MicroRNAs: As Critical Regulators of Tumor- Associated Macrophages
Abstract
:1. Introduction
2. Diverse Strategies Opted by TAMs to Promote Tumor Progression—A Sneak Peek into Molecular Mechanisms
2.1. Proliferation and Growth of Cancer Cells
2.2. Angiogenesis
2.3. Metastasis
2.4. Resistance to Chemotherapy
2.5. Immunosuppression
3. MiRNAs Involved in Regulation of Macrophage Differentiation and Maturation
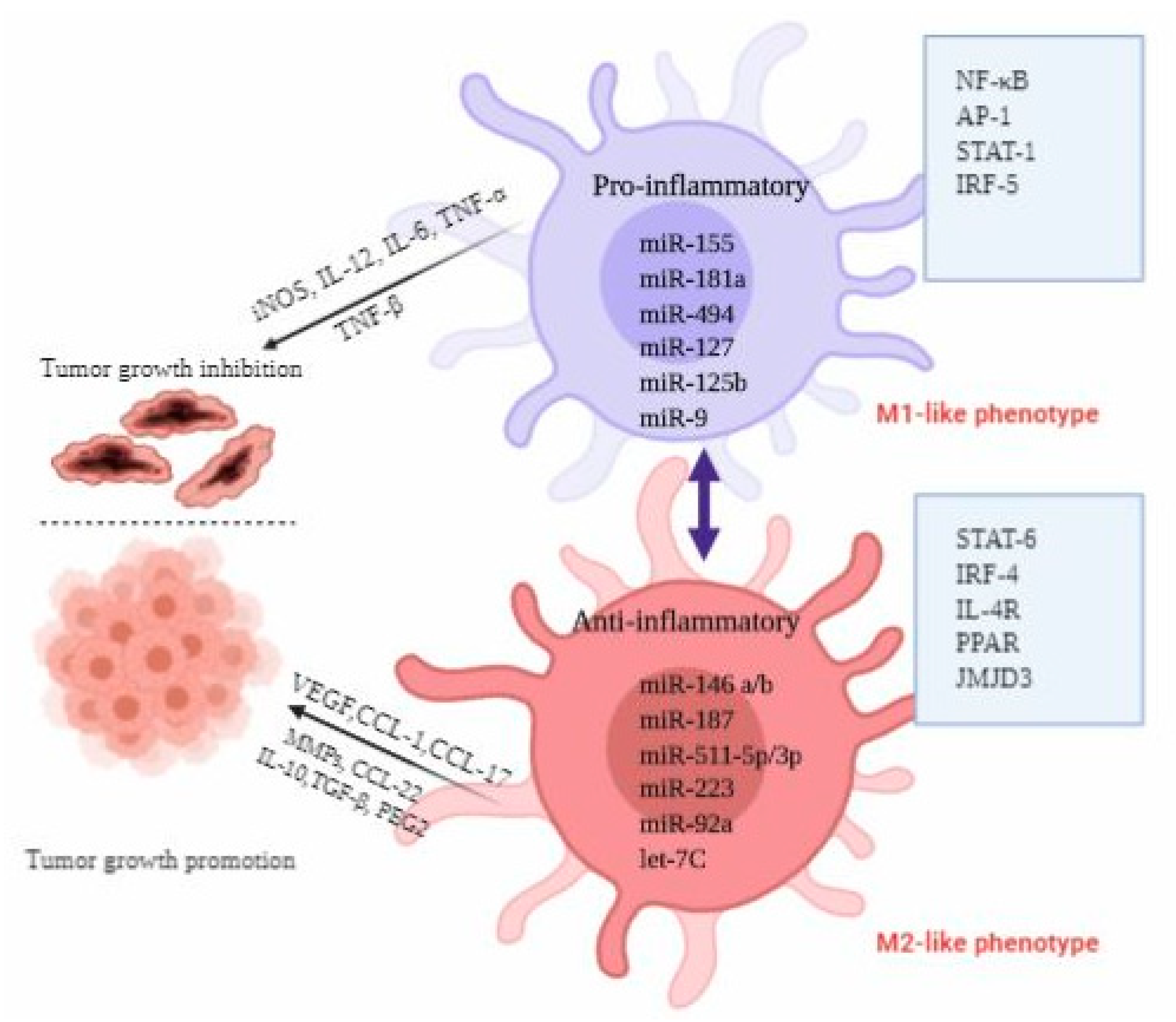
4. MiRNAs Involved in Macrophage Activation and Polarization
5. MiRNAs Involved in Regulation of Recruitment, Infiltration and Immunosuppressive Function of Macrophages
6. Extracellular MiRNAs Secreted from TAMs
7. Extracellular miRNAs Secreted from Cancer Cells
8. Conclusions and Future Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| A2AR | Adenosine A2A receptor |
| ANG2 | Angiopoietin-2 |
| BAK1 | BCL2 antagonist/Killer 1 |
| CCL2 | C-C motif chemokine ligand 2 |
| CRC | Colorectal cancer |
| CSF-1 | Colony-stimulating factor-1 |
| EGCG | Epigallocatechin gallate |
| EGF | Epidermal growth Factor |
| Egr2 | Early growth response protein 2 |
| EMT | Epithelial to mesenchymal transition |
| EOC | Epithelial ovarian cancer |
| EVs | Extracellular vesicles |
| GATA-3 | GATA binding protein-3 |
| GMP | Granulocyte–monocyte progenitor |
| GSK3β | Glycogen synthase kinase 3 beta |
| HCC | Hepatocellular carcinoma |
| HIF-2α | Hypoxia-inducible factor-2 alpha |
| ILs | Interleukins |
| MDSCs | Myeloid-derived suppressor cells |
| MERTK | MER proto-oncogene, tyrosine kinase |
| NF-kB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NSCLC | Non-small-cell lung carcinoma |
| OSCC | Oral squamous cells carcinoma |
| PDAC | Pancreatic ductal adenocarcinoma |
| PD-L-1 | Programmed death-ligand-1 |
| PI3K | Phosphatidylinositol 3-kinase |
| PPARδ | Peroxisome proliferator-activated receptor δ |
| SOCS3 | Suppressor of cytokine signaling 3 |
| STAT-3 | Signal transducer and activator of transcription-3 |
| TGCTs | Testicular germ cell tumors |
| TGFBR3 | Transforming Growth Factor Beta Receptor 3 |
| TLR | Toll like receptor |
| TNF-α | Tumor-necrosis factor-α |
| TWIST1 | Twist-related protein 1 |
| uPA | Urokinase-type plasminogen activator |
| VEGF | Vascular endothelial growth factor |
| WNT5B | Wnt family member 5B |
References
- Shishir, T.A.; Khan, R.; Nirzhor, S. The critical role of tumor microenvironment in cancer evolution and metastasis. Int. J. Bus. Res. 2018, 9, 244–258. [Google Scholar]
- Auffray, C.; Sieweke, M.H.; Geissmann, F. Blood monocytes: Development, heterogeneity, and relationship with dendritic cells. Annu. Rev. Immunol. 2009, 27, 669–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoeffel, G.; Ginhoux, F. Ontogeny of tissue-resident macrophages. Front. Immunol. 2015, 6, 486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perdiguero, E.G.; Klapproth, K.; Schulz, C.; Busch, K.; Azzoni, E.; Crozet, L.; Garner, H.; Trouillet, C.; De Bruijn, M.F.; Geissmann, F. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 2015, 518, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.-Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef] [Green Version]
- Lewis, C.E.; Pollard, J.W. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006, 66, 605–612. [Google Scholar] [CrossRef] [Green Version]
- Leek, R.D.; Lewis, C.E.; Whitehouse, R.; Greenall, M.; Clarke, J.; Harris, A.L. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res. 1996, 56, 4625–4629. [Google Scholar]
- Hanada, T.; Nakagawa, M.; Emoto, A.; Nomura, T.; Nasu, N.; Nomura, Y. Prognostic value of tumor-associated macrophage count in human bladder cancer. Int. J. Urol. 2000, 7, 263–269. [Google Scholar] [CrossRef]
- Lissbrant, I.F.; Stattin, P.; Wikstrom, P.; Damber, J.; Egevad, L.; Bergh, A. Tumor associated macrophages in human prostate cancer: Relation to clinicopathological variables and survival. Int. J. Oncol. 2000, 17, 445–496. [Google Scholar] [CrossRef]
- Cortez-Retamozo, V.; Etzrodt, M.; Newton, A.; Rauch, P.J.; Chudnovskiy, A.; Berger, C.; Ryan, R.J.; Iwamoto, Y.; Marinelli, B.; Gorbatov, R. Origins of tumor-associated macrophages and neutrophils. Proc. Natl. Acad. Sci. USA 2012, 109, 2491–2496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. 2016, 1, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Baer, C.; Squadrito, M.L.; Laoui, D.; Thompson, D.; Hansen, S.K.; Kiialainen, A.; Hoves, S.; Ries, C.H.; Ooi, C.-H.; De Palma, M. Suppression of microRNA activity amplifies IFN-γ-induced macrophage activation and promotes anti-tumour immunity. Nat. Cell Biol. 2016, 18, 790–802. [Google Scholar] [CrossRef]
- Xi, J.; Huang, Q.; Wang, L.; Ma, X.; Deng, Q.; Kumar, M.; Zhou, Z.; Li, L.; Zeng, Z.; Young, K.H. miR-21 depletion in macrophages promotes tumoricidal polarization and enhances PD-1 immunotherapy. Oncogene 2018, 37, 3151–3165. [Google Scholar] [CrossRef]
- Shidal, C.; Singh, N.P.; Nagarkatti, P.; Nagarkatti, M. MicroRNA-92 expression in CD133+ melanoma stem cells regulates immunosuppression in the tumor microenvironment via integrin-dependent activation of TGFβ. Cancer Res. 2019, 79, 3622–3635. [Google Scholar]
- Li, H.; Yang, B.B. Friend or foe: The role of microRNA in chemotherapy resistance. Acta Pharm. Sin. 2013, 34, 870–879. [Google Scholar] [CrossRef] [Green Version]
- Rutnam, Z.J.; Yang, B. The Involvement of microRNAs in Malignant Transformation. Histol Histopathol. 2012, 27, 1263–1270. [Google Scholar]
- Fang, L.; Du, W.W.; Yang, W.; Rutnam, Z.J.; Peng, C.; Li, H.; O’Malley, Y.Q.; Askeland, R.W.; Sugg, S.; Liu, M. MiR-93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle 2012, 11, 4352–4365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siragam, V.; Rutnam, Z.J.; Yang, W.; Fang, L.; Luo, L.; Yang, X.; Li, M.; Deng, Z.; Qian, J.; Peng, C. MicroRNA miR-98 inhibits tumor angiogenesis and invasion by targeting activin receptor-like kinase-4 and matrix metalloproteinase-11. Oncotarget 2012, 3, 1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, A.; Kuo, M.T.; Saunders, G.F. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell. Biol. 2006, 26, 8191–8201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syed, S.N.; Frank, A.-C.; Raue, R.; Brüne, B. Microrna—A tumor trojan horse for tumor-associated macrophages. Cells 2019, 8, 1482. [Google Scholar] [CrossRef] [Green Version]
- Mateescu, B.; Batista, L.; Cardon, M.; Gruosso, T.; De Feraudy, Y.; Mariani, O.; Nicolas, A.; Meyniel, J.-P.; Cottu, P.; Sastre-Garau, X. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat. Med. 2011, 17, 1627–1635. [Google Scholar] [CrossRef]
- Lan, H.; Lu, H.; Wang, X.; Jin, H. MicroRNAs as potential biomarkers in cancer: Opportunities and challenges. Biomed Res. Int. 2015, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Yang, N.; Zhu, S.; Lv, X.; Qiao, Y.; Liu, Y.-J.; Chen, J. MicroRNAs: Pleiotropic regulators in the tumor microenvironment. Front. Immunol. 2018, 9, 2491. [Google Scholar] [CrossRef] [Green Version]
- Petty, A.J.; Yang, Y. Tumor-associated macrophages: Implications in cancer immunotherapy. Immunotherapy 2017, 9, 289–302. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Zhang, Y. Tumor-associated macrophages: From basic research to clinical application. J. Hematol. Oncol. 2017, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Lohela, M.; Casbon, A.-J.; Olow, A.; Bonham, L.; Branstetter, D.; Weng, N.; Smith, J.; Werb, Z. Intravital imaging reveals distinct responses of depleting dynamic tumor-associated macrophage and dendritic cell subpopulations. Proc. Natl. Acad. Sci. USA 2014, 111, E5086–E5095. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Jin, Z.; Yuan, Y.; Liu, R.; Xu, T.; Wei, H.; Xu, X.; He, S.; Chen, S.; Shi, Z. New mechanisms of tumor-associated macrophages on promoting tumor progression: Recent research advances and potential targets for tumor immunotherapy. J. Immunol. Res. 2016, 9720912, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mu, X.; Shi, W.; Xu, Y.; Xu, C.; Zhao, T.; Geng, B.; Yang, J.; Pan, J.; Hu, S.; Zhang, C. Tumor-derived lactate induces M2 macrophage polarization via the activation of the ERK/STAT3 signaling pathway in breast cancer. Cell Cycle 2018, 17, 428–438. [Google Scholar] [CrossRef]
- El-Arabey, A.A.; Denizli, M.; Kanlikilicer, P.; Bayraktar, R.; Ivan, C.; Rashed, M.; Kabil, N.; Ozpolat, B.; Calin, G.A.; Salama, S.A. GATA3 as a master regulator for interactions of tumor-associated macrophages with high-grade serous ovarian carcinoma. Cell. Signal. 2020, 68, 109539. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, D.; Cang, H.; Guo, B. Crosstalk between cancer and immune cells: Role of tumor-associated macrophages in the tumor microenvironment. Cancer Med. 2019, 8, 4709–4721. [Google Scholar] [CrossRef]
- Soave, D.F.; Miguel, M.P.; Tomé, F.D.; de Menezes, L.B.; Nagib, P.R.A.; Celes, M.R.N. The fate of the tumor in the hands of microenvironment: Role of TAMs and mTOR pathway. Mediat. Inflamm. 2016, 8910520, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Leek, R.D.; Harris, A.L. Tumor-associated macrophages in breast cancer. J. Mammary Gland Biol. Neoplasia 2002, 7, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.; Landers, R.; Underwood, J.; Harris, A.; Lewis, C. Expression of vascular endothelial growth factor by macrophages is up-regulated in poorly vascularized areas of breast carcinomas. J. Pathol. A J. Pathol. Soc. Great Br. Irel. 2000, 192, 150–158. [Google Scholar] [CrossRef]
- Kitamura, T.; Qian, B.-Z.; Soong, D.; Cassetta, L.; Noy, R.; Sugano, G.; Kato, Y.; Li, J.; Pollard, J.W. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J. Exp. Med. 2015, 212, 1043–1059. [Google Scholar] [CrossRef]
- Low-Marchelli, J.M.; Ardi, V.C.; Vizcarra, E.A.; van Rooijen, N.; Quigley, J.P.; Yang, J. Twist1 induces CCL2 and recruits macrophages to promote angiogenesis. Cancer Res. 2013, 73, 662–671. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Chen, Y.-S.; Yao, Y.-D.; Chen, J.-Q.; Chen, J.-N.; Huang, S.-Y.; Zeng, Y.-J.; Yao, H.-R.; Zeng, S.-H.; Fu, Y.-S. CCL18 from tumor-associated macrophages promotes angiogenesis in breast cancer. Oncotarget 2015, 6, 34758. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.-D.; Zhou, B. TNF-α/NF-κ B/Snail pathway in cancer cell migration and invasion. Br. J. Cancer 2010, 102, 639–644. [Google Scholar] [CrossRef] [Green Version]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef] [Green Version]
- Dhanasekaran, R.; Baylot, V.; Kim, M.; Kuruvilla, S.; Bellovin, D.I.; Adeniji, N.; KD, A.R.; Lai, I.; Gabay, M.; Tong, L. MYC and Twist1 cooperate to drive metastasis by eliciting crosstalk between cancer and innate immunity. Elife 2020, 9, e50731. [Google Scholar] [CrossRef]
- Liu, C.-Y.; Xu, J.-Y.; Shi, X.-Y.; Huang, W.; Ruan, T.-Y.; Xie, P.; Ding, J.-L. M2-polarized tumor-associated macrophages promoted epithelial–mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab. Investig. 2013, 93, 844–854. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Song, Y.; Du, W.; Gong, L.; Chang, H.; Zou, Z. Tumor-associated macrophages: An accomplice in solid tumor progression. J. Biomed. Sci. 2019, 26, 1–13. [Google Scholar] [CrossRef]
- Paulus, P.; Stanley, E.R.; Schäfer, R.; Abraham, D.; Aharinejad, S. Colony-stimulating factor-1 antibody reverses chemoresistance in human MCF-7 breast cancer xenografts. Cancer Res. 2006, 66, 4349–4356. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Ye, J.; Huang, C.; Yan, Y.; Li, J. M2 macrophage-derived IL6 mediates resistance of breast cancer cells to hedgehog inhibition. Toxicol. Appl. Pharmacol. 2019, 364, 77–82. [Google Scholar] [CrossRef]
- Jinushi, M.; Chiba, S.; Yoshiyama, H.; Masutomi, K.; Kinoshita, I.; Dosaka-Akita, H.; Yagita, H.; Takaoka, A.; Tahara, H. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc. Natl. Acad. Sci. USA 2011, 108, 12425–12430. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Liu, R.; Su, X.; Pan, Y.; Han, X.; Shao, C.; Shi, Y. Harnessing tumor-associated macrophages as aids for cancer immunotherapy. Mol. Cancer 2019, 18, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Ruffell, B.; Chang-Strachan, D.; Chan, V.; Rosenbusch, A.; Ho, C.M.; Pryer, N.; Daniel, D.; Hwang, E.S.; Rugo, H.S.; Coussens, L.M. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell 2014, 26, 623–637. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.-J.; LI, J. The role of tumor-associated macrophages in remodeling tumor immune microenvironment. Tumor 2013, 33, 922–927. [Google Scholar]
- Han, Q.; Shi, H.; Liu, F. CD163+ M2-type tumor-associated macrophage support the suppression of tumor-infiltrating T cells in osteosarcoma. Int. Immunopharmacol. 2016, 34, 101–106. [Google Scholar] [CrossRef]
- Xiangrong, C.; Jingbo, L. Dendritic cells play a role in the specific cellular immunity. J. Immunol. 2001, 17, 231–234. [Google Scholar]
- Zheng, X.; Turkowski, K.; Mora, J.; Brüne, B.; Seeger, W.; Weigert, A.; Savai, R. Redirecting tumor-associated macrophages to become tumoricidal effectors as a novel strategy for cancer therapy. Oncotarget 2017, 8, 48436. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, P.C.; Quiceno, D.G.; Zabaleta, J.; Ortiz, B.; Zea, A.H.; Piazuelo, M.B.; Delgado, A.; Correa, P.; Brayer, J.; Sotomayor, E.M. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004, 64, 5839–5849. [Google Scholar] [CrossRef] [Green Version]
- Lu, T.; Ramakrishnan, R.; Altiok, S.; Youn, J.-I.; Cheng, P.; Celis, E.; Pisarev, V.; Sherman, S.; Sporn, M.B.; Gabrilovich, D. Tumor-infiltrating myeloid cells induce tumor cell resistance to cytotoxic T cells in mice. J. Clin. Investig. 2011, 121, 4015–4029. [Google Scholar] [CrossRef] [Green Version]
- Kierdorf, K.; Prinz, M.; Geissmann, F.; Perdiguero, E.G. Development and Function of Tissue Resident Macrophages in Mice. In Seminars in Immunology; Academic Press: Cambridge, MA, USA, 2015; Volume 27, pp. 369–378. [Google Scholar]
- Zhao, Y.; Zou, W.; Du, J.; Zhao, Y. The origins and homeostasis of monocytes and tissue-resident macrophages in physiological situation. J. Cell. Physiol. 2018, 233, 6425–6439. [Google Scholar] [CrossRef]
- Roy, S. miRNA in macrophage development and function. Antioxid. Redox Signal. 2016, 25, 795–804. [Google Scholar] [CrossRef] [Green Version]
- Gruber, J.J.; Zatechka, D.S.; Sabin, L.R.; Yong, J.; Lum, J.J.; Kong, M.; Zong, W.-X.; Zhang, Z.; Lau, C.-K.; Rawlings, J. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell 2009, 138, 328–339. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, R.M.; Chaudhuri, A.A.; Rao, D.S.; Gibson, W.S.; Balazs, A.B.; Baltimore, D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc. Natl. Acad. Sci. USA 2010, 107, 14235–14240. [Google Scholar] [CrossRef] [Green Version]
- Gentner, B.; Visigalli, I.; Hiramatsu, H.; Lechman, E.; Ungari, S.; Giustacchini, A.; Schira, G.; Amendola, M.; Quattrini, A.; Martino, S. Identification of hematopoietic stem cell–specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci. Transl. Med. 2010, 2, ra58–ra84. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Lu, J.; Schlanger, R.; Zhang, H.; Wang, J.Y.; Fox, M.C.; Purton, L.E.; Fleming, H.H.; Cobb, B.; Merkenschlager, M. MicroRNA miR-125a controls hematopoietic stem cell number. Proc. Natl. Acad. Sci. USA 2010, 107, 14229–14234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lechman, E.R.; Gentner, B.; Van Galen, P.; Giustacchini, A.; Saini, M.; Boccalatte, F.E.; Hiramatsu, H.; Restuccia, U.; Bachi, A.; Voisin, V. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell 2012, 11, 799–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klemsz, M.J.; McKercher, S.R.; Celada, A.; Van Beveren, C.; Maki, R.A. The macrophage and B cell-specific transcription factor PU. 1 is related to the ets oncogene. Cell 1990, 61, 113–124. [Google Scholar] [CrossRef]
- Ghani, S.; Riemke, P.; Schönheit, J.; Lenze, D.; Stumm, J.; Hoogenkamp, M.; Lagendijk, A.; Heinz, S.; Bonifer, C.; Bakkers, J. Macrophage development from HSCs requires PU. 1-coordinated microRNA expression. Blood 2011, 118, 2275–2284. [Google Scholar] [CrossRef] [Green Version]
- Pospisil, V.; Vargova, K.; Kokavec, J.; Rybarova, J.; Savvulidi, F.; Jonasova, A.; Necas, E.; Zavadil, J.; Laslo, P.; Stopka, T. Epigenetic silencing of the oncogenic miR-17-92 cluster during PU. 1-directed macrophage differentiation. Embo J. 2011, 30, 4450–4464. [Google Scholar] [CrossRef]
- Fazi, F.; Rosa, A.; Fatica, A.; Gelmetti, V.; De Marchis, M.L.; Nervi, C.; Bozzoni, I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell 2005, 123, 819–831. [Google Scholar] [CrossRef] [Green Version]
- Velu, C.S.; Baktula, A.M.; Grimes, H.L. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood J. Am. Soc. Hematol. 2009, 113, 4720–4728. [Google Scholar] [CrossRef] [Green Version]
- Cobos Jiménez, V.; Bradley, E.J.; Willemsen, A.M.; van Kampen, A.H.; Baas, F.; Kootstra, N.A. Next-generation sequencing of microRNAs uncovers expression signatures in polarized macrophages. Physiol. Genom. 2014, 46, 91–103. [Google Scholar] [CrossRef] [Green Version]
- Frank, A.-C.; Ebersberger, S.; Fink, A.F.; Lampe, S.; Weigert, A.; Schmid, T.; Ebersberger, I.; Syed, S.N.; Brüne, B. Apoptotic tumor cell-derived microRNA-375 uses CD36 to alter the tumor-associated macrophage phenotype. Nat. Commun. 2019, 10, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.; Liu, M.; Xu, Z.; Li, Y.; Guo, H.; Ge, Y.; Liu, Y.; Zheng, D.; Shi, J. A double feedback loop mediated by microRNA-23a/27a/24-2 regulates M1 versus M2 macrophage polarization and thus regulates cancer progression. Oncotarget 2016, 7, 13502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Li, X.; Zhang, Y.; Wang, H.; Rong, X.; Peng, J.; He, L.; Peng, Y. An miR-340-5p-macrophage feedback loop modulates the progression and tumor microenvironment of glioblastoma multiforme. Oncogene 2019, 38, 7399–7415. [Google Scholar] [CrossRef] [PubMed]
- Su, M.-J.; Aldawsari, H.; Amiji, M. Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of microRNAs 155 and 125b2 transfection using nanoparticle delivery systems. Sci. Rep. 2016, 6, 30110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, J.; Qiao, B.; Gao, N.; Lin, N.; He, W. Oral squamous cell carcinoma-derived exosomes promote M2 subtype macrophage polarization mediated by exosome-enclosed miR-29a-3p. Am. J. Physiol. Cell Physiol. 2019, 316, C731–C740. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Wu, Q.; Wu, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wang, X. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macrophages. Oncotarget 2016, 7, 43076. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Ying, X.; Wang, X.; Wu, X.; Zhu, Q.; Wang, X. Exosomes derived from hypoxic epithelial ovarian cancer deliver microRNA-940 to induce macrophage M2 polarization. Oncol. Rep. 2017, 38, 522–528. [Google Scholar] [CrossRef] [Green Version]
- Takano, Y.; Masuda, T.; Iinuma, H.; Yamaguchi, R.; Sato, K.; Tobo, T.; Hirata, H.; Kuroda, Y.; Nambara, S.; Hayashi, N. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget 2017, 8, 78598. [Google Scholar] [CrossRef] [Green Version]
- Shinohara, H.; Kuranaga, Y.; Kumazaki, M.; Sugito, N.; Yoshikawa, Y.; Takai, T.; Taniguchi, K.; Ito, Y.; Akao, Y. Regulated polarization of tumor-associated macrophages by mir-145 via colorectal cancer–derived extracellular vesicles. J. Immunol. 2017, 199, 1505–1515. [Google Scholar] [CrossRef] [Green Version]
- Jang, J.-Y.; Lee, J.-K.; Jeon, Y.-K.; Kim, C.-W. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. Bmc Cancer 2013, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Hsu, Y.-L.; Hung, J.-Y.; Chang, W.-A.; Jian, S.-F.; Lin, Y.-S.; Pan, Y.-C.; Wu, C.-Y.; Kuo, P.-L. Hypoxic lung-cancer-derived extracellular vesicle microRNA-103a increases the oncogenic effects of macrophages by targeting PTEN. Mol. Ther. 2018, 26, 568–581. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zhou, J.; Li, X.; Wang, X.; Lin, Y.; Wang, X. Exosomes derived from hypoxic epithelial ovarian cancer cells deliver microRNAs to macrophages and elicit a tumor-promoted phenotype. Cancer Lett. 2018, 435, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Luo, G.; Zhang, K.; Cao, J.; Huang, C.; Jiang, T.; Liu, B.; Su, L.; Qiu, Z. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to promote pancreatic cancer metastasis. Cancer Res. 2018, 78, 4586–4598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graff, J.W.; Dickson, A.M.; Clay, G.; McCaffrey, A.P.; Wilson, M.E. Identifying functional microRNAs in macrophages with polarized phenotypes. J. Biol. Chem. 2012, 287, 21816–21825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.E.; Dutta, B.; Tse, S.W.; Gupta, N.; Tan, C.F.; Low, J.K.; Yeoh, K.W.; Kon, O.L.; Tam, J.P.; Sze, S.K. Hypoxia-induced tumor exosomes promote M2-like macrophage polarization of infiltrating myeloid cells and microRNA-mediated metabolic shift. Oncogene 2019, 38, 5158–5173. [Google Scholar] [CrossRef]
- Fortunato, O.; Borzi, C.; Milione, M.; Centonze, G.; Conte, D.; Boeri, M.; Verri, C.; Moro, M.; Facchinetti, F.; Andriani, F. Circulating mir-320a promotes immunosuppressive macrophages M2 phenotype associated with lung cancer risk. Int. J. Cancer 2019, 144, 2746–2761. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhao, L.; Shi, B.; Ma, S.; Xu, Z.; Ge, Y.; Liu, Y.; Zheng, D.; Shi, J. Functions of miR-146a and miR-222 in tumor-associated macrophages in breast cancer. Sci. Rep. 2015, 5, 18648. [Google Scholar] [CrossRef]
- Chen, C.; Liu, J.-M.; Luo, Y.-P. MicroRNAs in tumor immunity: Functional regulation in tumor-associated macrophages. J. Zhejiang Univ. Sci. B 2019, 1–17. [Google Scholar] [CrossRef]
- Zhang, J.; Shan, W.-F.; Wu, G.-Q.; Xiong, X.-X.; Jin, H.-Y.; Zhu, S.-M. Propofol exerts anti-hepatocellular carcinoma by microvesicle-mediated transfer of miR-142-3p from macrophage to cancer cells. J. Transl. Med. 2014, 12, 279. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Xie, N.; Cui, H.; Tan, Z.; Yang, S.; Icyuz, M.; Abraham, E.; Liu, G. MicroRNA let-7c regulates macrophage polarization. J. Immunol. 2013, 190, 6542–6549. [Google Scholar] [CrossRef]
- Qian, M.; Wang, S.; Guo, X.; Wang, J.; Zhang, Z.; Qiu, W.; Gao, X.; Chen, Z.; Xu, J.; Zhao, R. Hypoxic glioma-derived exosomes deliver microRNA-1246 to induce M2 macrophage polarization by targeting TERF2IP via the STAT3 and NF-κB pathways. Oncogene 2020, 39, 428–442. [Google Scholar] [CrossRef]
- van der Vos, K.E.; Abels, E.R.; Zhang, X.; Lai, C.; Carrizosa, E.; Oakley, D.; Prabhakar, S.; Mardini, O.; Crommentuijn, M.H.; Skog, J. Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro-Oncol. 2015, 18, 58–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Lai, X.; Yu, S.; Chen, S.; Ma, Y.; Zhang, Y.; Li, H.; Zhu, X.; Yao, L.; Zhang, J. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res. Treat. 2014, 147, 423–431. [Google Scholar] [CrossRef]
- Kanlikilicer, P.; Bayraktar, R.; Denizli, M.; Rashed, M.H.; Ivan, C.; Aslan, B.; Mitra, R.; Karagoz, K.; Bayraktar, E.; Zhang, X. Exosomal miRNA confers chemo resistance via targeting Cav1/p-gp/M2-type macrophage axis in ovarian cancer. EBioMedicine 2018, 38, 100–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Challagundla, K.B.; Wise, P.M.; Neviani, P.; Chava, H.; Murtadha, M.; Xu, T.; Kennedy, R.; Ivan, C.; Zhang, X.; Vannini, I. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Fan, L.; Yu, H.; Zhang, J.; He, Y.; Feng, D.; Wang, F.; Li, X.; Liu, Q.; Li, Y. Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-regulate Programmed Death Ligand 1 Expression in Macrophages. Hepatology 2019, 70, 241–258. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Shen, H.; Yin, X.; Long, L.; Chen, X.; Feng, F.; Liu, Y.; Zhao, P.; Xu, Y.; Li, M. IL-6R/STAT3/miR-204 feedback loop contributes to cisplatin resistance of epithelial ovarian cancer cells. Oncotarget 2017, 8, 39154. [Google Scholar] [CrossRef] [Green Version]
- Dickman, C.T.; Lawson, J.; Jabalee, J.; MacLellan, S.A.; LePard, N.E.; Bennewith, K.L.; Garnis, C. Selective extracellular vesicle exclusion of miR-142-3p by oral cancer cells promotes both internal and extracellular malignant phenotypes. Oncotarget 2017, 8, 15252. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Cai, X.; Chen, X.; Liang, H.; Zhang, Y.; Li, J.; Wang, Z.; Chen, X.; Zhang, W.; Yokoyama, S. Tumor-secreted miR-214 induces regulatory T cells: A major link between immune evasion and tumor growth. Cell Res. 2014, 24, 1164–1180. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Li, D.; Wu, A.; Qiu, X.; Di, W.; Huang, L.; Qiu, L. TWEAK-stimulated macrophages inhibit metastasis of epithelial ovarian cancer via exosomal shuttling of microRNA. Cancer Lett. 2017, 393, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Binenbaum, Y.; Fridman, E.; Yaari, Z.; Milman, N.; Schroeder, A.; David, G.B.; Shlomi, T.; Gil, Z. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. 2018, 78, 5287–5299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Li, X.; Wu, X.; Zhang, T.; Zhu, Q.; Wang, X.; Wang, H.; Wang, K.; Lin, Y.; Wang, X. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol. Res. 2018, 6, 1578–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, J.; Sun, L.; Xu, F.; Liu, L.; Hu, F.; Song, D.; Hou, Z.; Wu, W.; Luo, X.; Wang, J. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019, 79, 146–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, B.; Xiao, S.; Li, Y.; Chen, Q. miR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J. Cell. Biochem. 2019, 120, 3046–3055. [Google Scholar] [CrossRef]
- Zheng, P.; Chen, L.; Yuan, X.; Luo, Q.; Liu, Y.; Xie, G.; Ma, Y.; Shen, L. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J. Exp. Clin. Cancer Res. 2017, 36, 53. [Google Scholar] [CrossRef] [Green Version]
- Yin, Z.; Ma, T.; Huang, B.; Lin, L.; Zhou, Y.; Yan, J.; Zou, Y.; Chen, S. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-β signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 310. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Shen, H.; Yin, X.; Yang, M.; Wei, H.; Chen, Q.; Feng, F.; Liu, Y.; Xu, W.; Li, Y. Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J. Exp. Clin. Cancer Res. 2019, 38, 1–14. [Google Scholar] [CrossRef]
- Roy, S.; Sen, C.K. miRNA in wound inflammation and angiogenesis. Microcirculation 2012, 19, 224–232. [Google Scholar] [CrossRef] [Green Version]
- Ke, M.; Zhang, Z.; Cong, L.; Zhao, S.; Li, Y.; Wang, X.; Lv, Y.; Zhu, Y.; Dong, J. MicroRNA-148b-colony-stimulating factor-1 signaling-induced tumor-associated macrophage infiltration promotes hepatocellular carcinoma metastasis. Biomed. Pharmacother. 2019, 120, 109523. [Google Scholar] [CrossRef]
- Batool, A.; Wang, Y.-Q.; Hao, X.-X.; Chen, S.-R.; Liu, Y.-X. A miR-125b/CSF1-CX3CL1/tumor-associated macrophage recruitment axis controls testicular germ cell tumor growth. Cell Death Dis. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Hao, N.-B.; Lü, M.-H.; Fan, Y.-H.; Cao, Y.-L.; Zhang, Z.-R.; Yang, S.-M. Macrophages in tumor microenvironments and the progression of tumors. Clin. Dev. Immunol. 2012, 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, P.J. Macrophage polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef] [PubMed]
- Curtale, G. MiRNAs at the crossroads between innate immunity and cancer: Focus on macrophages. Cells 2018, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Aras, S.; Zaidi, M.R. TAMeless traitors: Macrophages in cancer progression and metastasis. Br. J. Cancer 2017, 117, 1583–1591. [Google Scholar] [CrossRef] [Green Version]
- Vinchi, F.; Muckenthaler, M.U.; Da Silva, M.C.; Balla, G.; Balla, J.; Jeney, V. Atherogenesis and iron: From epidemiology to cellular level. Front. Pharmacol. 2014, 5, 94. [Google Scholar] [CrossRef] [Green Version]
- Gleissner, C.A.; Shaked, I.; Little, K.M.; Ley, K. CXC chemokine ligand 4 induces a unique transcriptome in monocyte-derived macrophages. J. Immunol. 2010, 184, 4810–4818. [Google Scholar] [CrossRef] [Green Version]
- Ostrand-Rosenberg, S.; Sinha, P. Myeloid-derived suppressor cells: Linking inflammation and cancer. J. Immunol. 2009, 182, 4499–4506. [Google Scholar] [CrossRef]
- Nahrendorf, M.; Swirski, F.K.; Aikawa, E.; Stangenberg, L.; Wurdinger, T.; Figueiredo, J.-L.; Libby, P.; Weissleder, R.; Pittet, M.J. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J. Exp. Med. 2007, 204, 3037–3047. [Google Scholar] [CrossRef] [Green Version]
- Hume, D.A. Macrophages as APC and the dendritic cell myth. J. Immunol. 2008, 181, 5829–5835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atochina, O.; Daly-Engel, T.; Piskorska, D.; McGuire, E.; Harn, D.A. A schistosome-expressed immunomodulatory glycoconjugate expands peritoneal Gr1+ macrophages that suppress naive CD4+ T cell proliferation via an IFN-γ and nitric oxide-dependent mechanism. J. Immunol. 2001, 167, 4293–4302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rőszer, T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediat. Inflamm. 2015, 816460, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhang, M.; Zhong, M.; Suo, Q.; Lv, K. Expression profiles of miRNAs in polarized macrophages. Int. J. Mol. Med. 2013, 31, 797–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponomarev, E.D.; Veremeyko, T.; Barteneva, N.; Krichevsky, A.M.; Weiner, H.L. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α–PU. 1 pathway. Nat. Med. 2011, 17, 64–70. [Google Scholar] [CrossRef]
- Li, D.; Duan, M.; Feng, Y.; Geng, L.; Li, X.; Zhang, W. MiR-146a modulates macrophage polarization in systemic juvenile idiopathic arthritis by targeting INHBA. Mol. Immunol. 2016, 77, 205–212. [Google Scholar] [CrossRef]
- Huang, C.; Liu, X.-J.; Xie, J.; Ma, T.-T.; Meng, X.-M.; Li, J. MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264. 7 macrophages. Int. Immunopharmacol. 2016, 32, 46–54. [Google Scholar] [CrossRef] [Green Version]
- Ying, H.; Kang, Y.; Zhang, H.; Zhao, D.; Xia, J.; Lu, Z.; Wang, H.; Xu, F.; Shi, L. MiR-127 modulates macrophage polarization and promotes lung inflammation and injury by activating the JNK pathway. J. Immunol. 2015, 194, 1239–1251. [Google Scholar] [CrossRef] [Green Version]
- Chaudhuri, A.A.; So, A.Y.-L.; Sinha, N.; Gibson, W.S.; Taganov, K.D.; O’Connell, R.M.; Baltimore, D. MicroRNA-125b potentiates macrophage activation. J. Immunol. 2011, 187, 5062–5068. [Google Scholar] [CrossRef]
- Squadrito, M.L.; Etzrodt, M.; De Palma, M.; Pittet, M.J. MicroRNA-mediated control of macrophages and its implications for cancer. Trends Immunol. 2013, 34, 350–359. [Google Scholar] [CrossRef] [Green Version]
- Petty, A.J.; Yang, Y. Tumor-Associated Macrophages in Hematologic Malignancies: New Insights and Targeted Therapies. Cells 2019, 8, 1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortezaee, K. CXCL12/CXCR4 axis in the microenvironment of solid tumors: A critical mediator of metastasis. Life Sci. 2020, 249, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bingle, L.; Brown, N.; Lewis, C.E. The role of tumour-associated macrophages in tumour progression: Implications for new anticancer therapies. J. Pathol. A J. Pathol. Soc. Great Br. Irel. 2002, 196, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Movahedi, K.; Laoui, D.; Gysemans, C.; Baeten, M.; Stangé, G.; Van den Bossche, J.; Mack, M.; Pipeleers, D.; In’t Veld, P.; De Baetselier, P. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C (high) monocytes. Cancer Res. 2010, 70, 5728–5739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, L.; Zhou, Y.; Bu, H.; Lv, T.; Shi, Y.; Yang, J. Deletion of interleukin-6 in monocytes/macrophages suppresses the initiation of hepatocellular carcinoma in mice. J. Exp. Clin. Cancer Res. 2016, 35, 131. [Google Scholar] [CrossRef] [Green Version]
- Chai, Z.-T.; Zhu, X.-D.; Ao, J.-Y.; Wang, W.-Q.; Gao, D.-M.; Kong, J.; Zhang, N.; Zhang, Y.-Y.; Ye, B.-G.; Ma, D.-N. microRNA-26a suppresses recruitment of macrophages by down-regulating macrophage colony-stimulating factor expression through the PI3K/Akt pathway in hepatocellular carcinoma. J. Hematol. Oncol. 2015, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- He, M.; Xu, Z.; Ding, T.; Kuang, D.-M.; Zheng, L. MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPβ. Cell. Mol. Immunol. 2009, 6, 343–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andaloussi, S.E.; Lakhal, S.; Mäger, I.; Wood, M.J. Exosomes for targeted siRNA delivery across biological barriers. Adv. Drug Deliv. Rev. 2013, 65, 391–397. [Google Scholar] [CrossRef]
- Muralidharan-Chari, V.; Clancy, J.W.; Sedgwick, A.; D'Souza-Schorey, C. Microvesicles: Mediators of extracellular communication during cancer progression. J. Cell Sci. 2010, 123, 1603–1611. [Google Scholar] [CrossRef] [Green Version]
- Moradi-Chaleshtori, M.; Hashemi, S.M.; Soudi, S.; Bandehpour, M.; Mohammadi-Yeganeh, S. Tumor-derived exosomal microRNAs and proteins as modulators of macrophage function. J. Cell. Physiol. 2019, 234, 7970–7982. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, X.; Li, L.; Wang, C.; Feng, M.; Zhao, K.; Zhao, R.; Liu, J.; Fang, Y. Tumor-associated macrophage-derived exosomal microRNA-155-5p stimulates intracranial aneurysm formation and macrophage infiltration. Clin. Sci. 2019, 133, 2265–2282. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Chen, J.; Su, F.; Yu, B.; Su, F.; Lin, L.; Liu, Y.; Huang, J.-D.; Song, E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 2011, 10, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frediani, J.N.; Fabbri, M. Essential role of miRNAs in orchestrating the biology of the tumor microenvironment. Mol. Cancer 2016, 15, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooks, T.; Pateras, I.S.; Jenkins, L.M.; Patel, K.M.; Robles, A.I.; Morris, J.; Forshew, T.; Appella, E.; Gorgoulis, V.G.; Harris, C.C. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, C.-H.; Tai, S.-K.; Yang, M.-H. Snail-overexpressing cancer cells promote M2-like polarization of tumor-associated macrophages by delivering MiR-21-abundant exosomes. Neoplasia 2018, 20, 775–788. [Google Scholar] [CrossRef]
- Xing, F.; Liu, Y.; Wu, S.-Y.; Wu, K.; Sharma, S.; Mo, Y.-Y.; Feng, J.; Sanders, S.; Jin, G.; Singh, R. Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. 2018, 78, 4316–4330. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Z.; Chen, C.; Liu, Y.; Si, Q.; Chuang, T.; Li, N.; Gomez-Cabrero, A.; Reisfeld, R.; Xiang, R. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene 2014, 33, 3014–3023. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Zhu, Y.; Li, L.; Zhou, S.; Yin, G.; Yu, G.; Cui, H. Breast cancer cell-derived exosomal miR-20a-5p promotes the proliferation and differentiation of osteoclasts by targeting SRCIN1. Cancer Med. 2019, 8, 5687–5701. [Google Scholar] [CrossRef] [Green Version]


| miRNAs Involved in Macrophage Polarization | Function | Reference |
|---|---|---|
| miR-375 | Facilitates macrophage recruitment, M2-like phenotype and tumor progression | [71] |
| miR-23a/miR-27a/miR24-2 cluster | Promote M1-like phenotype, inhibit M2-like phenotype in breast cancer | [72] |
| miR-340-5p | Promotes M2 polarization in glioblastoma | [73] |
| miR-155, miR-125b-2 | Macrophage re-programming to M1-like phenotype in pancreatic cancer | [74] |
| miR-29a-3p | Promotes M2 polarization in OSCC | [75] |
| miR-222-3p | Promotes M2-like phenotype in ovarian cancer | [76] |
| miR-940 | Promotes M2 polarization in epithelial ovarian cell carcinoma | [77] |
| miR-203 | Promotes M2 polarization and metastasis in CRC | [78] |
| miR-145 | Promotes M2-like phenotype in CRC | [79] |
| miR-16 | Promotes M1-like phenotype in breast cancer | [80] |
| miR-103a | Increases M2 polarization in lung cancer | [81] |
| miR-21-3p, miR-181d-5p, miR-125b -5p | Promote M2 polarization, cancer cell migration, proliferation in EOC | [82] |
| miR-301a-3p | Enriches M2-like macrophages via modulating PTEN/PI3Kγ axis in pancreatic cancer | [83] |
| miR-132, miR-29b-1, miR-27a, miR-146a, miR-222 | Higher expression of these miRNAs promote M2b-like phenotype | [59,84] |
| miR-let7a, miR-320a, miR-146a | Promote M2-like phenotype | [85,86,87] |
| miR-142-3p | Inhibit M2 polarization, reduces tumor growth in HCC | [88,89] |
| let-7c | Inhibits M1 polarization and promote M2-like phenotype | [90] |
| miR-1246 | Promote M2 polarization via modulating STAT3 and NF-κB axis. | [88,91] |
| let-7d-5p | Promote M2-like phenotype | [16] |
| miR-451, miR-21 | Influence macrophage polarization in glioblastoma | [92] |
| Extracellular miRNAs Secreted from Cancer Cells Regulating Cancer Progression or Chemoresistance | Function | Reference |
| miR-221, miR-222 | Promote tamoxifen resistance in breast tumor cells | [93] |
| miR-1246 | Imparts chemoresistance in ovarian cancer cells | [94] |
| miR-21 | Induces chemoresistance in neuroblastoma, enhances miR-155 expression | [95] |
| miR-23a-3p | Promotes tumor cell escape via impairing T-cell function | [96] |
| miR-204-5p | Regulates cisplatin resistance in EOC | [97] |
| miR-let7a | Creates an immunosuppressive environment by enhancing the expression of M2-like phenotype associated genes | [85] |
| miR-142-3p | Induces malignant phenotype in oral carcinoma | [98] |
| miR-105 | Promotes tumor progression and metastasis via degrading vascular endothelial barriers | [99] |
| miR-214 | Promotes tumor growth via deregulating PTEN and impairing T-cell function in mouse model | [100] |
| miR-21, miR-29a | Induces inflammatory response in NSCLC cells via activating NF-kB pathway | [101] |
| Extracellular miRNAs Derived from TAMs Regulating Cancer Progression or Chemoresistance | Function | Reference |
| miR-7 | Inhibits metastasis in EOC via modulating EGFR/AKT/ERK1/2 axis | [102] |
| miR-365 | Promotes gemcitabine chemoresistance in PDAC | [103] |
| miR-29a-3p, miR-21-5p | Establish an immunosuppressive environment in EOC | [104] |
| miR-155-5p, miR-21-5p | Increase migration and invasion of colon cancer cells via downregulating BRG 1 | [105] |
| miR-125a/b | Negatively influence tumor cell division and stemness properties of HCC via targeting CD90 | [106] |
| miR-21 | Induces chemoresistance in gastric cancer | [107] |
| miR-501-3p | Progression of PDAC via modulating TGF-β | [108] |
| miR-223 | Imparts cisplatin resistance in EOC | [109] |
| miRNAs Involved in Macrophage Differentiation and Maturation | Function | Reference |
| miR-155, miR-146a, miR-338, and miR-342 | Facilitate the progression of HSCs differentiation process | [66] |
| miR-17-92 cluster: miR-18a, miR-17, miR-92a, miR-19a, miR-19b-1, miR-20a | Inhibition of their expression by PU.1 promote HSCs differentiation | [67] |
| miR-146a, miR-126, miR-29a, miR-155, miR-130a, miR-125a/b, miR-338, miR-342, miR-21, miR-196b | Mediate differentiation and maturation of HSCs by regulating expression of various target genes | [61,63,64,66,69,110] |
| miRNAs Regulating Recruitment of Macrophages at Tumor Site | Function | Reference |
| miR148b | Inhibits TAM infiltration in tumor | [111] |
| miR-375 | Induces TAM infiltration in breast cancer | [71] |
| miR-125b | Reduces recruitment of macrophages at tumor site | [112] |
| Subtype | Functions | Key Activating Stimuli | Markers | References |
|---|---|---|---|---|
| M2a | Anti-inflammatory and tissue repair, killing of the infectious parasites | M-CSF, IL-13, IL-14 | CD206, MHC-II, FZZI, CD163, Arg-1, IL-10, TGF-β, WNT5b | [117,118,119,120,121,122,123,124] |
| M2b | Increases infection, immunoregulation, tumor growth and progression | TLR, IL-1R antagonist, immunocomplexes | CD206, CD86, IL-6, IL-1, IL-10 TNF-α | |
| M2c | Immunosuppression, phagocytosis, tissue remodeling, matrix deposition, and efferocytosis | IL-10, glucocorticoids | CD206, CD163, IL-10, MERTK, ECM, TGF-β | |
| M2d | Angiogenesis, anti and pro-tumoral properties | A2AR ligands, TLR, IL-6 | IL-10, IL-12, VEGF, TGF-β |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatterjee, B.; Saha, P.; Bose, S.; Shukla, D.; Chatterjee, N.; Kumar, S.; Tripathi, P.P.; Srivastava, A.K. MicroRNAs: As Critical Regulators of Tumor- Associated Macrophages. Int. J. Mol. Sci. 2020, 21, 7117. https://doi.org/10.3390/ijms21197117
Chatterjee B, Saha P, Bose S, Shukla D, Chatterjee N, Kumar S, Tripathi PP, Srivastava AK. MicroRNAs: As Critical Regulators of Tumor- Associated Macrophages. International Journal of Molecular Sciences. 2020; 21(19):7117. https://doi.org/10.3390/ijms21197117
Chicago/Turabian StyleChatterjee, Bilash, Priyanka Saha, Subhankar Bose, Devendra Shukla, Nabanita Chatterjee, Sanjay Kumar, Prem Prakash Tripathi, and Amit Kumar Srivastava. 2020. "MicroRNAs: As Critical Regulators of Tumor- Associated Macrophages" International Journal of Molecular Sciences 21, no. 19: 7117. https://doi.org/10.3390/ijms21197117
APA StyleChatterjee, B., Saha, P., Bose, S., Shukla, D., Chatterjee, N., Kumar, S., Tripathi, P. P., & Srivastava, A. K. (2020). MicroRNAs: As Critical Regulators of Tumor- Associated Macrophages. International Journal of Molecular Sciences, 21(19), 7117. https://doi.org/10.3390/ijms21197117





