The Interplay between Transcriptional Factors and MicroRNAs as an Important Factor for Th17/Treg Balance in RA Patients
Abstract
1. Introduction
2. Results
2.1. Increased Expression Levels of miR-146a and miR-155 in Treg Cells from RA and OA
2.2. Lack of Correlation between microRNA and mRNA in Th17 Cells in Control Groups: OA Patients and Healthy Subjects
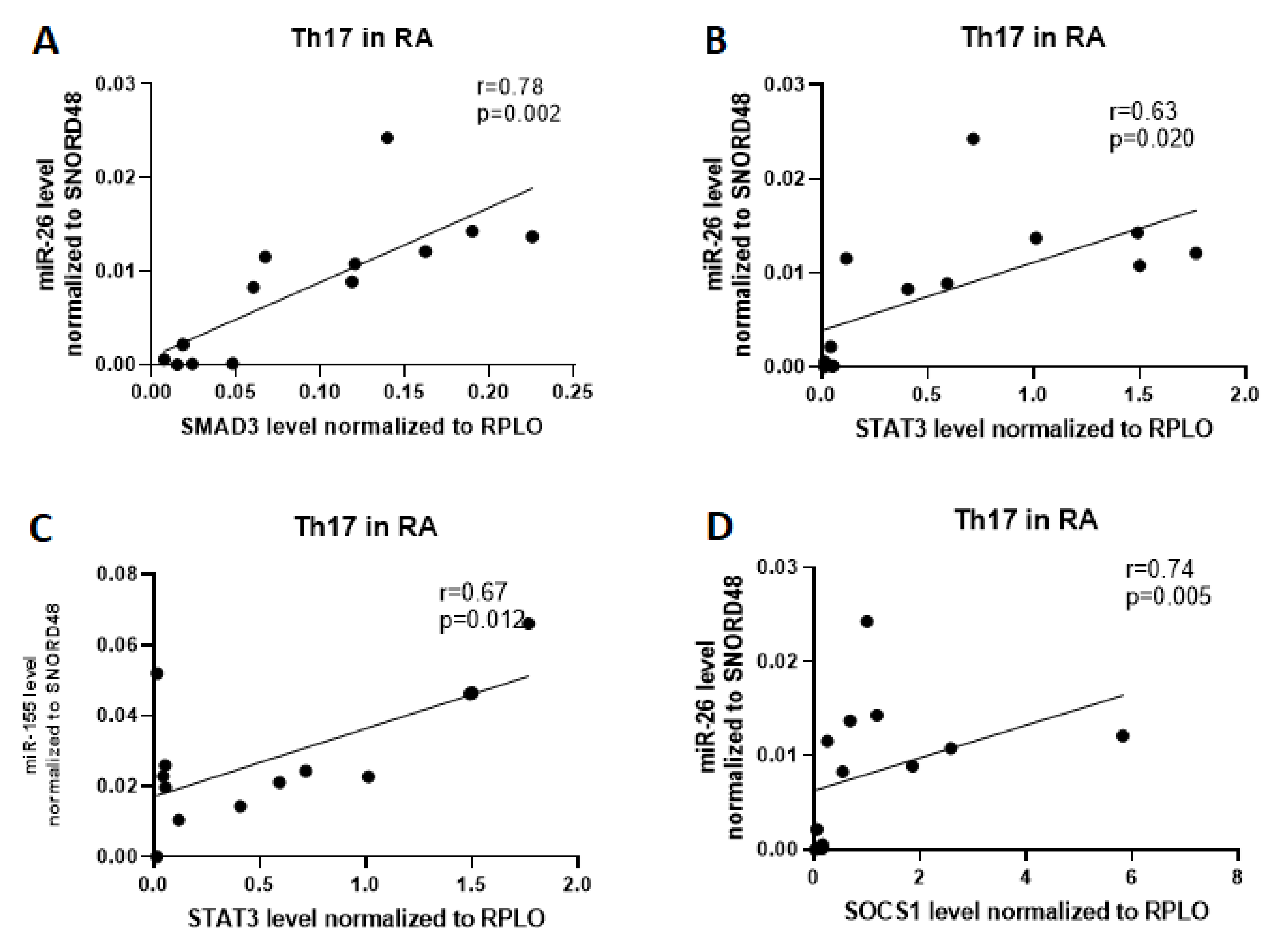
2.3. Correlation Analysis Of Related Target Genes Based on the mRNA–miRNA Interaction Network
2.4. MicroRNAs Are Significantly Correlated with Each Other in Th17 and Treg Cells in RA and OA Patients
2.5. DAS-28, Anti-CCP and Rheumatoid Factor (RF) Parameters in Relation to miR-24, -26, -31, -146a, and -155 Expression Levels in Th17 and Treg Cells from RA Patients
2.6. Multivariable Logistic Regression
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Detection of Th17 and Treg Cells Using Flow Cytometry.
4.3. Isolation of Total RNA from Th17 and Treg Cells
4.4. Quantitative Real-Time PCR
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| aCCP | Anti-cyclin citrullinated peptide autoantibodies |
| AUC | Area under curve |
| CD | Cluster of differentiation |
| CNS1 | Conserved noncoding sequence 1 |
| CRP | C-reactive protein |
| DAS | Disease activity score |
| DNMT 1 | DNA methyltransferase 1 |
| ESR | Erythrocyte sedimentation ratio |
| FBS | Fetal bovine serum |
| FOXP3 | Forkhead box P3 |
| GCs | Glucocorticoids |
| HCs | Healthy controls |
| IL-2 | Interleukin-2 |
| Il-6 | Interleukin-6 |
| JAK/STAT | Janus kinase/signal transducers and activators of transcription |
| miRNAs, miRs | MicroRNAs |
| MS | Multiple sclerosis |
| MTX | Methotrexate |
| OA | Osteoarthritis |
| PBC | Primary biliary cholangitis |
| PBMCs | Peripheral blood mononuclear cells |
| PLT | Platelets |
| pTreg | Peripherally derived Treg |
| RA | Rheumatoid arthritis |
| RF | Rheumatoid factor |
| ROC | Receiver operating characteristic |
| SLE | Systemic lupus erythematosus |
| SMAD2 | SMAD family member 2 |
| SMAD3 | SMAD family member 3 |
| SMAD4 | SMAD family member 4 |
| SOCS1 | Suppressor of cytokine signaling 1 |
| SSA | Somatostatin analogue |
| STAT3 | Signal transducer and activator of transcription 3 |
| STAT5 | Signal transducer and activator of transcription 5 |
| Th cells | T helper cells, helper T cells |
| Treg cells | T regulatory cells, regulatory T cells |
| VAS | Visual analog scale. |
References
- Tateiwa, D.; Yoshikawa, H.; Kaito, T. Cartilage and bone destruction in arthritis: Pathogenesis and treatment strategy: A literature review. Cells 2019, 8, 818. [Google Scholar] [CrossRef] [PubMed]
- Glyn-Jones, S.; Palmer, A.J.R.; Agricola, R.; Price, A.J.; Vincent, T.L.; Weinans, H.; Carr, A.J. Osteoarthritis. Lancet 2015, 386, 376–387. [Google Scholar] [CrossRef]
- Nugent, M. MicroRNAs: Exploring new horizons in osteoarthritis. Osteoarthr. Cart. 2016, 24, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Sondag, G.R.; Haqqi, T.M. The role of MicroRNAs and their targets in osteoarthritis. Curr. Rheumatol. Rep. 2016, 18, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wu, H.; Zhao, M.; Chang, C.; Lu, Q. Clinical significance of MiRNAs in autoimmunity. J. Autoimmun. 2020, 109, 102438. [Google Scholar] [CrossRef]
- Yang, G.; Wu, D.; Zeng, G.; Jiang, O.; Yuan, P.; Huang, S.; Zhu, J.; Tian, J.; Weng, Y.; Rao, Z. Correlation between MiR-126 expression and DNA hypomethylation of CD4+ T cells in rheumatoid arthritis patients. Int. J. Clin. Exp. Pathol. 2015, 8, 8929–8936. [Google Scholar]
- Murata, K.; Furu, M.; Yoshitomi, H.; Ishikawa, M.; Shibuya, H.; Hashimoto, M.; Imura, Y.; Fujii, T.; Ito, H.; Mimori, T.; et al. Comprehensive MicroRNA analysis identifies MiR-24 and MiR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS ONE 2013, 8, e69118. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, H.; Zhao, M.; Lu, Q. Identifying the differentially expressed MicroRNAs in autoimmunity: A systemic review and meta-analysis. Autoimmunity 2020, 53, 122–136. [Google Scholar] [CrossRef]
- Lu, L.F.; Boldin, M.P.; Chaudhry, A.; Lin, L.L.; Taganov, K.D.; Hanada, T.; Yoshimura, A.; Baltimore, D.; Rudensky, A.Y. Function of MiR-146a in controlling treg cell-mediated regulation of Th1 responses. Cell 2010, 142, 914–929. [Google Scholar] [CrossRef]
- Yao, R.; Ma, Y.; Liang, W.; Li, H.; Ma, Z.; Yu, X.; Liao, Y. MicroRNA-155 modulates treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS ONE 2012, 7, e46082. [Google Scholar] [CrossRef]
- Li, X.; Tian, F.; Wang, F. Rheumatoid arthritis-associated microrna-155 targets Socs1 and upregulates TNF-α and IL-1β in PBMCs. Int. J. Mol. Sci. 2013, 14, 23910–23921. [Google Scholar] [CrossRef] [PubMed]
- Samson, M.; Audia, S.; Janikashvili, N.; Ciudad, M.; Trad, M.; Fraszczak, J.; Ornetti, P.; Maillefert, J.F.; Miossec, P.; Bonnotte, B. Brief report: Inhibition of interleukin-6 function corrects Th17/Treg cell imbalance in patients with rheumatoid arthritis. Arthritis Rheum. 2012, 64, 2499–2503. [Google Scholar] [CrossRef] [PubMed]
- Evangelatos, G.; Fragoulis, G.E.; Koulouri, V.; Lambrou, G.I. MicroRNAs in rheumatoid arthritis: From pathogenesis to clinical impact. Autoimm. Rev. 2019, 18, 102391. [Google Scholar] [CrossRef] [PubMed]
- Dudics, S.; Venkatesha, S.H.; Moudgil, K.D. The Micro-RNA expression profiles of autoimmune arthritis reveal novel biomarkers of the disease and therapeutic response. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef]
- Kurowska-Stolarska, M.; Alivernini, S.; Ballantine, L.E.; Asquith, D.L.; Millar, N.L.; Gilchrist, D.S.; Reilly, J.; Ierna, M.; Fraser, A.R.; Stolarski, B.; et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc. Natl. Acad. Sci. USA 2011, 108, 11193–11198. [Google Scholar] [CrossRef]
- Che, X.M.; Huang, Q.C.; Yang, S.L.; Chu, Y.L.; Yan, Y.H.; Han, L.; Huang, Y.; Huang, R.Y. Role of Micro RNAs in pathogenesis of rheumatoid arthritis. Medicine 2015, 94, e1326. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Blüml, S.; Bonelli, M.; Niederreiter, B.; Puchner, A.; Mayr, G.; Hayer, S.; Koenders, M.I.; van den Berg, W.B.; Smolen, J.; Redlich, K. Essential role of microRNA-155 in the pathogenesis of autoimmune arthritis in mice. Arthritis Rheum. 2011, 63, 1281–1288. [Google Scholar] [CrossRef]
- Churov, A.V.; Oleinik, E.K.; Knip, M. MicroRNAs in rheumatoid arthritis: Altered expression and diagnostic potential. Autoimmun. Rev. 2015, 14, 1029–1037. [Google Scholar] [CrossRef]
- Hu, J.; Chen., C.; Liu, Q.; Liu, B.; Song, C.; Zhu, S.; Wu, C.; Liu, S.; Yu, H.; Yao, D.; et al. The role of the miR-31/FIH1 pathway in TGF-β-induced liver fibrosis. Clin. Sci. 2015, 129, 305–317. [Google Scholar] [CrossRef]
- Monticelli, S.; Ansel, K.M.; Xiiia, C.; Socci, N.D.; Krichevsky, A.M.; Thai, T.; Rajewsky, N.; Marks, D.S.; Sander, C.; Rajewsky, K.; et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005, 6, R71. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Kong, R.; Zhou, X.; Ji, L.; Zhang, J.; Zhao, D. MiRNA-126 expression inhibits IL-23R mediated TNF-α or IFN-γ production in fibroblast-like synoviocytes in a mice model of collagen-induced rheumatoid arthritis. Apoptosis 2018, 23, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Pesce, B.; Soto, L.; Sabugo, F.; Wurmann, P.; Cuchacivuch, M.; Lopez, M.N.; Sotelo, P.H.; Molina, M.C.; Aguillon, J.C.; Catalan, D. Effect of interleukin-6 receptor blockade on the balance between regulatory T cells and T helper type 17 cells in rheumatoid arthritis patients. Clin. Exp. Immunol. 2013, 171, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Cai, B.; Huand, Z.C.; Shi, Y.Y.; Wang, L.I. Disturbed Th17/Treg balance in patients with rheumatoid arthritis. Rheumatol. Int. 2012, 32, 2731–2736. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hong, X.; Lin, D.; Luo, X.; Zhu, M.; Mo, H. Artesunate influences Th17/Treg lymphocyte balance by modulating Treg apoptosis and Th17 proliferation in a murine model of rheumatoid arthritis. Exp. Ther. Med. 2017, 13, 2267–2273. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.C.; Lee, Y.H. MiR-146a Levels in rheumatoid arthritis and their correlation with disease activity: A meta-analysis. Int. J. Rheum. Dis. 2018, 21, 1335–1342. [Google Scholar] [CrossRef] [PubMed]
- Abou-Zeid, A.; Saad, M.; Soliman, E. MicroRNA 146a expression in rheumatoid arthritis: Association with tumor necrosis factor-alpha and disease activity. Genet. Test. Mol. Biomarkers 2011, 15, 807–812. [Google Scholar] [CrossRef]
- Niimoto, T.; Nakasa, T.; Ishikawa, M.; Okuhara, A.; Izumi, B.; Deie, M.; Suzuki, O.; Adachi, N.; Ochi, M. MicroRNA-146a expresses in interleukin-17 producing T cells in rheumatoid arthritis patients. BMC Musculoskelet. Disord. 2010, 11. [Google Scholar] [CrossRef]
- Su, L.C.; Huang, A.F.; Jia, H.; Liu, Y.; Xu, W.D. Role of microRNA-155 in rheumatoid arthritis. Int. J. Rheum. Dis. 2017, 20, 1631–1637. [Google Scholar] [CrossRef]
- Singh, A.; Patro, P.S.; Aggarwal, A. MicroRNA-132, miR-146a, and miR-155 as potential biomarkers of methotrexate response in patients with rheumatoid arthritis. Clin. Rheumatol. 2019, 38, 877–884. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Frédérique, L.; Jean-Charles, S.; Markus, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]











| microRNA | Group | AUC | Th17 p | Treg AUC | p |
|---|---|---|---|---|---|
| miR-26 | HC RA | 0.75 | 0.02 | 0.92 | 0.0002 |
| HC OA | 0.66 | 0.16 | 0.86 | 0.0013 | |
| RA OA | 0.55 | 0.66 | 0.66 | 0.17 | |
| miR-155 | HC RA | 0.80 | 0.006 | 0.73 | 0.048 |
| HC OA | 0.75 | 0.03 | 0.51 | 0.94 | |
| RA OA | 0.52 | 0.88 | 0.68 | 0.12 |
| Parameter | RA n = 14 | OA n = 11 | HCs n = 15 |
|---|---|---|---|
| Age | 52 ± 19 64 (21–75) | 70 ± 10 66.5 (56–85) | 48 ± 6.6 45 (41–63) |
| Female/Male | 13/1 | 7/4 | 9/6 |
| ESR (mm/h) | 14 (9–64) | 15 (3–26) | |
| CRP (mg/L) | 12.5 (5–73) | 5 (5–10) |
| Clinical Characteristics of Patients with RA | |
|---|---|
| Disease duration (years) | 6.5 (0.5–18) |
| DAS-28 > 5.1 n (%) | 6 (42%) |
| Larsen 1–3 n (%) | 9 (64%) |
| RF Positivity n (%) | 5 (35%) |
| Anti-CCP Positivity n (%) | 7 (50%) |
| Methotrexate (MTX) n (%) | 9 (64%) |
| Somatostatin Analogue (SSA) n (%) | 3 (21%) |
| Antimalarials Drug n (%) | 7 (50%) |
| Glucocorticoids (GCs) n (%) | 4 (28%) |
| IL-6 Inhibitor n (%) | 1 (7.1%) |
| No Treatment n (%) | 1 (7.1%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kmiołek, T.; Rzeszotarska, E.; Wajda, A.; Walczuk, E.; Kuca-Warnawin, E.; Romanowska-Próchnicka, K.; Stypinska, B.; Majewski, D.; Jagodzinski, P.P.; Pawlik, A.; et al. The Interplay between Transcriptional Factors and MicroRNAs as an Important Factor for Th17/Treg Balance in RA Patients. Int. J. Mol. Sci. 2020, 21, 7169. https://doi.org/10.3390/ijms21197169
Kmiołek T, Rzeszotarska E, Wajda A, Walczuk E, Kuca-Warnawin E, Romanowska-Próchnicka K, Stypinska B, Majewski D, Jagodzinski PP, Pawlik A, et al. The Interplay between Transcriptional Factors and MicroRNAs as an Important Factor for Th17/Treg Balance in RA Patients. International Journal of Molecular Sciences. 2020; 21(19):7169. https://doi.org/10.3390/ijms21197169
Chicago/Turabian StyleKmiołek, Tomasz, Ewa Rzeszotarska, Anna Wajda, Ewa Walczuk, Ewa Kuca-Warnawin, Katarzyna Romanowska-Próchnicka, Barbara Stypinska, Dominik Majewski, Pawel Piotr Jagodzinski, Andrzej Pawlik, and et al. 2020. "The Interplay between Transcriptional Factors and MicroRNAs as an Important Factor for Th17/Treg Balance in RA Patients" International Journal of Molecular Sciences 21, no. 19: 7169. https://doi.org/10.3390/ijms21197169
APA StyleKmiołek, T., Rzeszotarska, E., Wajda, A., Walczuk, E., Kuca-Warnawin, E., Romanowska-Próchnicka, K., Stypinska, B., Majewski, D., Jagodzinski, P. P., Pawlik, A., & Paradowska-Gorycka, A. (2020). The Interplay between Transcriptional Factors and MicroRNAs as an Important Factor for Th17/Treg Balance in RA Patients. International Journal of Molecular Sciences, 21(19), 7169. https://doi.org/10.3390/ijms21197169







