diao, Wei Wang, Dewei Yang, Shengping Li, and Dingzhong Tang. 2020. "OsExo70B1 Positively Regulates Disease Resistance to Magnaporthe oryzae in Rice" International Journal of Molecular Sciences 21, no. 19: 7049. https://doi.org/10.3390/ijms21197049
APA StyleHou, H., Fang, J., Liang, J., Diao, Z., Wang, W., Yang, D., Li, S., & Tang, D. (2020). OsExo70B1 Positively Regulates Disease Resistance to Magnaporthe oryzae in Rice. International Journal of Molecular Sciences, 21(19), 7049. https://doi.org/10.3390/ijms21197049




