Regenerative Response of Degenerate Human Nucleus Pulposus Cells to GDF6 Stimulation
Abstract
:1. Introduction
2. Results
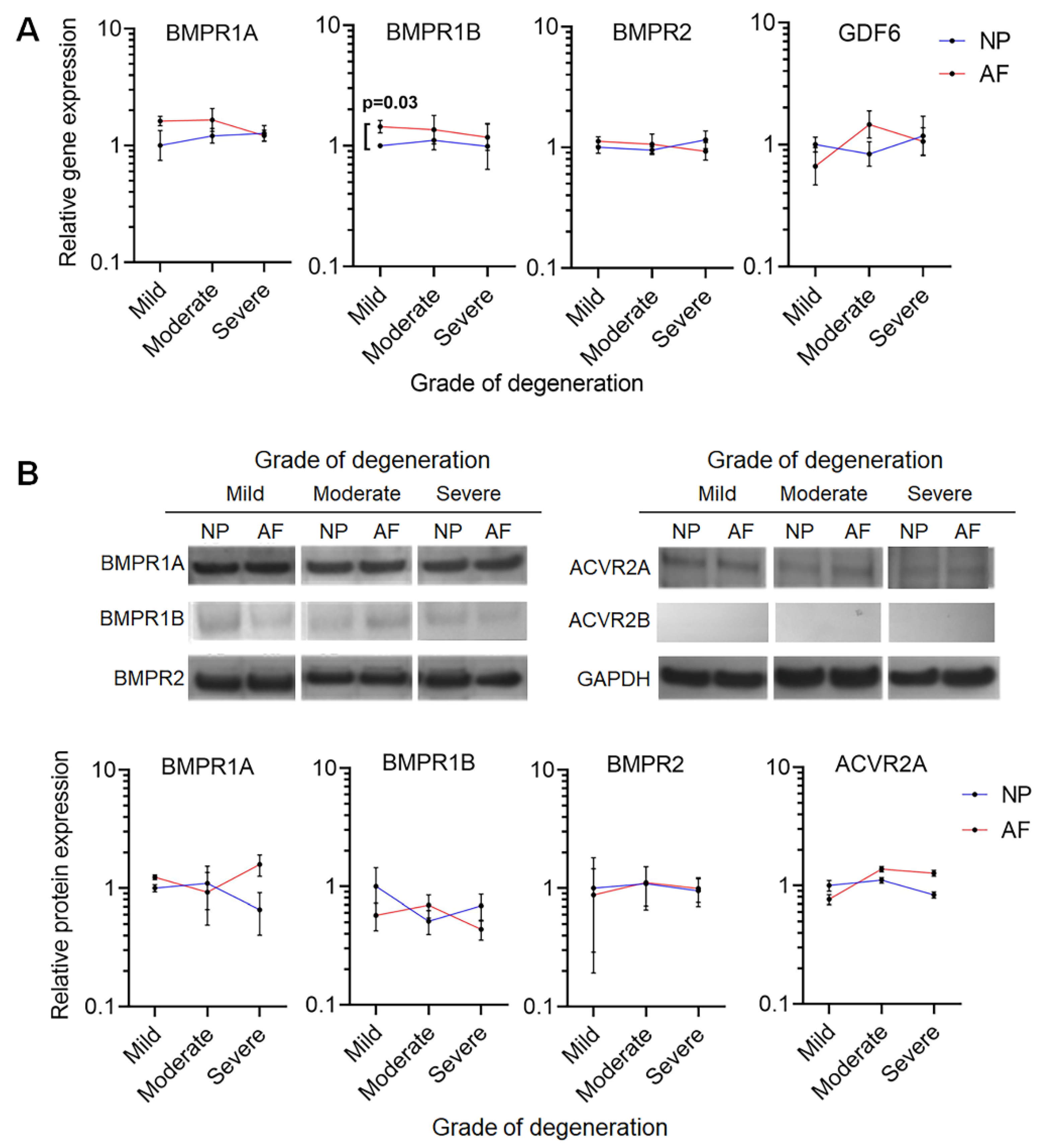
2.1. GDF6 Receptor Expression in NP and AF Cells
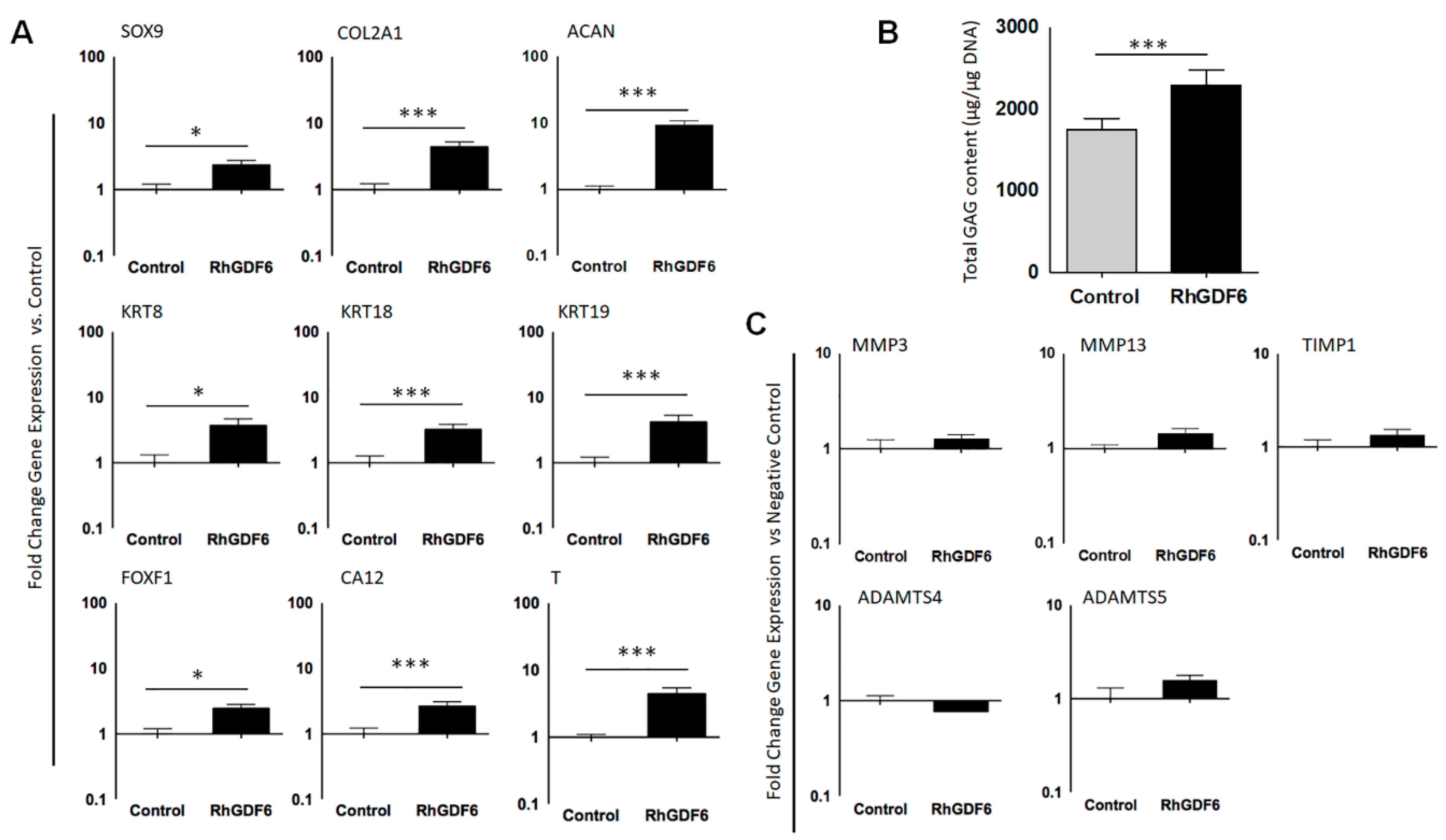
2.2. GDF6 Stimulation Upregulates the Expression of Healthy NP Marker Genes in NP Cells in Culture
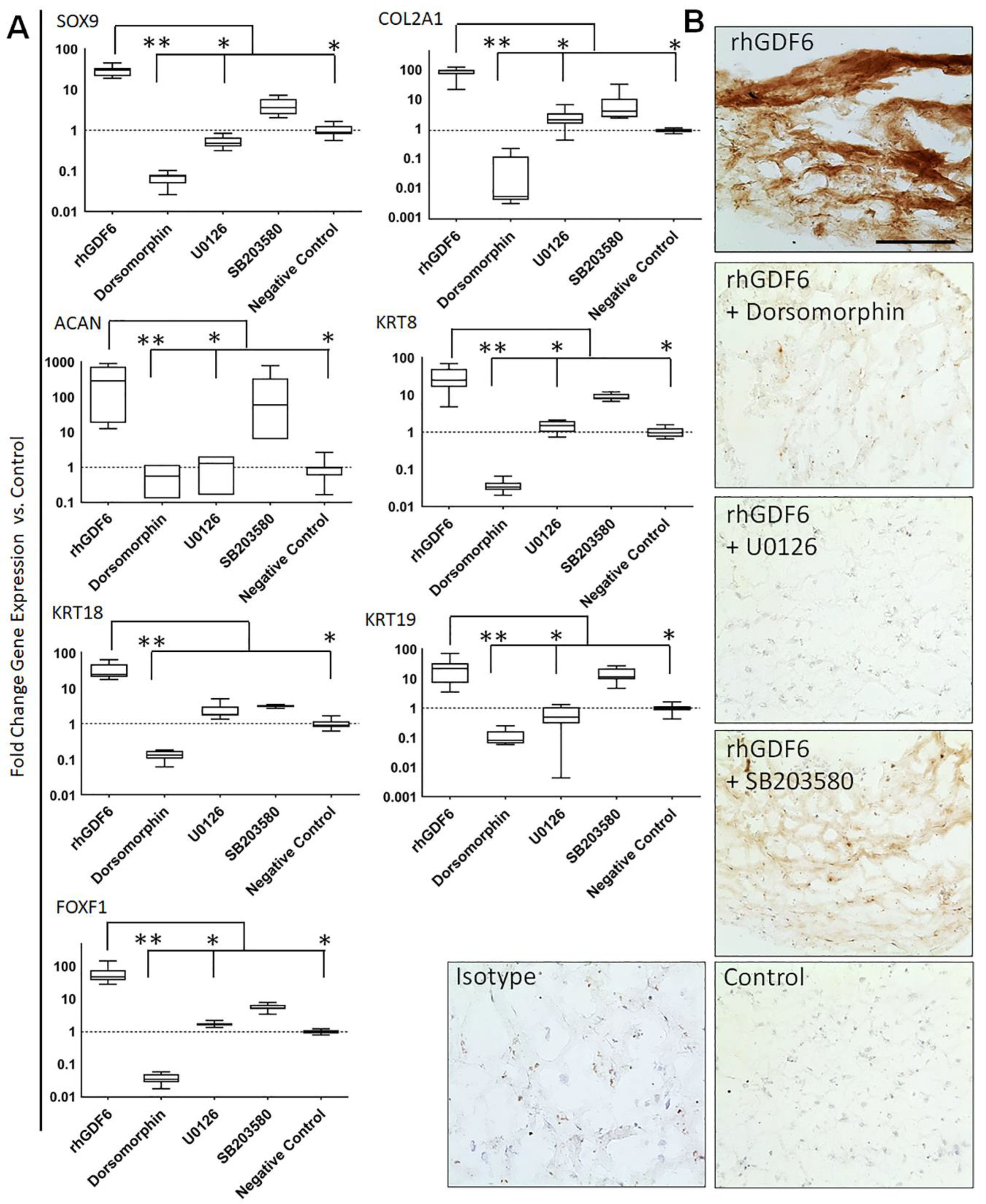
2.3. Determination of GDF6 Intracellular Signalling and Importance to Healthy NP Phenotype
3. Discussion
4. Materials and Methods
4.1. Extraction of NP and AF Cells
4.2. RNA Extraction and Quantitative Real-Time PCR
4.3. GDF Receptor Protein Profiles
4.4. ELISA and Western Blot Analysis of rhGDF6 Signalling Pathway Activation
4.5. Pathway-Specific Inhibition in 3D Collagen Gel Culture
4.6. Immunohistochemical Staining of NP Cell Seeded Type I Collagen Gels
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Freemont, A.J.; Watkins, A.; Le Maitre, C.; Jeziorska, M.; Hoyland, J.A. Current understanding of cellular and molecular events in intervertebral disc degeneration: Implications for therapy. J. Pathol. 2002, 196, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Stefanakis, M.; Al-Abbasi, M.; Harding, I.; Pollintine, P.; Dolan, P.; Tarlton, J.F.; Adams, M.A. Annulus Fissures Are Mechanically and Chemically Conducive to the Ingrowth of Nerves and Blood Vessels. Spine 2012, 37, 1883–1891. [Google Scholar] [CrossRef] [PubMed]
- Freemont, T.; Watkins, A.; Le Maitre, C.; Baird, P.; Jeziorska, M.; Knight, M.T.N.; Ross, E.R.S.; O’Brien, J.P.; Hoyland, J.A. Nerve growth factor expression and innervation of the painful intervertebral disc. J. Pathol. 2002, 197, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.P.; Oegema, T.R.; Bradford, D. Stimulation of Mature Canine Intervertebral Disc by Growth Factors. Spine 1991, 16, 253–260. [Google Scholar] [CrossRef]
- Gruber, H.E.; Fisher, J.E.; Desai, B.; Stasky, A.A.; Hoelscher, G.; Hanley, J.E.N. Human Intervertebral Disc Cells from the Annulus: Three-Dimensional Culture in Agarose or Alginate and Responsiveness to TGF-β1. Exp. Cell Res. 1997, 235, 13–21. [Google Scholar] [CrossRef]
- Osada, R.; Ohshima, H.; Ishihara, H.; Yudoh, K.; Sakai, K.; Matsui, H.; Tsuji, H. Autocrine/paracrine mechanism of insulin-like growth factor-1 secretion, and the effect of insulin-like growth factor-1 on proteoglycan synthesis in bovine intervertebral discs. J. Orthop. Res. 1996, 14, 690–699. [Google Scholar] [CrossRef]
- Risbud, M.V.; Albert, T.J.; Guttapalli, A.; Vresilovic, E.J.; Hillibrand, A.S.; Vaccaro, A.R.; Shapiro, I.M. Differentiation of Mesenchymal Stem Cells Towards a Nucleus Pulposus-like Phenotype In Vitro: Implications for Cell-Based Transplantation Therapy. Spine 2004, 29, 2627–2632. [Google Scholar] [CrossRef]
- Ehlicke, F.; Freimark, D.; Heil, B.; Dorresteijn, A.; Czermak, P. Intervertebral disc regeneration: Influence of growth factors on differentiation of human mesenchymal stem cells (hMSC). Int. J. Artif. Organs 2010, 33, 244–252. [Google Scholar] [CrossRef] [Green Version]
- Le Maitre, C.; Richardson, S.M.; Baird, P.; Freemont, A.J.; Hoyland, J.A. Expression of receptors for putative anabolic growth factors in human intervertebral disc: Implications for repair and regeneration of the disc. J. Pathol. 2005, 207, 445–452. [Google Scholar] [CrossRef]
- Hodgkinson, T.; Shen, B.; Diwan, A.D.; Hoyland, J.A.; Richardson, S.M. Therapeutic potential of growth differentiation factors in the treatment of degenerative disc diseases. JOR Spine 2019, 2, e1045. [Google Scholar] [CrossRef] [Green Version]
- Clendenning, D.E.; Mortlock, D.P. The BMP Ligand Gdf6 Prevents Differentiation of Coronal Suture Mesenchyme in Early Cranial Development. PLoS ONE 2012, 7, e36789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Settle, S.H.; Rountree, R.B.; Sinha, A.; Thacker, A.; Higgins, K.; Kingsley, D.M. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev. Biol. 2003, 254, 116–130. [Google Scholar] [CrossRef] [Green Version]
- Wei, A.; Shen, B.; Williams, L.A.; Bhargav, D.; Gulati, T.; Fang, Z.; Pathmanandavel, S.; Diwan, A.D. Expression of growth differentiation factor 6 in the human developing fetal spine retreats from vertebral ossifying regions and is restricted to cartilaginous tissues. J. Orthop. Res. 2015, 34, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Le Maitre, C.; Freemont, A.J.; Hoyland, J.A. Expression of cartilage-derived morphogenetic protein in human intervertebral discs and its effect on matrix synthesis in degenerate human nucleus pulposus cells. Arthritis Res. Ther. 2009, 11, R137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tassabehji, M.; Fang, Z.M.; Hilton, E.; McGaughran, J.; Zhao, Z.; De Bock, C.E.; Howard, E.; Malass, M.; Donnai, D.; Diwan, A.D.; et al. Mutations in GDF6 are associated with vertebral segmentation defects in Klippel-Feil syndrome. Hum. Mutat. 2008, 29, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Clarke, L.; McConnell, J.C.; Sherratt, M.J.; Derby, B.; Richardson, S.M.; Hoyland, J.A. Growth differentiation factor 6 and transforming growth factor-beta differentially mediate mesenchymal stem cell differentiation, composition, and micromechanical properties of nucleus pulposus constructs. Arthritis Res. Ther. 2014, 16, R67. [Google Scholar] [CrossRef] [Green Version]
- Shen, B.; Bhargav, D.; Wei, A.; Williams, L.A.; Tao, H.; Ma, D.D.; Diwan, A.D. BMP-13 emerges as a potential inhibitor of bone formation. Int. J. Biol. Sci. 2009, 5, 192. [Google Scholar] [CrossRef] [Green Version]
- Gulati, T.; Chung, S.A.; Wei, A.-Q.; Diwan, A.D.; And, A.W. Localization of bone morphogenetic protein 13 in human intervertebral disc and its molecular and functional effects in vitro in 3D culture. J. Orthop. Res. 2015, 33, 1769–1775. [Google Scholar] [CrossRef]
- Wei, A.; Williams, L.A.; Bhargav, D.; Shen, B.; Kishen, T.; Duffy, N.; Diwan, A.D. BMP13 prevents the effects of annular injury in an ovine model. Int. J. Biol. Sci. 2009, 5, 388. [Google Scholar] [CrossRef]
- Cui, H.; Zhang, J.; Li, Z.; Long, J.; Chen, F.; Cui, H.; Du, X.; Liu, H.; Wang, J.; Wang, H.; et al. Growth differentiation factor-6 attenuates inflammatory and pain-related factors and degenerated disc-induced pain behaviors in rat model. J. Orthop. Res. 2020. [Google Scholar] [CrossRef]
- Cheng, H.; Jiang, W.; Phillips, F.M.; Haydon, R.C.; Peng, Y.; Zhou, L.; Luu, H.H.; An, N.; Breyer, B.; Vanichakarn, P.; et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs). J. Bone Joint Surg. Am. 2003, 85, 1544–1552. [Google Scholar] [CrossRef] [PubMed]
- Balemans, W.; Van Hul, W. Extracellular regulation of BMP signaling in vertebrates: A cocktail of modulators. Dev. Biol. 2002, 250, 231–250. [Google Scholar] [CrossRef] [PubMed]
- Miyazono, K.; Kamiya, Y.; Morikawa, M. Bone morphogenetic protein receptors and signal transduction. J. Biochem. 2009, 147, 35–51. [Google Scholar] [CrossRef] [Green Version]
- Mazerbourg, S.; Sangkuhl, K.; Luo, C.-W.; Sudo, S.; Klein, C.; Hsueh, A.J.W. Identification of Receptors and Signaling Pathways for Orphan Bone Morphogenetic Protein/Growth Differentiation Factor Ligands Based on Genomic Analyses. J. Biol. Chem. 2005, 280, 32122–32132. [Google Scholar] [CrossRef] [Green Version]
- Erlacher, L.; McCartney, J.; Piek, E.; Dijke, P.T.; Yanagishita, M.; Oppermann, H.; Luyten, F.P. Cartilage-Derived Morphogenetic Proteins and Osteogenic Protein-1 Differentially Regulate Osteogenesis. J. Bone Miner. Res. 1998, 13, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, Y.; Ishikawa, T.; Shimabara, M.; Tanda, N.; Enomoto-Iwamoto, M.; Kuwana, T.; Ueki, A.; Noji, S.; Nohno, T. BMP signaling during bone pattern determination in the developing limb. Development 1996, 122, 3557–3566. [Google Scholar]
- Zhang, X.-Y.; Chang, H.-M.; Taylor, E.L.; Liu, R.-Z.; Leung, P.C.K. BMP6 Downregulates GDNF Expression Through SMAD1/5 and ERK1/2 Signaling Pathways in Human Granulosa-Lutein Cells. Endocrinology 2018, 159, 2926–2938. [Google Scholar] [CrossRef] [Green Version]
- Gallea, S.; Lallemand, F.; Atfi, A.; Rawadi, G.; Ramez, V.; Spinella-Jaegle, S.; Kawai, S.; Faucheu, C.; Huet, L.; Baron, R.; et al. Activation of mitogen-activated protein kinase cascades is involved in regulation of bone morphogenetic protein-2-induced osteoblast differentiation in pluripotent C2C12 cells. Bone 2001, 28, 491–498. [Google Scholar] [CrossRef]
- Guicheux, J.; Lemonnier, J.; Ghayor, C.; Suzuki, A.; Palmer, G.; Caverzasio, J. Activation of p38 Mitogen-Activated Protein Kinase and c-Jun-NH2-Terminal Kinase by BMP-2 and Their Implication in the Stimulation of Osteoblastic Cell Differentiation. J. Bone Miner. Res. 2003, 18, 2060–2068. [Google Scholar] [CrossRef]
- Wuertz, K.; Vo, N.; Kletsas, D.; Boos, N. Inflammatory and catabolic signalling in intervertebral discs: The roles of NF-κB and MAP Kinases. Eur. Cells Mater. 2012, 23, 102–120. [Google Scholar] [CrossRef]
- Herlaar, E.; Brown, Z. p38 MAPK signalling cascades in inflammatory disease. Mol. Med. Today 1999, 5, 439–447. [Google Scholar] [CrossRef]
- Le Maitre, C.; Freemont, A.J.; Hoyland, J.A. The role of interleukin-1 in the pathogenesis of human Intervertebral disc degeneration. Arthritis Res. Ther. 2005, 7, R732–R745. [Google Scholar] [CrossRef] [Green Version]
- Sive, J.; Baird, P.; Jeziorsk, M.; Watkins, A.; Hoyland, J.; Freemont, A. Expression of chondrocyte markers by cells of normal and degenerate intervertebral discs. Mol. Pathol. 2002, 55, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodgkinson, T.; Wignall, F.; Hoyland, J.A.; Richardson, S.M. High BMPR2 expression leads to enhanced SMAD1/5/8 signalling and GDF6 responsiveness in human adipose-derived stem cells: Implications for stem cell therapies for intervertebral disc degeneration. J. Tissue Eng. 2020, 11. [Google Scholar] [CrossRef]
- Miyazaki, S.; Diwan, A.D.; Kato, K.; Cheng, K.; Bae, W.C.; Sun, Y.; Yamada, J.; Muehleman, C.; Lenz, M.E.; Inoue, N.; et al. ISSLS PRIZE IN BASIC SCIENCE 2018: Growth differentiation factor-6 attenuated pro-inflammatory molecular changes in the rabbit anular-puncture model and degenerated disc-induced pain generation in the rat xenograft radiculopathy model. Eur. Spine J. 2018, 27, 739–751. [Google Scholar] [CrossRef]
- Frauchiger, D.; Heeb, S.R.; May, R.D.; Wöltje, M.; Benneker, L.M.; Gantenbein, B. Differentiation of MSC and annulus fibrosus cells on genetically engineered silk fleece-membrane-composites enriched for GDF-6 or TGF-β3. J. Orthop. Res. 2017, 36, 1324–1333. [Google Scholar] [CrossRef] [PubMed]
- Hodgkinson, T.; Stening, J.Z.; White, L.J.; Shakesheff, K.M.; Hoyland, J.A.; Richardson, S.M. Microparticles for controlled growth differentiation factor 6 delivery to direct adipose stem cell-based nucleus pulposus regeneration. J. Tissue Eng. Regen. Med. 2019, 13, 1406–1417. [Google Scholar] [CrossRef] [Green Version]
- Daniels, J.; Binch, A.A.A.L.; Le Maitre, C. Inhibiting IL-1 signaling pathways to inhibit catabolic processes in disc degeneration. J. Orthop. Res. 2016, 35, 74–85. [Google Scholar] [CrossRef]
- Le Maitre, C.; Hoyland, J.A.; Freemont, A.J. Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1β and TNFα expression profile. Arthritis Res. Ther. 2007, 9, R77. [Google Scholar] [CrossRef] [Green Version]
- Hoyland, J.A.; Le Maitre, C.; Freemont, A.J. Investigation of the role of IL-1 and TNF in matrix degradation in the intervertebral disc. Rheumatology 2008, 47, 809–814. [Google Scholar] [CrossRef] [Green Version]
- Gruber, H.E.; Hoelscher, G.L.; Ingram, J.A.; Bethea, S.; Hanley, E.N. Growth and differentiation factor-5 (GDF-5) in the human intervertebral annulus cells and its modulation by IL-1ß and TNF-α in vitro. Exp. Mol. Pathol. 2014, 96, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, Y.; Xia, P.; Li, S.; Feng, X.; Gao, Y.; Wang, K.; Song, Y.; Duan, Z.; Yang, S.; et al. MicroRNA-7 regulates IL-1β-induced extracellular matrix degeneration by targeting GDF5 in human nucleus pulposus cells. Biomed. Pharmacother. 2016, 83, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.-L.; Yuan, Y.; Tu, J.; Zou, G.-M.; Li, Q. Opposing TNF-α/IL-1β- and BMP-2-activated MAPK signaling pathways converge on Runx2 to regulate BMP-2-induced osteoblastic differentiation. Cell Death Dis. 2014, 5, e1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Burrow, K.L.; Hoyland, J.A.; Richardson, S.M. Human Adipose-Derived Stem Cells Exhibit Enhanced Proliferative Capacity and Retain Multipotency Longer than Donor-Matched Bone Marrow Mesenchymal Stem Cells during Expansion In Vitro. Stem Cells Int. 2017, 2017, 1–15. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hodgkinson, T.; Gilbert, H.T.J.; Pandya, T.; Diwan, A.D.; Hoyland, J.A.; Richardson, S.M. Regenerative Response of Degenerate Human Nucleus Pulposus Cells to GDF6 Stimulation. Int. J. Mol. Sci. 2020, 21, 7143. https://doi.org/10.3390/ijms21197143
Hodgkinson T, Gilbert HTJ, Pandya T, Diwan AD, Hoyland JA, Richardson SM. Regenerative Response of Degenerate Human Nucleus Pulposus Cells to GDF6 Stimulation. International Journal of Molecular Sciences. 2020; 21(19):7143. https://doi.org/10.3390/ijms21197143
Chicago/Turabian StyleHodgkinson, Tom, Hamish T. J. Gilbert, Tej Pandya, Ashish D. Diwan, Judith A. Hoyland, and Stephen M. Richardson. 2020. "Regenerative Response of Degenerate Human Nucleus Pulposus Cells to GDF6 Stimulation" International Journal of Molecular Sciences 21, no. 19: 7143. https://doi.org/10.3390/ijms21197143
APA StyleHodgkinson, T., Gilbert, H. T. J., Pandya, T., Diwan, A. D., Hoyland, J. A., & Richardson, S. M. (2020). Regenerative Response of Degenerate Human Nucleus Pulposus Cells to GDF6 Stimulation. International Journal of Molecular Sciences, 21(19), 7143. https://doi.org/10.3390/ijms21197143




