Spectroscopic Properties of Two 5′-(4-Dimethylamino)Azobenzene Conjugated G-Quadruplex Forming Oligonucleotides
Abstract
1. Introduction
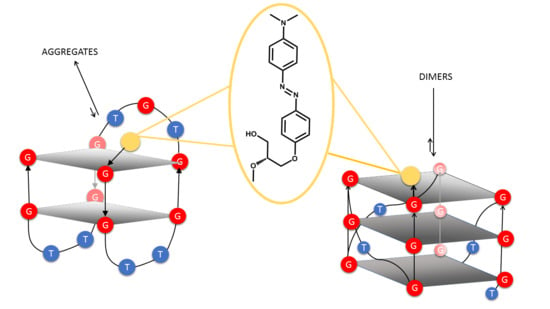
2. Results
2.1. Chemistry
2.2. UV and CD Spectroscopy
2.3. Fluorescence Results
2.4. Native PAGE
2.5. Spectroscopic Characterization of the Free and Conjugated DMAzo
3. Discussion
4. Materials and Methods
4.1. General
4.2. Chemistry
4.3. UV Experiments
4.4. CD and CD Melting Experiments
4.5. Fluorescence Spectroscopy
4.6. Electrophoresis Gel Shift Assay
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Q | Guanine Quadruplex |
| GRO | G-quadruplex rich oligonucleotide |
| CPG | Controlled pore glass |
| ON | oligonucleotide |
| DMAzo | (R) 4-(dimethylamino)azobenzene glycerol derivative |
| DMT | 4,4′-dimethoxytrityl protecting group |
| TBA | thrombin binding aptamer |
| TEA | triethylamine |
| Py | pyridine |
| 10 K | 10 mM of potassium phosphate at pH = 7.4 |
| 100 K | 10 mM of phosphate containing 90 mM of KCl at pH = 7.4 |
| 5KEW | EtOH: H2O 1:1 v/v containing 5 mM of KOH |
| 5KEW-TEA | 5KEW blend containing 0.1 mM of TEA |
| TEAA | triethylammonium acetate |
| DIPEA | N,N-Diisopropylethylamine |
| DCM | dichloromethane |
References
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. TrAC Trends Anal. Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Oikonomidi, I.; Burbridge, E.; Cavadas, M.; Sullivan, G.; Clancy, D.; Collis, B.; Březinová, .J.; Humpolickova, J.; Hu, T.; Bileck, A.; et al. iTAP, a novel iRhom interactor, controls TNF secretion by policing the stability of iRhom/TACE. In Structural Features of Nucleoprotein CST/Shelterin Complex Involved in the Telomere Maintenance and Its Association with Disease Mutations. Cells 2020; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2018; Volume 9, p. e35032. [Google Scholar]
- Wang, Q.; Liu, J.-Q.; Chen, Z.; Zheng, K.-W.; Chen, C.-Y.; Hao, Y.-H.; Tan, Z. G-Quadruplex Formation at the 3′ End of Telomere DNA Inhibits Its Extension by Telomerase, Polymerase and Unwinding by Helicase. Nucleic Acids Res. 2011, 39, 6229–6237. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Tawani, A.; Mishra, A.; Kumar, A. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci. Rep. 2016, 6, 38144. [Google Scholar] [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Boil. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.-Y.; Wang, X.-N.; Cheng, S.-Q.; Su, X.-X.; Ou, T.-M. Developing Novel G-Quadruplex Ligands: From Interaction with Nucleic Acids to Interfering with Nucleic Acid–Protein Interaction. Molecules 2019, 24, 396. [Google Scholar] [CrossRef] [PubMed]
- Asamitsu, S.; Obata, S.; Yu, Z.; Bando, T.; Sugiyama, H. Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy. Molecules 2019, 24, 429. [Google Scholar] [CrossRef] [PubMed]
- Asamitsu, S.; Takeuchi, M.; Ikenoshita, S.; Imai, Y.; Kashiwagi, H.; Shioda, N. Perspectives for Applying G-Quadruplex Structures in Neurobiology and Neuropharmacology. Int. J. Mol. Sci. 2019, 20, 2884. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.-L.; Wang, W.; Mao, Z.; Kang, T.-S.; Han, Q.-B.; Chan, P.W.H.; Leung, C.-H. Utilization of G-Quadruplex-Forming Aptamers for the Construction of Luminescence Sensing Platforms. ChemPlusChem 2016, 82, 8–17. [Google Scholar] [CrossRef]
- Stefan, L.; Monchaud, D. Applications of guanine quartets in nanotechnology and chemical biology. Nat. Rev. Chem. 2019, 3, 650–668. [Google Scholar] [CrossRef]
- Kaur, J.; Vergara, A.; Rossi, M.; Gravagnuolo, A.M.; Valadan, M.; Corrado, F.; Conte, M.; Gesuele, F.; Giardina, P.; Altucci, C. Electrostatically driven scalable synthesis of MoS2–graphene hybrid films assisted by hydrophobins. RSC Adv. 2017, 7, 50166–50175. [Google Scholar] [CrossRef]
- Kaur, J.; Singh, M.; Dell’Aversana, C.; Benedetti, R.; Giardina, P.; Rossi, M.; Valadan, M.; Vergara, A.; Cutarelli, A.; Montone, A.M.I.; et al. Biological interactions of biocompatible and water-dispersed MoS2 nanosheets with bacteria and human cells. Sci. Rep. 2018, 8, 16386. [Google Scholar] [CrossRef] [PubMed]
- Koutsoudakis, G.; De León, A.P.; Herrera, C.; Dorner, M.; Pérez-Vilaró, G.; Lyonnais, S.; Grijalvo, S.; Eritja, R.; Meyerhans, A.; Mirambeau, G.; et al. Oligonucleotide-Lipid Conjugates Forming G-Quadruplex Structures Are Potent and Pangenotypic Hepatitis C Virus Entry Inhibitors In Vitro and Ex Vivo. Antimicrob. Agents Chemother. 2017, 61, e02354-16. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Reznichenko, O.; Chaput, L.; Martin, P.; Teulade-Fichou, M.-P.; Granzhan, A. Topology-Selective, Fluorescent “Light-Up” Probes for G-Quadruplex DNA Based on Photoinduced Electron Transfer. Chem. Eur. J. 2018, 24, 12638–12651. [Google Scholar] [CrossRef] [PubMed]
- Umar, M.I.; Ji, D.-Y.; Chan, C.-Y.; Kwok, C.K. G-Quadruplex-Based Fluorescent Turn-On Ligands and Aptamers: From Development to Applications. Molecules 2019, 24, 2416. [Google Scholar] [CrossRef] [PubMed]
- Franceschin, M.; Borbone, N.; Oliviero, G.; Casagrande, V.; Scuotto, M.; Coppola, T.; Borioni, S.; Mayol, L.; Ortaggi, G.; Bianco, A.; et al. Synthesis of a Dibromoperylene Phosphoramidite Building Block and Its Incorporation at the 5′ End of a G-Quadruplex Forming Oligonucleotide: Spectroscopic Properties and Structural Studies of the Resulting Dibromoperylene Conjugate. Bioconjug. Chem. 2011, 22, 1309–1319. [Google Scholar] [CrossRef]
- Lubbe, A.S.; Szymanski, W.; Feringa, B.L. Recent Developments in Reversible Photoregulation of Oligonucleotide Structure and Function. Chem. Soc. Rev. 2017, 46, 1052–1079. [Google Scholar] [CrossRef]
- Wu, L.; Koumoto, K.; Sugimoto, N. Reversible stability switching of a hairpin DNA via a photo-responsive linker unit. Chem. Commun. 2009, 14, 1915–1917. [Google Scholar] [CrossRef]
- Mondal, P.; Granucci, G.; Rastädter, D.; Persico, M.; Burghardt, I. Azobenzene as a photoregulator covalently attached to RNA: A quantum mechanics/molecular mechanics-surface hopping dynamics study† †Electronic supplementary information (ESI) available. Chem. Sci. 2018, 9, 4671–4681. [Google Scholar] [CrossRef]
- Thevarpadam, J.; Bessi, I.; Binas, O.; Goncalves, D.P.N.; Slavov, C.; Jonker, H.R.A.; Richter, C.; Wachtveitl, J.; Schwalbe, H.; Heckel, A. Photoresponsive Formation of an Intermolecular Minimal G-Quadruplex Motif. Angew. Chem. Int. Ed. 2016, 55, 2738–2742. [Google Scholar] [CrossRef]
- Mo, M.; Kong, D.; Ji, H.; Lin, D.; Tang, X.; Yang, Z.-J.; He, Y.; Wu, L. Reversible Photocontrol of Thrombin Activity by Replacing Loops of Thrombin Binding Aptamer using Azobenzene Derivatives. Bioconjug. Chem. 2018, 30, 231–241. [Google Scholar] [CrossRef]
- Kedar, U.; Phutane, P.; Shidhaye, S.; Kadam, V. Advances in polymeric micelles for drug delivery and tumor targeting. Nanomed. Nanotech. Boil. Med. 2010, 6, 714–729. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Wang, H.; Camden, J.P. Utilizing light-triggered plasmon-driven catalysis reactions as a template for molecular delivery and release. Chem. Sci. 2017, 8, 5902–5908. [Google Scholar] [CrossRef] [PubMed]
- Do, N.Q.; Lim, K.W.; Teo, M.H.; Heddi, B.; Phan, A.T. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011, 39, 9448–9457. [Google Scholar] [CrossRef] [PubMed]
- Krauss, I.R.; Merlino, A.; Randazzo, A.; Novellino, E.; Mazzarella, L.; Sica, F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012, 40, 8119–8128. [Google Scholar] [CrossRef]
- Padmanabhan, K.; Tulinsky, A. An Ambiguous Structure of a DNA 15-mer Thrombin Complex. Acta Crystallogr. Sect. D Boil. Crystallogr. 1996, 52, 272–282. [Google Scholar] [CrossRef]
- Scuotto, M.; Rivieccio, E.; Varone, A.; Corda, D.; Bucci, M.; Vellecco, V.; Cirino, G.; Virgilio, A.; Esposito, V.; Galeone, A.; et al. Site specific replacements of a single loop nucleoside with a dibenzyl linker may switch the activity of TBA from anticoagulant to antiproliferative. Nucleic Acids Res. 2015, 43, 7702–7716. [Google Scholar] [CrossRef]
- Virgilio, A.; Petraccone, L.; Vellecco, V.; Bucci, M.; Varra, M.; Irace, C.; Santamaria, R.; Pepe, A.; Mayol, L.; Esposito, V.; et al. Site-specific replacement of the thymine methyl group by fluorine in thrombin binding aptamer significantly improves structural stability and anticoagulant activity. Nucleic Acids Res. 2015, 43, 10602–10611. [Google Scholar] [CrossRef][Green Version]
- Mukundan, V.T.; Do, N.Q.; Phan, A.T. HIV-1 integrase inhibitor T30177 forms a stacked dimeric G-quadruplex structure containing bulges. Nucleic Acids Res. 2011, 39, 8984–8991. [Google Scholar] [CrossRef]
- Rivieccio, E.; Tartaglione, L.; Esposito, V.; Dell’Aversano, C.; Koneru, P.; Scuotto, M.; Virgilio, A.; Mayol, L.; Kvaratskhelia, M.; Varra, M. Structural studies and biological evaluation of T30695 variants modified with single chiral glycerol-T reveal the importance of LEDGF/p75 for the aptamer anti-HIV-integrase activities. Biochim. Biophys. Acta (BBA) Gen. Subj. 2019, 1863, 351–361. [Google Scholar] [CrossRef]
- Li, W.-X.; Kaplan, A.V.; Grant, G.W.; Toole, J.J.; Leun, L.L.K. A Novel Nucleotide-Based Thrombin Inhibitor Inhibits Clot-Bound Thrombin and Reduces Arterial Platelet Thrombus Formation. Blood 1994, 83, 677–682. [Google Scholar] [CrossRef]
- Coppola, T.; Varra, M.; Oliviero, G.; Galeone, A.; D’Isa, G.; Mayol, L.; Morelli, E.; Bucci, M.; Vellecco, V.; Cirino, G.; et al. Synthesis, structural studies and biological properties of new TBA analogues containing an acyclic nucleotide. Bioorganic Med. Chem. 2008, 16, 8244–8253. [Google Scholar] [CrossRef] [PubMed]
- Borbone, N.; Bucci, M.; Oliviero, G.; Morelli, E.; Amato, J.; D’Atri, V.; D’Errico, S.; Vellecco, V.; Cirino, G.; Piccialli, G.; et al. Investigating the Role of T7 and T12 Residues on the Biological Properties of Thrombin-Binding Aptamer: Enhancement of Anticoagulant Activity by a Single Nucleobase Modification. J. Med. Chem. 2012, 55, 10716–10728. [Google Scholar] [CrossRef] [PubMed]
- Petrera, N.S.; Stafford, A.R.; Leslie, B.A.; Kretz, C.A.; Fredenburgh, J.C.; Weitz, J.I. Long Range Communication between Exosites 1 and 2 Modulates Thrombin Function. J. Boil. Chem. 2009, 284, 25620–25629. [Google Scholar] [CrossRef]
- Imperatore, C.; Scuotto, M.; Valadan, M.; Rivieccio, E.; Saide, A.; Russo, A.; Altucci, C.; Menna, M.; Ramunno, A.; Mayol, L.; et al. Photo-control of cancer cell growth by benzodiazo N-substituted pyrrole derivatives. J. Photochem. Photobiol. Chem. 2019, 377, 109–118. [Google Scholar] [CrossRef]
- Imperatore, C.; Valadan, M.; Tartaglione, L.; Persico, M.; Ramunno, A.; Menna, M.; Casertano, M.; Dell’Aversano, C.; Singh, M.; Garigliota, M.L.D.; et al. Exploring the Photodynamic Properties of Two Antiproliferative Benzodiazopyrrole Derivatives. Int. J. Mol. Sci. 2020, 21, 1246. [Google Scholar] [CrossRef] [PubMed]
- Scuotto, M.; Persico, M.; Bucci, M.; Vellecco, V.; Borbone, N.; Morelli, E.; Oliviero, G.; Novellino, E.; Piccialli, G.; Cirino, G.; et al. Outstanding effects on antithrombin activity of modified TBA diastereomers containing an optically pure acyclic nucleotide analogue. Org. Biomol. Chem. 2014, 12, 5235–5242. [Google Scholar] [CrossRef]
- Del Villar-Guerra, R.; Trent, J.O.; Chaires, J.B. G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy. Angew. Chem. Int. Ed. 2018, 57, 7171–7175. [Google Scholar] [CrossRef] [PubMed]
- Kwok, C.K.; Sherlock, M.E.; Bevilacqua, P.C. Effect of Loop Sequence and Loop Length on the Intrinsic Fluorescence of G-Quadruplexes. Biochemistry 2013, 52, 3019–3021. [Google Scholar] [CrossRef]
- Chan, C.-Y.; Umar, M.I.; Kwok, C.K. Spectroscopic analysis reveals the effect of a single nucleotide bulge on G-quadruplex structures. Chem. Commun. 2019, 55, 2616–2619. [Google Scholar] [CrossRef]
- Jia, X.-Q.; Li, Y.; Zhang, C.-X.; Gao, Y.-C.; Wu, Y. Supramolecular clusters clarification in ethanol-water mixture by using fluorescence spectroscopy and 2D correlation analysis. J. Mol. Struct. 2020, 1219, 128569. [Google Scholar] [CrossRef]
- Changenet-Barret, P.; Emanuele, E.; Gustavsson, T.; Improta, R.; Kotlyar, A.; Markovitsi, D.; Vayá, I.; Zakrzewska, K.; Zikich, D. Optical Properties of Guanine Nanowires: Experimental and Theoretical Study. J. Phys. Chem. 2010, 114, 14339–14346. [Google Scholar] [CrossRef]
- Majerová, E.; Streckerová, T.; Bednárová, L.; Curtis, E.A. Sequence Requirements of Intrinsically Fluorescent G-Quadruplexes. Biochemistry 2018, 57, 4052–4062. [Google Scholar] [CrossRef] [PubMed]
- Onidas, D.; Markovitsi, D.; Marguet, S.; Sharonov, A.; Gustavsson, T. Fluorescence Properties of DNA Nucleosides and Nucleotides: A Refined Steady-State and Femtosecond Investigation. J. Phys. Chem. 2002, 106, 11367–11374. [Google Scholar] [CrossRef]
- Sherlock, M.E.; Rumble, C.; Kwok, C.K.; Breffke, J.; Maroncelli, M.; Bevilacqua, P.C. Steady-State and Time-Resolved Studies into the Origin of the Intrinsic Fluorescence of G-Quadruplexes. J. Phys. Chem. 2016, 120, 5146–5158. [Google Scholar] [CrossRef] [PubMed]
- Zuffo, M.; Gandolfini, A.; Heddi, B.; Granzhan, A. Harnessing intrinsic fluorescence for typing of secondary structures of DNA. Nucleic Acids Res. 2020, 48, e61. [Google Scholar] [CrossRef] [PubMed]
- Dao, N.T.; Haselsberger, R.; Michel-Beyerle, M.-E.; Phan, A.T. Following G-quadruplex formation by its intrinsic fluorescence. FEBS Lett. 2011, 585, 3969–3977. [Google Scholar] [CrossRef]
- Méndez, M.A.; Szalai, V.A. Fluorescence of unmodified oligonucleotides: A tool to probe G-quadruplex DNA structure. Biopolymers 2009, 91, 841–850. [Google Scholar] [CrossRef]
- Dao, N.T.; Haselsberger, R.; Michel-Beyerle, M.; Phan, A.T. Excimer Formation by Stacking G-Quadruplex Blocks. ChemPhysChem 2013, 14, 2667–2671. [Google Scholar] [CrossRef]
- Marturano, V.; Ambrogi, V.; Bandeira, N.A.G.; Tylkowski, B.; Giamberini, M.; Cerruti, P. Modeling of Azobenzene-Based Compounds. Phys. Sci. Rev. 2017, 2, 2. [Google Scholar] [CrossRef]
- Sánchez, A.; De Rossi, R.H. Strong inhibition of cis-trans isomerization of azo compounds by hydroxide ion. J. Org. Chem. 1993, 58, 2094–2096. [Google Scholar] [CrossRef]
- Sanchez, A.M.; Barra, M.; De Rossi, R.H. On the Mechanism of the Acid/Base-Catalyzed ThermalCis−TransIsomerization of Methyl Orange. J. Org. Chem. 1999, 64, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Azuki, M.; Morihashi, K.; Watanabe, T.; Takahashi, O.; Kikuchi, O. Ab Initio GB Study of the Acid-Catalyzed Cis Trans Isomerization of Methyl Yellow and Methyl Orange in Aqueous Solution. J. Mol. Struc-Theochem. 2001, 542, 255–262. [Google Scholar] [CrossRef]
- Sanchez, A.M.; Rossi, R.H. Effect of Hydroxide Ion on the Cis-Trans Thermal Isomerization of Azobenzene Derivatives. J. Org. Chem. 1995, 60, 2974–2976. [Google Scholar] [CrossRef]
- Dunn, N.J.; Humphries, W.H.; Offenbacher, A.R.; King, T.L.; Gray, J.A. PH-Dependent Cis-Trans Isomerization Rates for Azobenzene Dyes in Aqueous Solution. J. Phys. Chem. A 2009, 113, 13144–13151. [Google Scholar] [CrossRef]
- Kamei, T.; Kudo, M.; Akiyama, H.; Wada, M.; Nagasawa, J.; Funahashi, M.; Tamaoki, N.; Uyeda, T.Q.P. Visible-Light Photoresponsivity of a 4-(Dimethylamino)Azobenzene Unit Incorporated into Single-Stranded DNA: Demonstration of a Large Spectral Change Accompanying Isomerization in DMSO and Detection of Rapid (Z ) to (E) Isomerization in Aqueous Solution. Eur. J. Org. Chem. 2007, 11, 1846–1853. [Google Scholar] [CrossRef]
- Kamei, T.; Akiyama, H.; Morii, H.; Tamaoki, N.; Uyeda, T.Q.P. Visible-Light Photocontrol of (E)/(Z) Isomerization of the 4-(Dimethylamino)Azobenzene Pseudo-Nucleotide Unit Incorporated Into an Oligonucleotide and DNA Hybridization in Aqueous Media. Nucleos. Nucleot. Nucl. 2009, 28, 12–28. [Google Scholar] [CrossRef]
- Vorlíčková, M.; Bednářová, K.; Kypr, J. Ethanol is a better inducer of DNA guanine tetraplexes than potassium cations. Biopolymers 2006, 82, 253–260. [Google Scholar] [CrossRef]
- Vorlíčková, M.; Bednářová, K.; Kejnovska, I.; Kypr, J. Intramolecular and intermolecular guanine quadruplexes of DNA in aqueous salt and ethanol solutions. Biopolymers 2007, 86, 1–10. [Google Scholar] [CrossRef]
- Kejnovská, I.; Renčiuk, D.; Palacký, J.; Vorlíčková, M. CD Study of the G-Quadruplex Conformation. Methods Mol. Biol. 2019, 2035, 25–44. [Google Scholar] [CrossRef]
- Bullock, A.N.; Henckel, J.; Fersht, A.R. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: Definition of mutant states for rescue in cancer therapy. Oncogene 2000, 19, 1245–1256. [Google Scholar] [CrossRef]
- Aviñó, A.; Portella, G.; Ferreira, R.; Gargallo, R.; Mazzini, S.; Gabelica, V.; Orozco, M.; Eritja, R. Specific loop modifications of the thrombin-binding aptamer trigger the formation of parallel structures. FEBS J. 2014, 281, 1085–1099. [Google Scholar] [CrossRef]
- Debray, J.; Zeghida, W.; Jourdan, M.; Monchaud, D.; Dheu-Andries, M.-L.; Dumy, P.; Teulade-Fichou, M.-P.; Demeunynck, M. Synthesis and evaluation of fused bispyrimidinoacridines as novel pentacyclic analogues of quadruplex-binder BRACO-19. Org. Biomol. Chem. 2009, 7, 5219. [Google Scholar] [CrossRef]
- Marchand, A.; Granzhan, A.; Iida, K.; Tsushima, Y.; Ma, Y.; Nagasawa, K.; Teulade-Fichou, M.-P.; Gabelica, V. Ligand-Induced Conformational Changes with Cation Ejection upon Binding to Human Telomeric DNA G-Quadruplexes. J. Am. Chem. Soc. 2015, 137, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, J.; Zhou, Y.; Yan, S.; Weng, X.; Wu, X.; Deng, M.; Zhou, X. Conformational Switching of G-Quadruplex DNA by Photoregulation. Angew. Chem. Int. Ed. 2010, 49, 5305–5309. [Google Scholar] [CrossRef]
- Pieraccini, S.; Campitiello, M.; Carducci, F.; Davis, J.T.; Mariani, P.; Masiero, S. Playing supramolecular dominoes with light: Building and breaking a photoreversible G-quadruplex made from guanosine, boric acid and an azobenzene. Org. Biomol. Chem. 2019, 17, 2759–2769. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Wang, X.; Xu, L.; Tai, Y.; Dai, L.; Zheng, X.; Mao, W.; Xu, X.; Zhou, X. Light-driven conformational regulation of human telomeric G-quadruplex DNA in physiological conditions. Org. Biomol. Chem. 2011, 9, 6639–6645. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, M.P.; Ramos-Soriano, J.; Haldar, S.; Sheikh, S.; Morales, J.C.; Mulholland, A.J.; Galan, M.C. Visible-light photoswitching of ligand binding mode suggests G-quadruplex DNA as a target for photopharmacology. Chem. Commun. 2020, 56, 5186–5189. [Google Scholar] [CrossRef]
- Liu, M.; Liu, Y.; Wu, F.; Du, Y.; Zhou, X. Specific stabilization of DNA G-quadruplex structures with a chemically modified complementary probe. Bioorganic Med. Chem. 2019, 27, 1962–1965. [Google Scholar] [CrossRef]
- Wu, T.-Y.; Huang, Q.; Huang, Z.-S.; Hu, M.-H.; Tan, J.-H. A drug-like imidazole-benzothiazole conjugate inhibits malignant melanoma by stabilizing the c-MYC G-quadruplex. Bioorganic Chem. 2020, 99, 103866. [Google Scholar] [CrossRef]











© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imperatore, C.; Varriale, A.; Rivieccio, E.; Pennacchio, A.; Staiano, M.; D’Auria, S.; Casertano, M.; Altucci, C.; Valadan, M.; Singh, M.; et al. Spectroscopic Properties of Two 5′-(4-Dimethylamino)Azobenzene Conjugated G-Quadruplex Forming Oligonucleotides. Int. J. Mol. Sci. 2020, 21, 7103. https://doi.org/10.3390/ijms21197103
Imperatore C, Varriale A, Rivieccio E, Pennacchio A, Staiano M, D’Auria S, Casertano M, Altucci C, Valadan M, Singh M, et al. Spectroscopic Properties of Two 5′-(4-Dimethylamino)Azobenzene Conjugated G-Quadruplex Forming Oligonucleotides. International Journal of Molecular Sciences. 2020; 21(19):7103. https://doi.org/10.3390/ijms21197103
Chicago/Turabian StyleImperatore, Concetta, Antonio Varriale, Elisa Rivieccio, Angela Pennacchio, Maria Staiano, Sabato D’Auria, Marcello Casertano, Carlo Altucci, Mohammadhassan Valadan, Manjot Singh, and et al. 2020. "Spectroscopic Properties of Two 5′-(4-Dimethylamino)Azobenzene Conjugated G-Quadruplex Forming Oligonucleotides" International Journal of Molecular Sciences 21, no. 19: 7103. https://doi.org/10.3390/ijms21197103
APA StyleImperatore, C., Varriale, A., Rivieccio, E., Pennacchio, A., Staiano, M., D’Auria, S., Casertano, M., Altucci, C., Valadan, M., Singh, M., Menna, M., & Varra, M. (2020). Spectroscopic Properties of Two 5′-(4-Dimethylamino)Azobenzene Conjugated G-Quadruplex Forming Oligonucleotides. International Journal of Molecular Sciences, 21(19), 7103. https://doi.org/10.3390/ijms21197103