Characterization of Fecal Microbiota with Clinical Specimen Using Long-Read and Short-Read Sequencing Platform
Abstract
1. Introduction
2. Results
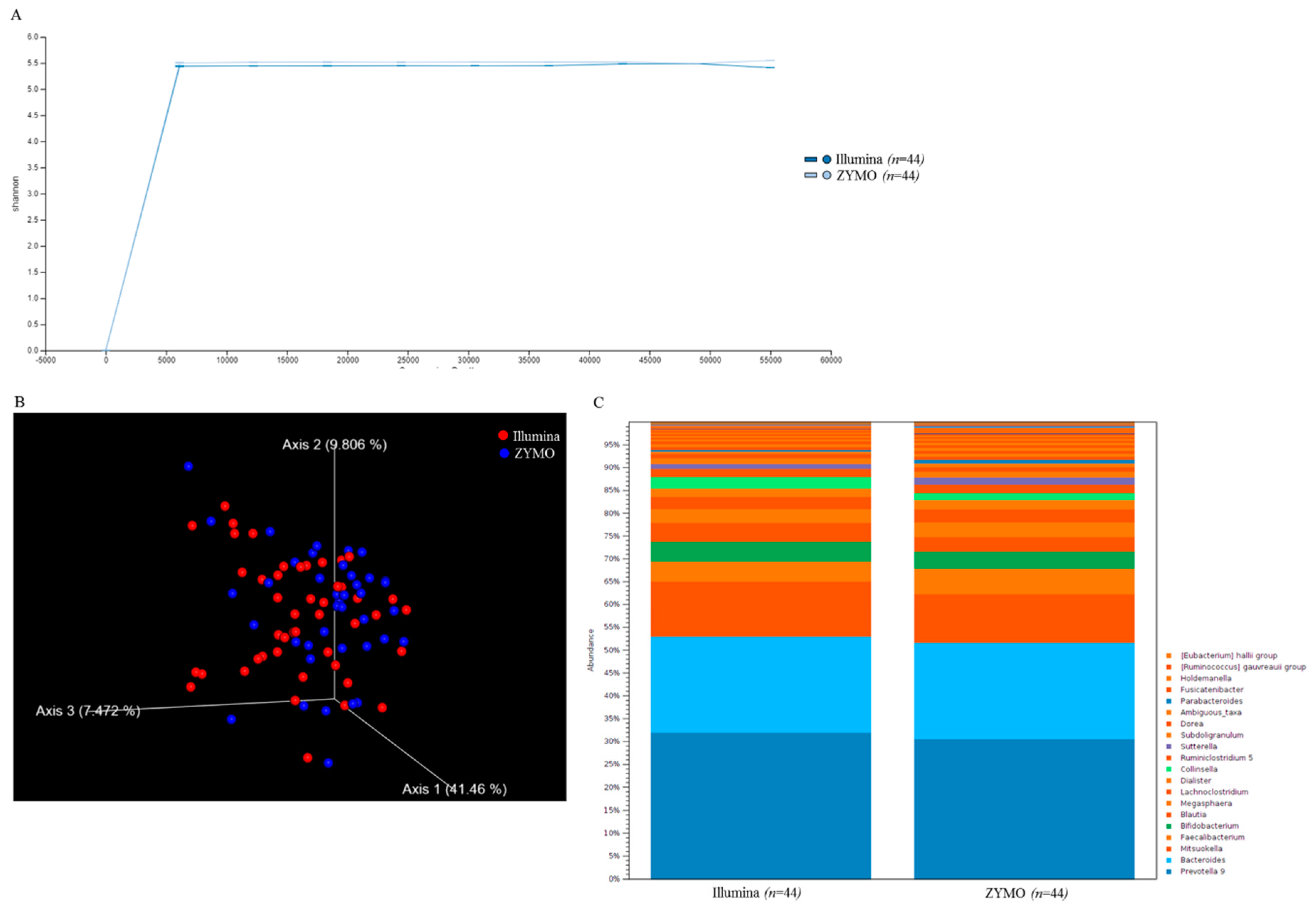
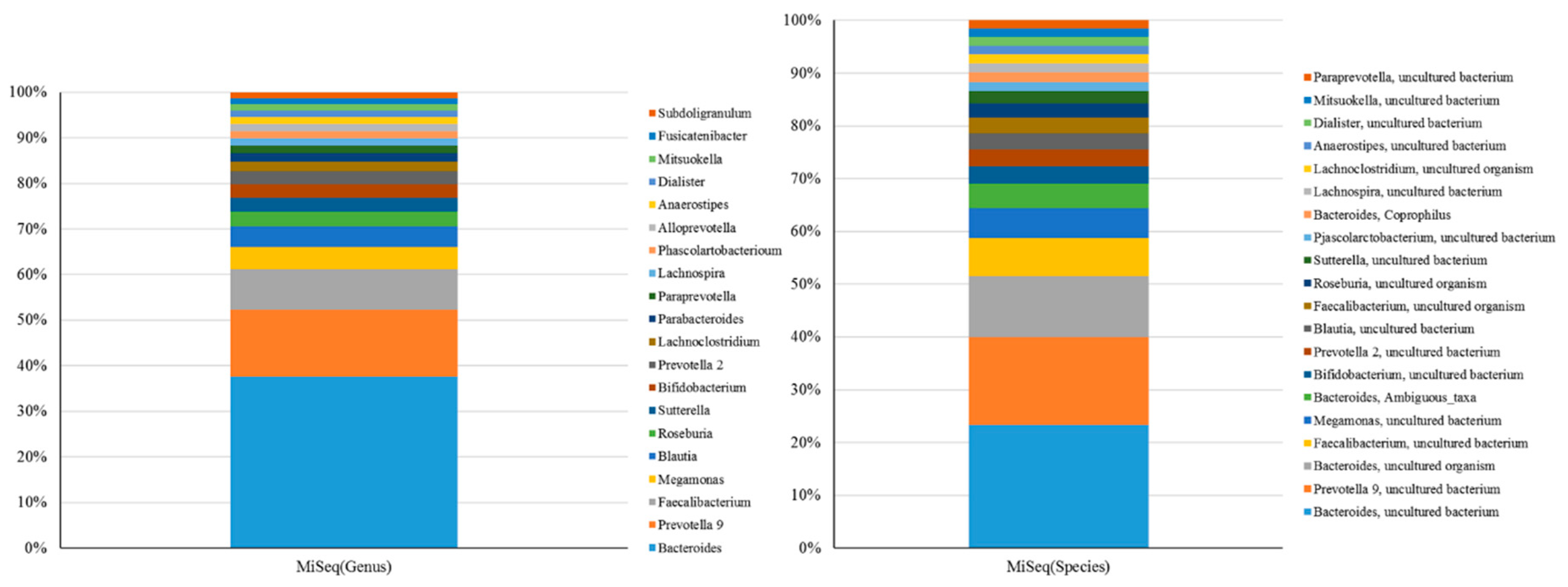
2.1. Short-Read Sequencing Consistently Classifies Taxonomic Profiles of Gut Microbiota Using 16S rRNA Sequences
2.2. Long-Read Sequencing is Practicable for Taxonomic Assignment of Microbial Communities
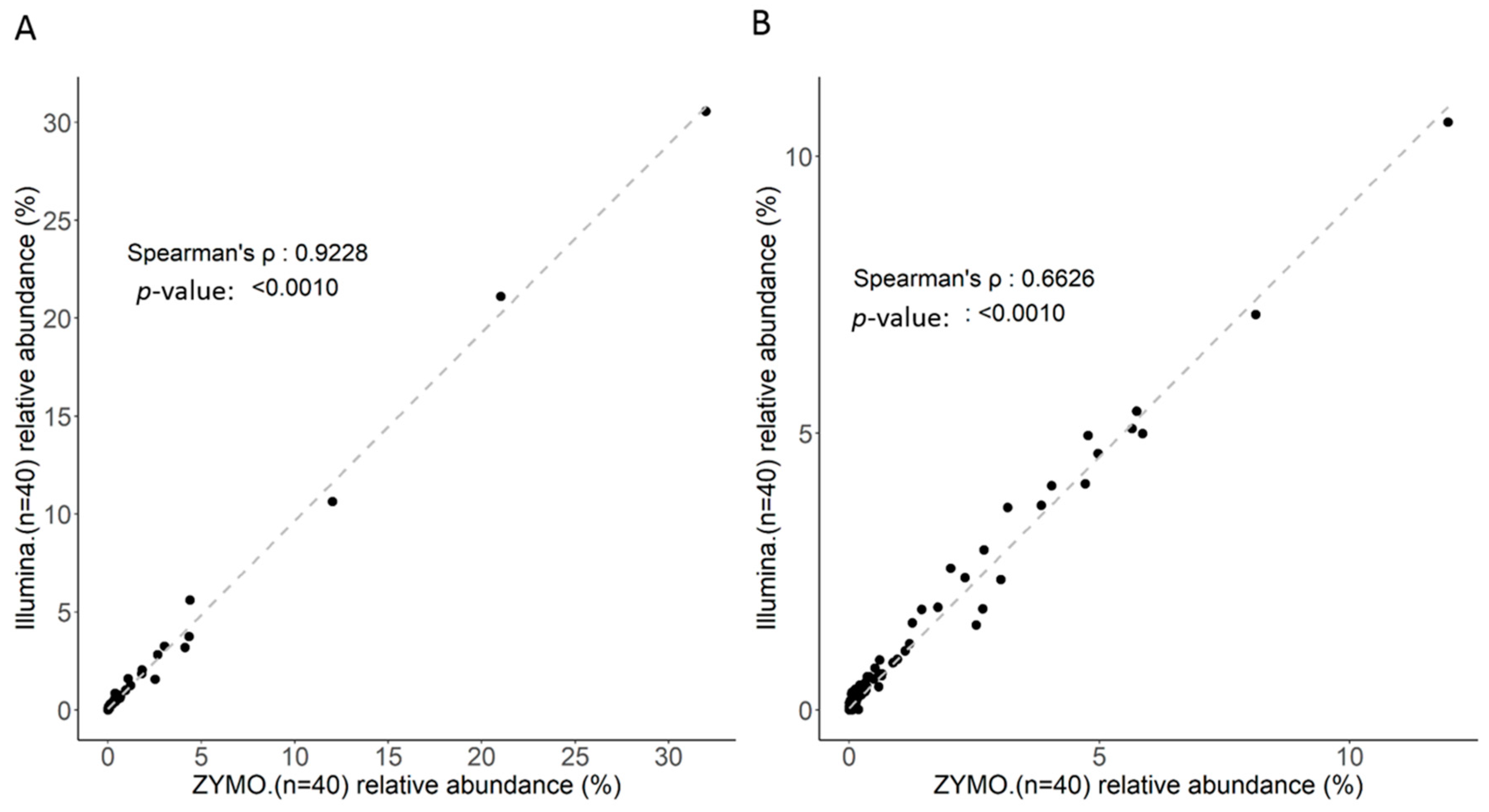
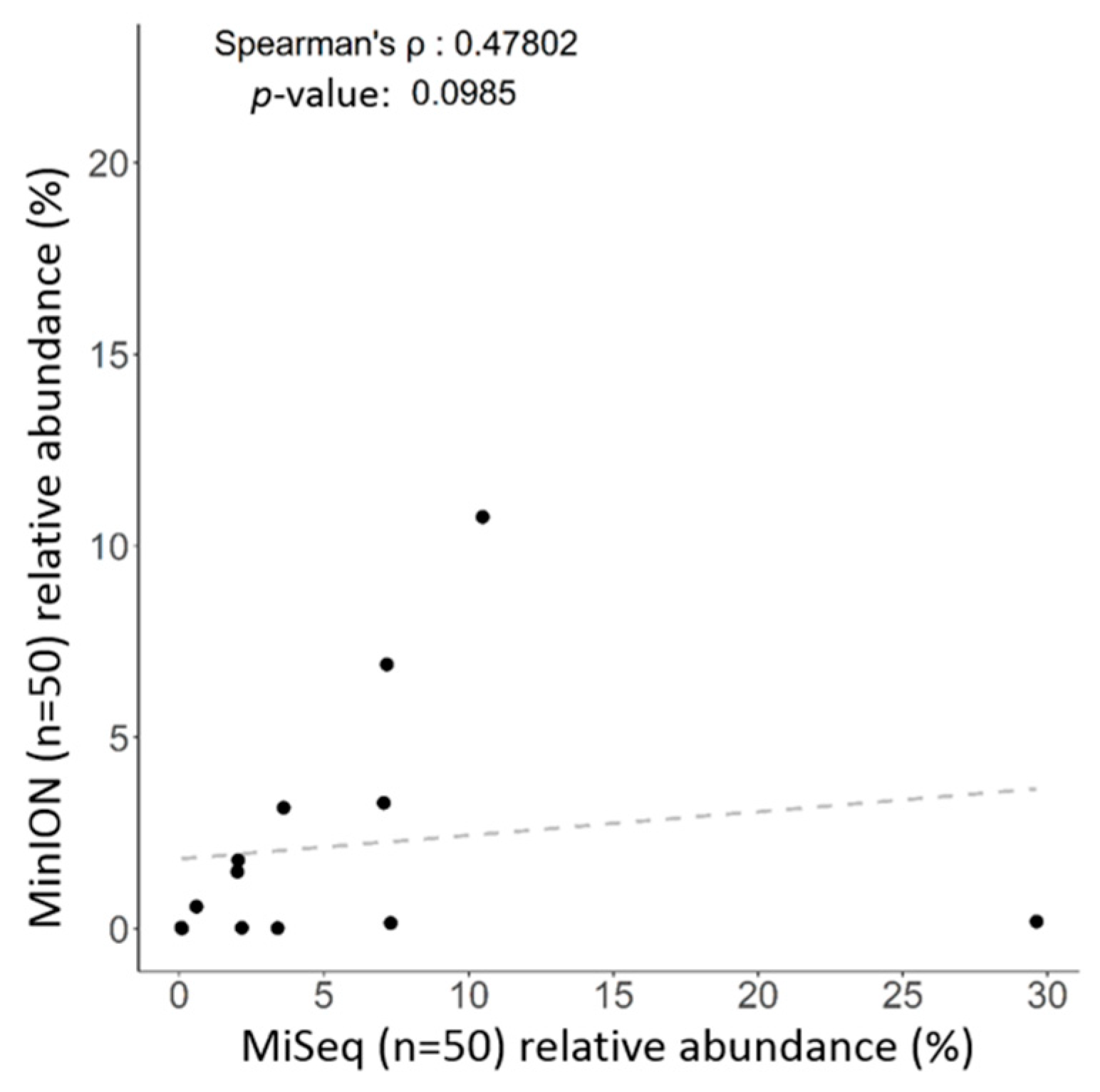
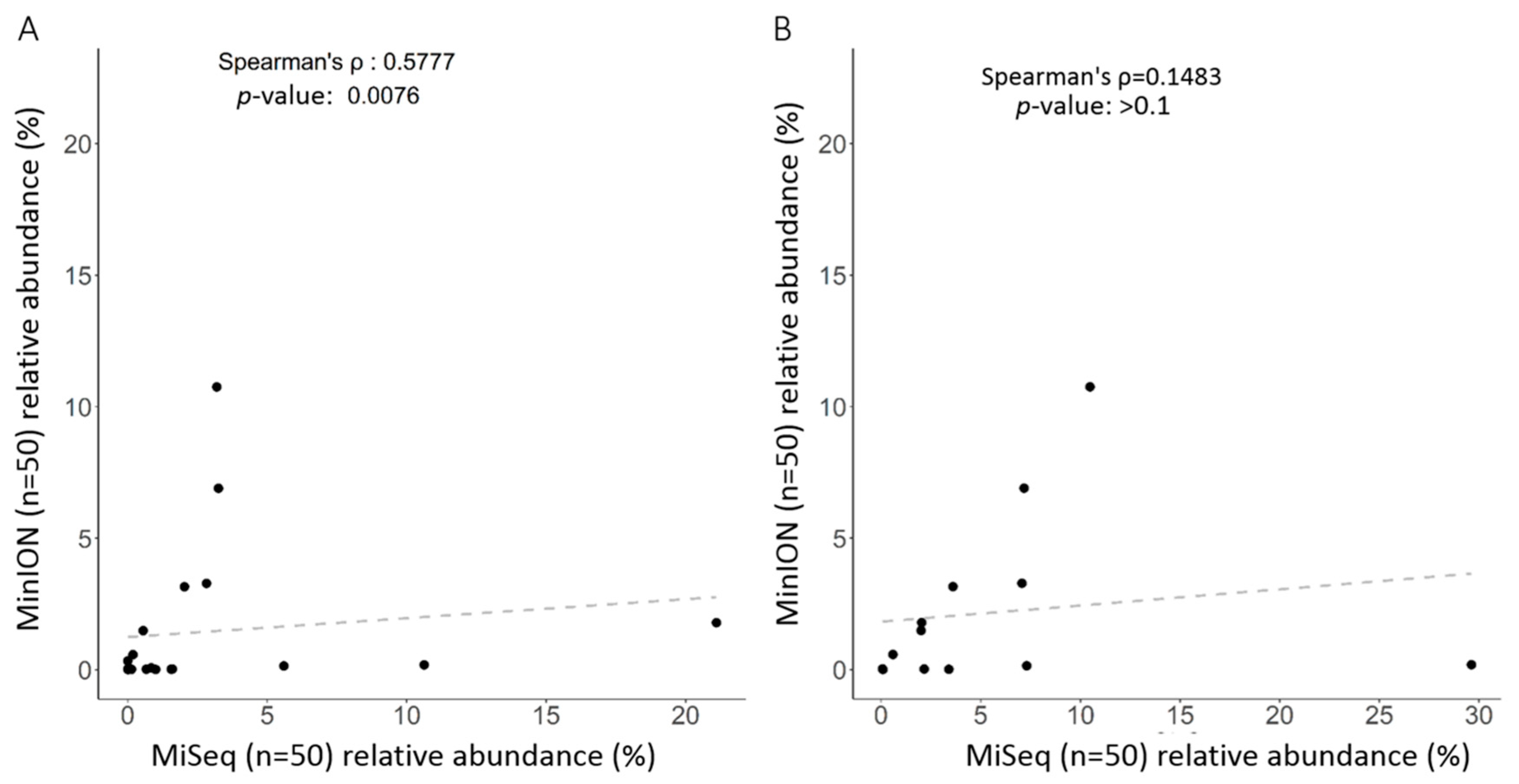
2.3. Correlation between Long Read and Short Read Sequencing toward Taxonomic Assignment of Gut Microbiota
3. Discussion
4. Materials and Methods
4.1. Ethics Statement for Use of Clinical Samples
4.2. Bacterial DNA Extraction
4.3. 16S rRNA Gene Sequencing
4.4. Bioinformatic Analysis
4.5. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NGS | Next generation sequencing |
| ONT | Oxford nanopore technology |
References
- Olsson, L.M.; Poitou, C.; Tremaroli, V.; Coupaye, M.; Aron-Wisnewsky, J.; Bäckhed, F.; Clément, K.; Caesar, R. Gut microbiota of obese subjects with Prader-Willi syndrome is linked to metabolic health. Gut 2020, 69, 1229–1238. [Google Scholar]
- Li, F.; Wang, M.; Wang, J.; Li, R.; Zhang, Y. Alterations to the gut microbiota and their correlation with inflammatory factors in chronic kidney disease. Front. Cell Infect. Microbiol. 2019, 9, 206. [Google Scholar]
- Kaźmierczak-Siedlecka, K.; Daca, A.; Fic, M.; van de Wetering, T.; Folwarski, M.; Makarewicz, W. Therapeutic methods of gut microbiota modification in colorectal cancer management—Fecal microbiota transplantation, prebiotics, probiotics, and synbiotics. Gut Microbes 2020, 11, 1518–1530. [Google Scholar] [PubMed]
- Boers, S.A.; Jansen, R.; Hays, J.P. Understanding and overcoming the pitfalls and biases of next-generation sequencing (NGS) methods for use in the routine clinical microbiological diagnostic laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1059–1070. [Google Scholar] [PubMed]
- Zhulin, I.B. Classic spotlight: 16s rRNA redefines microbiology. J. Bacteriol. 2016, 198, 2764–2765. [Google Scholar] [PubMed]
- Virtanen, S.; Kalliala, I.; Nieminen, P.; Salonen, A. Comparative analysis of vaginal microbiota sampling using 16S rRNA gene analysis. PLoS ONE 2017, 12, e0181477. [Google Scholar]
- Schriefer, A.E.; Cliften, P.F.; Hibberd, M.C.; Sawyer, C.; Brown-Kennerly, V.; Burcea, L.; Klotz, E.; Crosby, S.D.; Gordon, J.I.; Head, R.D. A multi-amplicon 16S rRNA sequencing and analysis method for improved taxonomic profiling of bacterial communities. J. Microbiol. Methods 2018, 154, 6–13. [Google Scholar] [PubMed]
- Laudadio, I.; Fulci, V.; Palone, F.; Stronati, L.; Cucchiara, S.; Carissimi, C. Quantitative assessment of shotgun metagenomics and 16s rdna amplicon sequencing in the study of human gut microbiome. OMICS 2018, 22, 248–254. [Google Scholar]
- Shin, J.; Lee, S.; Go, M.J.; Lee, S.Y.; Kim, S.C.; Lee, C.H.; Cho, B.K. Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Sci. Rep. 2016, 6, 29681. [Google Scholar]
- Somerville, V.; Lutz, S.; Schmid, M.; Frei, D.; Moser, A.; Irmler, S.; Frey, J.E.; Ahrens, C.H. Long-read based de novo assembly of low-complexity metagenome samples results in finished genomes and reveals insights into strain diversity and an active phage system. BMC Microbiol. 2019, 19, 143. [Google Scholar]
- Bainomugisa, A.; Duarte, T.; Lavu, E.; Pandey, S.; Coulter, C.; Marais, B.J.; Coin, L.M. A complete high-quality MinION nanopore assembly of an extensively drug-resistant Mycobacterium tuberculosis Beijing lineage strain identifies novel variation in repetitive PE/PPE gene regions. Microb. Genom. 2018, 4, e000188. [Google Scholar] [CrossRef] [PubMed]
- De Marchi, B.R.; Kinene, T.; Wainaina, J.M.; Krause-Sakate, R.; Boykin, L. Comparative transcriptome analysis reveals genetic diversity in the endosymbiont Hamiltonella between native and exotic populations of Bemisia tabaci from Brazil. PLoS ONE 2018, 13, e0201411. [Google Scholar]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Balvočiūtė, M.; Huson, D.H. SILVA, RDP, Greengenes, NCBI and OTT—How do these taxonomies compare? BMC Genom. 2017, 18, 114. [Google Scholar] [CrossRef]
- Nicholls, S.M.; Quick, J.C.; Tang, S.; Loman, N.J. Ultra-deep, long-read nanopore sequencing of mock microbial community standards. Gigascience 2019, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Wong, J.; Heiner, C.; Oh, S.; Theriot, C.M.; Gulati, A.S.; McGill, S.K.; Dougherty, M.K. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019, 47, e103. [Google Scholar] [CrossRef]
- McIntyre, A.B.R.; Rizzardi, L.; Yu, A.M.; Alexander, N.; Rosen, G.L.; Botkin, D.J.; Stahl, S.E.; John, K.K.; Castro-Wallace, S.L.; McGrath, K.; et al. Nanopore sequencing in microgravity. NPJ Micrograv. 2016, 2, 16035. [Google Scholar] [CrossRef]
- Johnson, S.S.; Zaikova, E.; Goerlitz, D.S.; Bai, Y.; Tighe, S.W. Real-time DNA sequencing in the antarctic dry valleys using the oxford nanopore sequencer. J. Biomol. Tech. 2017, 28, 2–7. [Google Scholar] [CrossRef]
- Castro-Wallace, S.L.; Chiu, C.Y.; John, K.K.; Stahl, S.E.; Rubins, K.H.; McIntyre, A.B.R.; Dworkin, J.P.; Lupisella, M.L.; Smith, D.J.; Botkin, D.J.; et al. Nanopore DNA Sequencing and Genome Assembly on the International Space Station. Sci. Rep. 2017, 7, 18022. [Google Scholar] [CrossRef]
- Kerkhof, L.J.; Dillon, K.P.; Häggblom, M.M.; McGuinness, L.R. Profiling bacterial communities by MinION sequencing of ribosomal operons. Microbiome 2017, 5, 116. [Google Scholar] [CrossRef]
- Winand, R.; Bogaerts, B.; Hoffman, S.; Lefevre, L.; Delvoye, M.; Van Braekel, J.; Fu, Q.; Roosens, N.H.; De Keersmaecker, S.C.; Vanneste, K. Targeting the 16s rRNA gene for bacterial identification in complex mixed samples: Comparative evaluation of second (illumina) and third (oxford nanopore technologies) generation sequencing technologies. Int. J. Mol. Sci. 2019, 21, 298. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Won, S. Evaluation of 16S rRNA Databases for Taxonomic Assignments Using a Mock Community. Genom. Inform. 2018, 16, e24. [Google Scholar] [CrossRef] [PubMed]
- Helmersen, K.; Aamot, H.V. DNA extraction of microbial DNA directly from infected tissue: An optimized protocol for use in nanopore sequencing. Sci. Rep. 2020, 10, 2985. [Google Scholar] [CrossRef] [PubMed]
- Catozzi, C.; Ceciliani, F.; Lecchi, C.; Talenti, A.; Vecchio, D.; De Carlo, E.; Grassi, C.; Sánchez, A.; Francino, O.; Cuscó, A. Short communication: Milk microbiota profiling on water buffalo with full-length 16S rRNA using nanopore sequencing. J. Dairy Sci. 2020, 103, 2693–2700. [Google Scholar] [CrossRef]

| Workflow for Library Construction | Illumina | ZYMO | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of Raw reads (n = 44) | Number of classified reads (n = 44) | Genus | Species | Number of Raw reads (n = 44) | Number of classified reads (n = 44) | Genus | Species | ||||
| CC | UC | CC | UC | CC | UC | CC | UC | ||||
| 4,937,768 | 2,482,744 | 97.34% | 2.66% | 63.27% | 36.73% | 2,269,916 | 1,014,156 | 98.02% | 1.98% | 66.45% | 33.55% |
| MinION Sequencing | EPI2ME | CLC Genomics Workbench | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of Raw reads (n = 50) | Number of classified reads (n = 50) | Genus | Species | Genus | Species | |||||
| CC | UC | CC | UC | CC | UC | CC | UC | |||
| 5,033,641 | 5,027,091 | 97.21% | 2.79% | 89.74% | 11.26% | 96.04% | 3.96% | 72.15% | 27.85% | |
| Assigned OTU | 257 (Classified reads > 10) | 729 (Classified reads > 10) | Assigned OTU | 228 (Classified reads > 10) | 52 (Classified reads > 10) | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, P.-L.; Hung, C.-S.; Kao, Y.-W.; Lin, Y.-C.; Lee, C.-Y.; Chang, T.-H.; Shia, B.-C.; Lin, J.-C. Characterization of Fecal Microbiota with Clinical Specimen Using Long-Read and Short-Read Sequencing Platform. Int. J. Mol. Sci. 2020, 21, 7110. https://doi.org/10.3390/ijms21197110
Wei P-L, Hung C-S, Kao Y-W, Lin Y-C, Lee C-Y, Chang T-H, Shia B-C, Lin J-C. Characterization of Fecal Microbiota with Clinical Specimen Using Long-Read and Short-Read Sequencing Platform. International Journal of Molecular Sciences. 2020; 21(19):7110. https://doi.org/10.3390/ijms21197110
Chicago/Turabian StyleWei, Po-Li, Ching-Sheng Hung, Yi-Wei Kao, Ying-Chin Lin, Cheng-Yang Lee, Tzu-Hao Chang, Ben-Chang Shia, and Jung-Chun Lin. 2020. "Characterization of Fecal Microbiota with Clinical Specimen Using Long-Read and Short-Read Sequencing Platform" International Journal of Molecular Sciences 21, no. 19: 7110. https://doi.org/10.3390/ijms21197110
APA StyleWei, P.-L., Hung, C.-S., Kao, Y.-W., Lin, Y.-C., Lee, C.-Y., Chang, T.-H., Shia, B.-C., & Lin, J.-C. (2020). Characterization of Fecal Microbiota with Clinical Specimen Using Long-Read and Short-Read Sequencing Platform. International Journal of Molecular Sciences, 21(19), 7110. https://doi.org/10.3390/ijms21197110







