Malic Enzyme 1 Is Associated with Tumor Budding in Oral Squamous Cell Carcinomas
Abstract
1. Introduction
2. Results
2.1. Relationship between Tumor Budding and ME1 Expression in OSCC
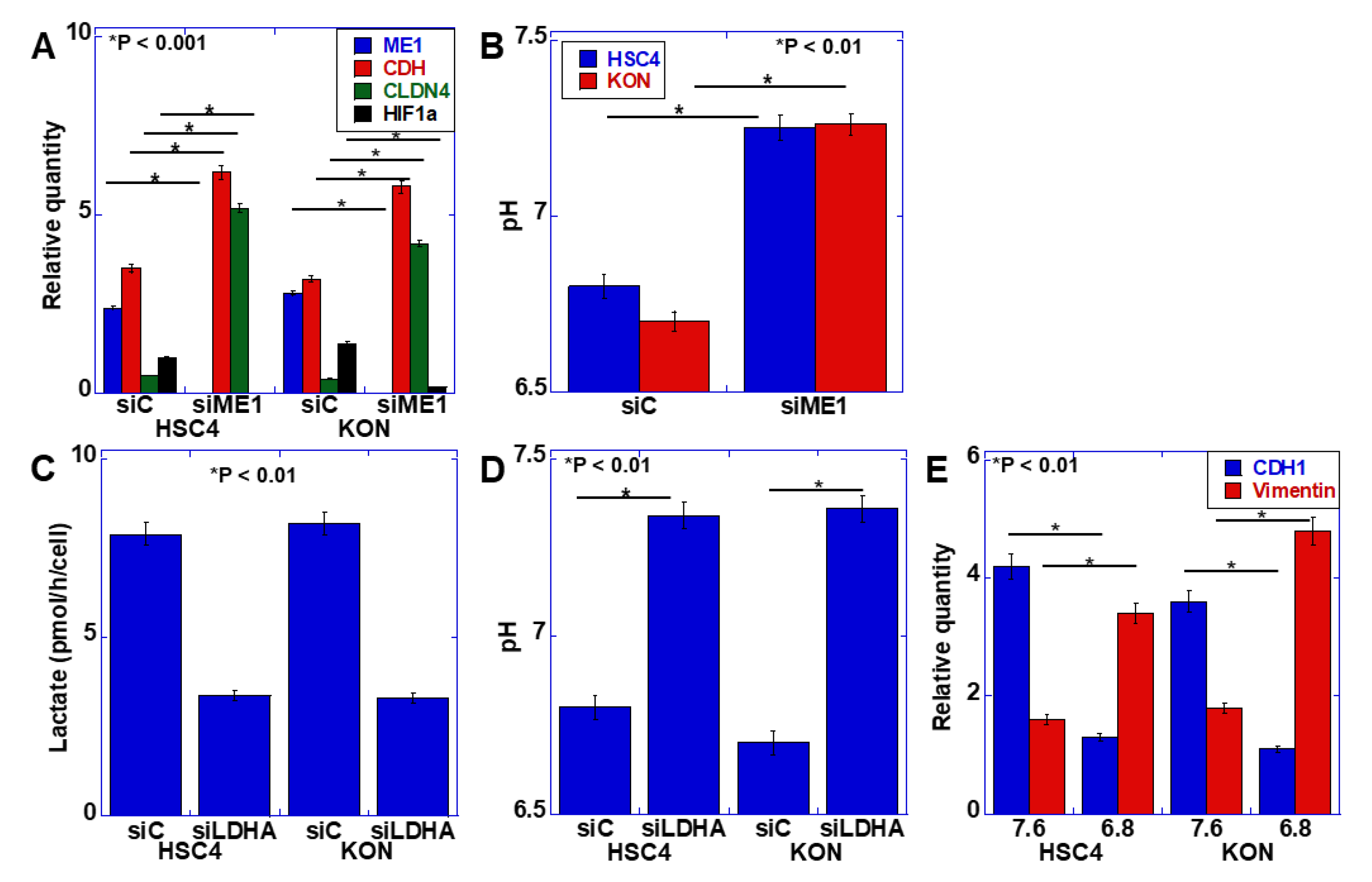
2.2. Effect of ME1 on Extracellular pH
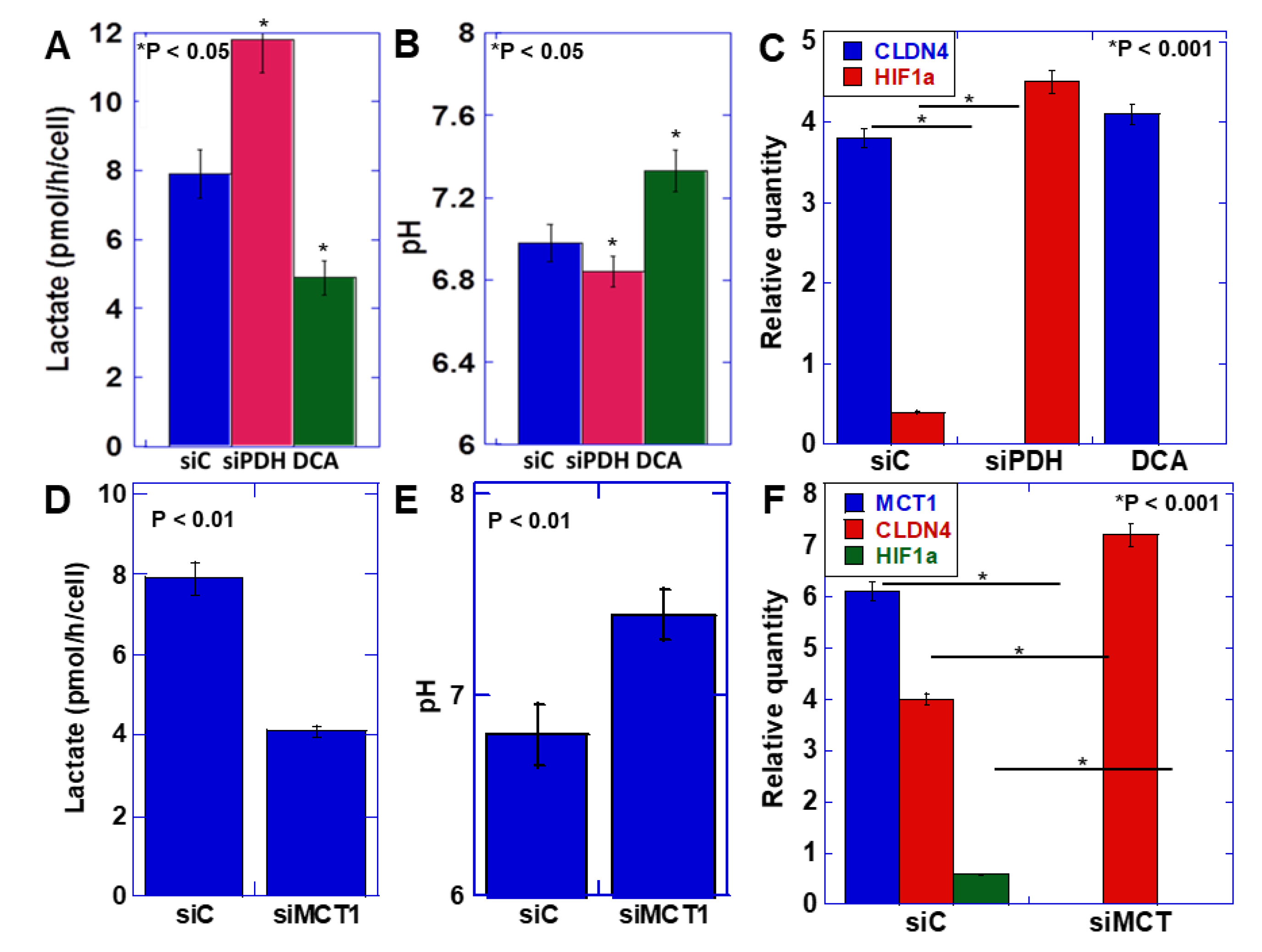
2.3. Relationship between the Warburg Effect and Lactate Secretion
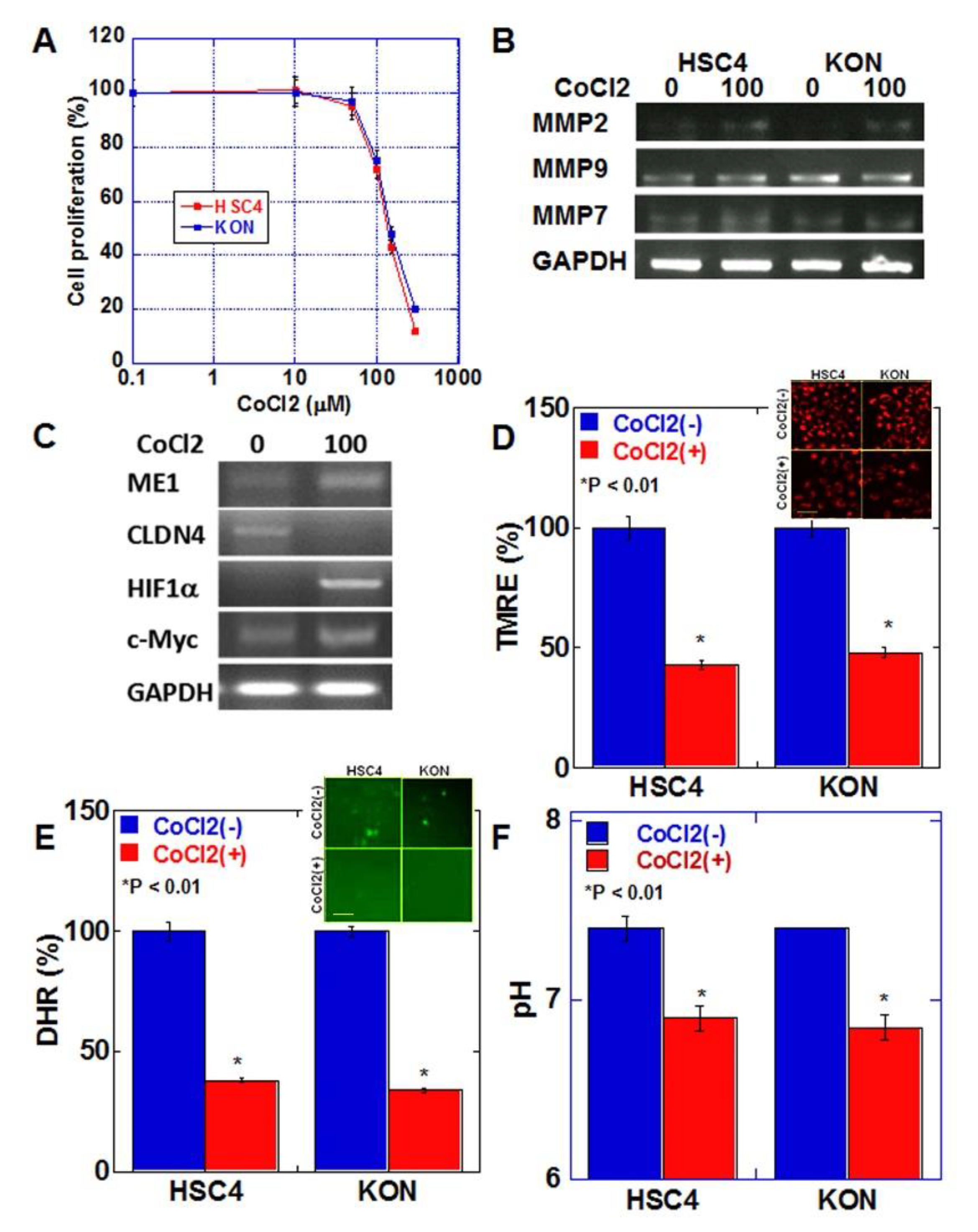
2.4. Effects of Hypoxic Environment on Cancer Cells
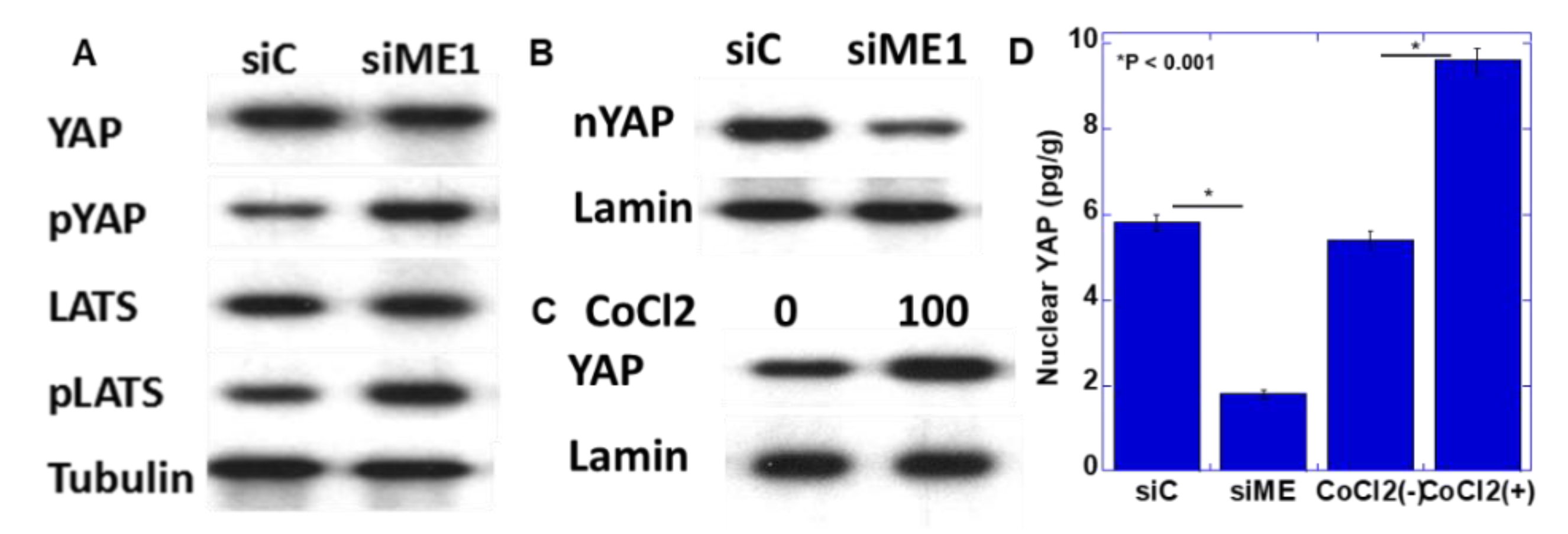
2.5. Yes-Associated Protein (YAP) Activation by ME1
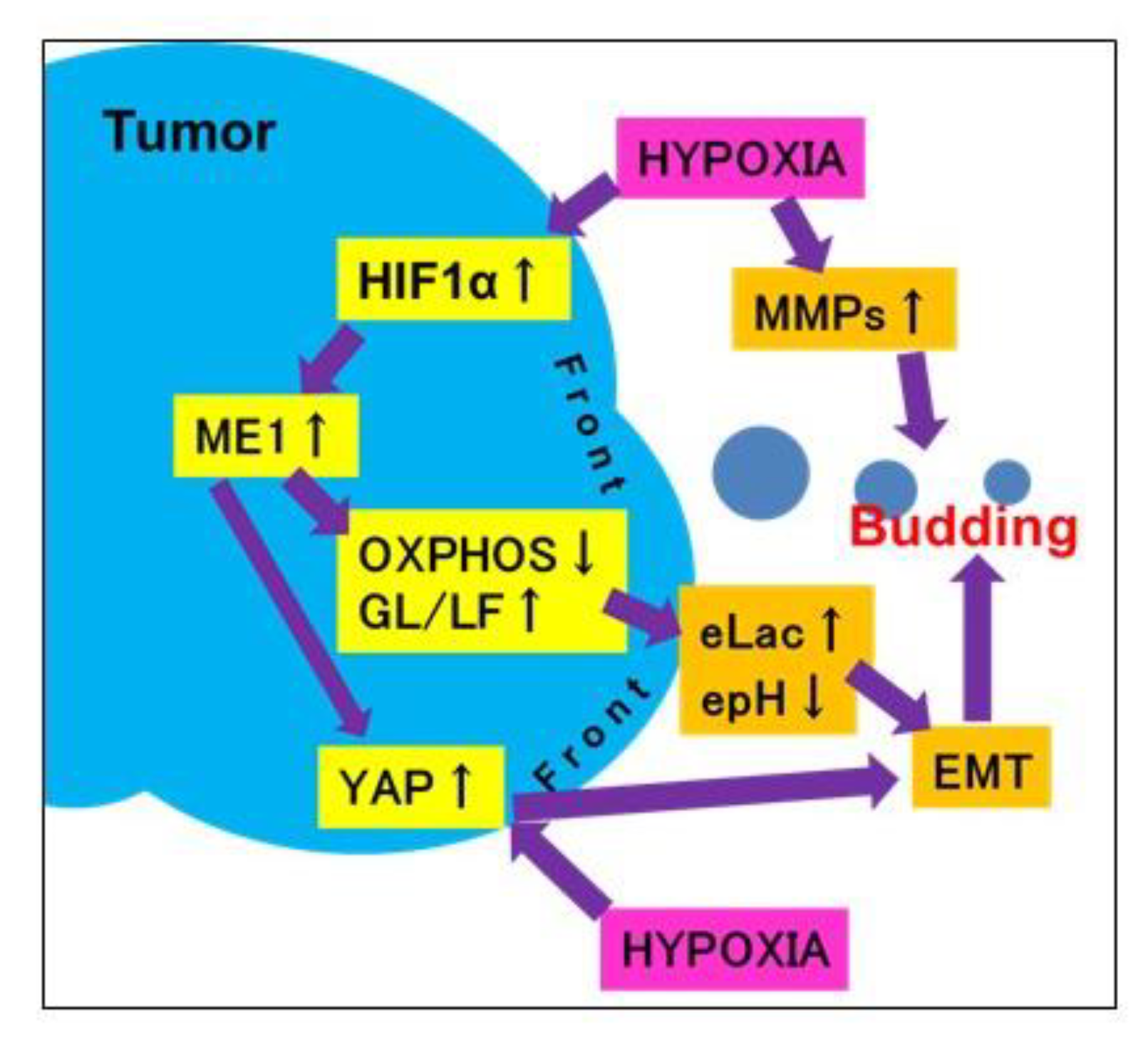
3. Discussion
4. Materials and Methods
4.1. Tissue Arrays
4.2. Immunohistochemistry
4.3. Human OSCC Cell Lines
4.4. Extracellular pH
4.5. Small Interfering RNA
4.6. Reverse Transcription Polymerase Chain Reaction (PCR)
4.7. Protein Extraction
4.8. Immunoblot Analysis
4.9. Mitochondrial Staining
4.10. Determination of Concentrations of Lactate and Nucelar YAP
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| EMT | epithelial–mesenchymal transition |
| HIF | hypoxia-inducible factor |
| ME | malic enzyme |
| OSCC | oral squamous cell carcinoma |
| CLDN | claudin |
| LDHA | lactate dehydrogenase A |
| PDH | pyruvate dehydrogenase |
| DCA | dichloroacetate |
| GPCR | G protein-coupled receptor |
References
- Prall, F. Tumour budding in colorectal carcinoma. Histopathology 2007, 50, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, F.; Li, S.; Zhong, A.; Meng, X.; Lai, M. The tumor microenvironment: An irreplaceable element of tumor budding and epithelial-mesenchymal transition-mediated cancer metastasis. Cell Adh. Migr. 2016, 10, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, H.; Mitomi, H.; Nishiyama, Y.; Kishimoto, I.; Fukui, N.; Nakamura, T.; Watanabe, M. Tumour budding at invasive margins and outcome in colorectal cancer. Colorectal. Dis. 2008, 10, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Mitrovic, B.; Schaeffer, D.F.; Riddell, R.H.; Kirsch, R. Tumor budding in colorectal carcinoma: Time to take notice. Mod. Pathol. 2012, 25, 1315–1325. [Google Scholar] [CrossRef]
- Almangush, A.; Pirinen, M.; Heikkinen, I.; Mäkitie, A.A.; Salo, T.; Leivo, I. Tumour budding in oral squamous cell carcinoma: A meta-analysis. Br. J. Cancer 2018, 118, 577–586. [Google Scholar] [CrossRef]
- Yamakawa, N.; Kirita, T.; Umeda, M.; Yanamoto, S.; Ota, Y.; Otsuru, M.; Okura, M.; Kurita, H.; Yamada, S.; Hasegawa, T.; et al. Tumor budding and adjacent tissue at the invasive front correlate with delayed neck metastasis in clinical early-stage tongue squamous cell carcinoma. J. Surg. Oncol. 2019, 119, 370–378. [Google Scholar] [CrossRef]
- Elseragy, A.; Salo, T.; Coletta, R.D.; Kowalski, L.P.; Haglund, C.; Nieminen, P.; Mäkitie, A.A.; Leivo, I.; Almangush, A. A Proposal to Revise the Histopathologic Grading System of Early Oral Tongue Cancer Incorporating Tumor Budding. Am. J. Surg. Pathol. 2019, 43, 703–709. [Google Scholar] [CrossRef]
- Georges, L.M.; Verset, L.; Zlobec, I.; Demetter, P.; De Wever, O. Impact of the Microenvironment on Tumour Budding in Colorectal Cancer. Adv. Exp. Med. Biol. 2018, 1110, 101–111. [Google Scholar]
- Wong, C.C.; Kai, A.K.; Ng, I.O. The impact of hypoxia in hepatocellular carcinoma metastasis. Front. Med. 2014, 8, 33–41. [Google Scholar] [CrossRef]
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.K.; Forsyth, N.R. Hypoxia-Modified Cancer Cell Metabolism. Front. Cell Dev. Biol. 2019, 7, 4. [Google Scholar] [CrossRef]
- Nakashima, C.; Yamamoto, K.; Fujiwara-Tani, R.; Luo, Y.; Matsushima, S.; Fujii, K.; Ohmori, H.; Sasahira, T.; Sasaki, T.; Kitadai, Y.; et al. Expression of cytosolic malic enzyme (ME1) is associated with disease progression in human oral squamous cell carcinoma. Cancer Sci. 2018, 109, 2036–2045. [Google Scholar] [CrossRef] [PubMed]
- Frenkel, R. Regulation and physiological functions of malic enzymes. Curr. Top Cell Regul. 1975, 9, 157–181. [Google Scholar] [PubMed]
- Rose, I.A. How fumarase recycles after the malate ⟶ fumarate reaction. Insights into the reaction mechanism. Biochemistry 1998, 37, 17651–17658. [Google Scholar] [CrossRef] [PubMed]
- Wehrle, J.P.; Ng, C.E.; McGovern, K.A.; Aiken, N.R.; Shungu, D.C.; Chance, E.M.; Glickson, J.D. Metabolism of alternative substrates and the bioenergetic status of EMT6 tumor cell spheroids. NMR Biomed. 2000, 13, 349–360. [Google Scholar] [CrossRef]
- Liao, R.; Ren, G.; Liu, H.; Chen, X.; Cao, Q.; Wu, X.; Li, J.; Dong, C. ME1 promotes basal-like breast cancer progression and associates with poor prognosis. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Sobin, L.H.; Gospodarowicz, M.; Wittekind, C. UICC TNM Classification of Malignant Tumours, 7th ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2009. [Google Scholar]
- Simonetti, O.; Lucarini, G.; Rubini, C.; Goteri, G.; Zizzi, A.; Staibano, S.; Campanati, A.; Ganzetti, G.; Di Primio, R.; Offidani, A. Microvessel density and VEGF, HIF-1α expression in primary oral melanoma: Correlation with prognosis. Oral. Dis. 2013, 19, 620–627. [Google Scholar] [CrossRef]
- Nakashima, C.; Yamamoto, K.; Kishi, S.; Sasaki, T.; Ohmori, H.; Fujiwara-Tani, R.; Mori, S.; Kawahara, I.; Nishiguchi, Y.; Mori, T.; et al. Clostridium perfringens enterotoxin induces claudin-4 to activate YAP in oral squamous cell carcinomas. Oncotarget 2020, 11, 309–321. [Google Scholar] [CrossRef]
- Elvidge, G.P.; Glenny, L.; Appelhoff, R.J.; Ratcliffe, P.J.; Ragoussis, J.; Gleadle, J.M. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: The role of HIF-1α, HIF-2α, and other pathways. J. Biol. Chem. 2006, 281, 15215–15226. [Google Scholar] [CrossRef]
- Li, Y.Y.; Zheng, Y.L. Hypoxia promotes invasion of retinoblastoma cells in vitro by upregulating HIF-1α/MMP9 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 5361–5369. [Google Scholar]
- Liu, H.L.; Liu, D.; Ding, G.R.; Liao, P.F.; Zhang, J.W. Hypoxia-inducible factor-1α and Wnt/β-catenin signaling pathways promote the invasion of hypoxic gastric cancer cells. Mol. Med. Rep. 2015, 12, 3365–3373. [Google Scholar] [CrossRef]
- Sullivan, R.; Graham, C.H. Hypoxia-driven selection of the metastatic phenotype. Cancer Metastasis Rev. 2007, 26, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Okuda, H.; Watabe, M.; Kobayashi, A.; Pai, S.K.; Liu, W.; Pandey, P.R.; Fukuda, K.; Hirota, S.; Sugai, T.; et al. Hypoxia-induced Jagged2 promotes breast cancer metastasis and self-renewal of cancer stem-like cells. Oncogene 2011, 30, 4075–4086. [Google Scholar] [CrossRef] [PubMed]
- Hockel, M.; Schlenger, K.; Aral, B.; Mitze, M.; Schaffer, U.; Vaupel, P. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Cancer Res. 1996, 56, 4509–4515. [Google Scholar] [PubMed]
- Mezheyeuski, A.; Nerovnya, A.; Bich, T.; Tur, G.; Ostman, A.; Portyanko, A. Inter- and intra-tumoral relationships between vasculature characteristics, GLUT1 and budding in colorectal carcinoma. Histol. Histopathol. 2015, 30, 1203–1211. [Google Scholar] [PubMed]
- Dong, C.; Yuan, T.; Wu, Y.; Wang, Y.; Fan, T.W.; Miriyala, S.; Lin, Y.; Yao, J.; Shi, J.; Kang, T.; et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 2013, 23, 316–331. [Google Scholar] [CrossRef]
- Justus, C.R.; Dong, L.; Yang, L.V. Acidic tumor microenvironment and pH-sensing G protein-coupled receptors. Front. Physiol. 2013, 4, 354. [Google Scholar] [CrossRef]
- Luo, Y.; Kishi, S.; Sasaki, T.; Ohmori, H.; Fujiwara-Tani, R.; Mori, S.; Goto, K.; Nishiguchi, Y.; Mori, T.; Kawahara, I.; et al. Targeting claudin-4 enhances chemosensitivity in breast cancer. Cancer Sci. 2020, 111, 1840–1850. [Google Scholar] [CrossRef]
- McDonald, P.C.; Chafe, S.C.; Dedhar, S. Overcoming Hypoxia-Mediated Tumor Progression: Combinatorial Approaches Targeting pH Regulation, Angiogenesis and Immune Dysfunction. Front. Cell Dev. Biol. 2016, 4, 27. [Google Scholar] [CrossRef]
- Greenhough, A.; Bagley, C.; Heesom, K.J.; Gurevich, D.B.; Gay, D.; Bond, M.; Collard, T.J.; Paraskeva, C.; Martin, P.; Sansom, O.J.; et al. Cancer cell adaptation to hypoxia involves a HIF-GPRC5A-YAP axis. EMBO Mol. Med. 2018, 10. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Ma, Y.; Yang, L.; Wang, T.; Meng, X.; Zong, Z.; Sun, X.; Hua, X.; Li, H. Yes-associated protein (YAP) binds to HIF-1α and sustains HIF-1α protein stability to promote hepatocellular carcinoma cell glycolysis under hypoxic stress. J. Exp. Clin. Cancer Res. 2018, 37, 216. [Google Scholar] [CrossRef]
- Bhattacharya, D.; Azambuja, A.P.; Simoes-Costa, M. Metabolic Reprogramming Promotes Neural Crest Migration via Yap/Tead Signaling. Dev. Cell 2020, 53, 199–211.e6. [Google Scholar] [CrossRef] [PubMed]
- Kuniyasu, H.; Yano, S.; Sasaki, T.; Sasahira, T.; Sone, S.; Ohmori, H. Colon cancer cell-derived high mobility group 1/amphoterin induces growth inhibition and apoptosis in macrophages. Am. J. Pathol. 2005, 166, 751–760. [Google Scholar] [CrossRef]
- Kuniyasu, H.; Oue, N.; Wakikawa, A.; Shigeishi, H.; Matsutani, N.; Kuraoka, K.; Ito, R.; Yokozaki, H.; Yasui, W. Expression of receptors for advanced glycation end-products (RAGE) is closely associated with the invasive and metastatic activity of gastric cancer. J. Pathol. 2002, 196, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Matsushima-Otsuka, S.; Fujiwara-Tani, R.; Sasaki, T.; Ohmori, H.; Nakashima, C.; Kishi, S.; Nishiguchi, Y.; Fujii, K.; Luo, Y.; Kuniyasu, H. Significance of intranuclear angiotensin-II type 2 receptor in oral squamous cell carcinoma. Oncotarget 2018, 9, 36561–36574. [Google Scholar] [CrossRef] [PubMed]

| Parameter | Case Number | ME1 Expression | p * | Budding (/mm2) | p * |
|---|---|---|---|---|---|
| Age (years) | |||||
| ≤59 | 59 | 1.28 ± 0.81 | 14.2 ± 15.1 | ||
| ≤60 | 37 | 1.12 ± 0.89 | NS | 10.0 ± 14.1 | NS |
| Sex | |||||
| F | 14 | 1.29 ± 0.96 | 10.8 ± 8.2 | ||
| M | 82 | 1.21 ± 0083 | NS | 12.9 ± 15.7 | NS |
| Site | |||||
| Gingiva | 26 | 1.19 ± 0.71 | 15.6 ± 17.5 | ||
| Tongue | 35 | 1.41 ± 0.94 | 13.1 ± 10.0 | ||
| Pharynx | 17 | 1.09 ± 0.69 | 12.1 ± 16.1 | ||
| Floor | 18 | 1.56 ± 1.00 | NS | 11.5 ± 18.1 | NS |
| Grade | |||||
| 1 | 38 | 1.5 ± 0.8 | 12.5 ± 15.4 | ||
| 2 | 29 | 1.3 ± 0.9 | 13.8 ± 16.4 | ||
| 3 | 29 | 0.8 ± 0.7 | 0.0199 | 11.6 ± 12.3 | NS |
| pT | |||||
| 1 | 13 | 0.9 ± 1.0 | 2.8 ± 6.2 | ||
| 2 | 37 | 1.3 ± 0.8 | 5.1 ± 4.4 | ||
| 3 | 39 | 1.2 ± 0.8 | 19.4 ± 11.1 | ||
| 4 | 7 | 2.1 ± 0.3 | 0.0175 | 22.1 ± 24.8 | <0.0001 |
| pN | |||||
| 0 | 57 | 1.3 ± 0.9 | 10.9 ± 12.1 | ||
| 1 | 28 | 1.1 ± 0.7 | 14.9 ± 13.9 | ||
| 2 | 11 | 1.4 ± 0.8 | NS | 18.8 ± 11.5 | <0.0001 |
| pStage | |||||
| 1 | 7 | 1.0 ± 1.2 | 1.6 ± 2.9 | ||
| 2 | 22 | 1.4 ± 0.9 | 3.7 ± 4.4 | ||
| 3 | 60 | 1.1 ± 0.8 | 14.8 ± 11.3 | ||
| 4 | 7 | 2.1 ± 0.3 | 0.0156 | 22.1 ± 24.8 | <0.0001 |
| Parameters | Case Number | Spearman r | p * |
|---|---|---|---|
| All cases | 96 | 0.284 | 0.005 |
| pT1–2 | 50 | 0.3633 | 0.0103 |
| pT3–4 | 46 | 0.4783 | 0.0004 |
| pN0 | 57 | 0.3085 | 0.0132 |
| pN1–2 | 39 | 0.5624 | 0.0016 |
| pStage1–2 | 29 | 0.3023 | 0.0129 |
| pStage3–4 | 67 | 0.5599 | <0.0001 |
| Gene Name | Gene Bank ID | Upper/Lower | Primer Sequence |
|---|---|---|---|
| ME1 | NM_002395.5 | U | GGATTGCACACCTGATTGTG |
| L | TCTTCATGTTCATGGGCAAA | ||
| CDH | Z13009.1 | U | TGCCCAGAAAATGAAAAAGG |
| L | GTGTATGTGGCAATGCGTTC | ||
| CLDN4 | NM_001305.4 | U | CTCCATGGGGCTACAGGTAA |
| L | AGCAGCGAGTCGTACACCTT | ||
| HIF1α | AF208487.1 | U | GAAAGCGCAAGTCCTCAAAG |
| L | TGGGTAGGAGATGGAGATGC | ||
| MCT1 | AY364258.1 | U | TCCTTTTATCCTGCCACACC |
| L | GCATGCTGTTTTCCTTCTGC | ||
| MMP2 | KR710613.1 | U | ATGACAGCTGCACCACTGAG |
| L | ATTTGTTGCCCAGGAAAGTG | ||
| MMP9 | NM_004994.3 | U | TTGACAGCGACAAGAAGTGG |
| L | GCCATTCACGTCGTCCTTAT | ||
| MMP7 | NM_002423.5 | U | GAGTGCCAGATGTTGCAGAA |
| L | AAATGCAGGGGGATCTCTTT | ||
| c-Myc | NM_002467.4 | U | TTCGGGTAGTGGAAAACCAG |
| L | CAGCAGCTCGAATTTCTTCC | ||
| GAPDH | BC025925.1 | U | GAGTCAACGGATTTGGTCGT |
| L | TTGATTTTGGAGGGATCTCG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakashima, C.; Kirita, T.; Yamamoto, K.; Mori, S.; Luo, Y.; Sasaki, T.; Fujii, K.; Ohmori, H.; Kawahara, I.; Mori, T.; et al. Malic Enzyme 1 Is Associated with Tumor Budding in Oral Squamous Cell Carcinomas. Int. J. Mol. Sci. 2020, 21, 7149. https://doi.org/10.3390/ijms21197149
Nakashima C, Kirita T, Yamamoto K, Mori S, Luo Y, Sasaki T, Fujii K, Ohmori H, Kawahara I, Mori T, et al. Malic Enzyme 1 Is Associated with Tumor Budding in Oral Squamous Cell Carcinomas. International Journal of Molecular Sciences. 2020; 21(19):7149. https://doi.org/10.3390/ijms21197149
Chicago/Turabian StyleNakashima, Chie, Tadaaki Kirita, Kazuhiko Yamamoto, Shiori Mori, Yi Luo, Takamitsu Sasaki, Kiyomu Fujii, Hitoshi Ohmori, Isao Kawahara, Takuya Mori, and et al. 2020. "Malic Enzyme 1 Is Associated with Tumor Budding in Oral Squamous Cell Carcinomas" International Journal of Molecular Sciences 21, no. 19: 7149. https://doi.org/10.3390/ijms21197149
APA StyleNakashima, C., Kirita, T., Yamamoto, K., Mori, S., Luo, Y., Sasaki, T., Fujii, K., Ohmori, H., Kawahara, I., Mori, T., Goto, K., Kishi, S., Fujiwara-Tani, R., & Kuniyasu, H. (2020). Malic Enzyme 1 Is Associated with Tumor Budding in Oral Squamous Cell Carcinomas. International Journal of Molecular Sciences, 21(19), 7149. https://doi.org/10.3390/ijms21197149





