Dicing the Disease with Dicer: The Implications of Dicer Ribonuclease in Human Pathologies
Abstract
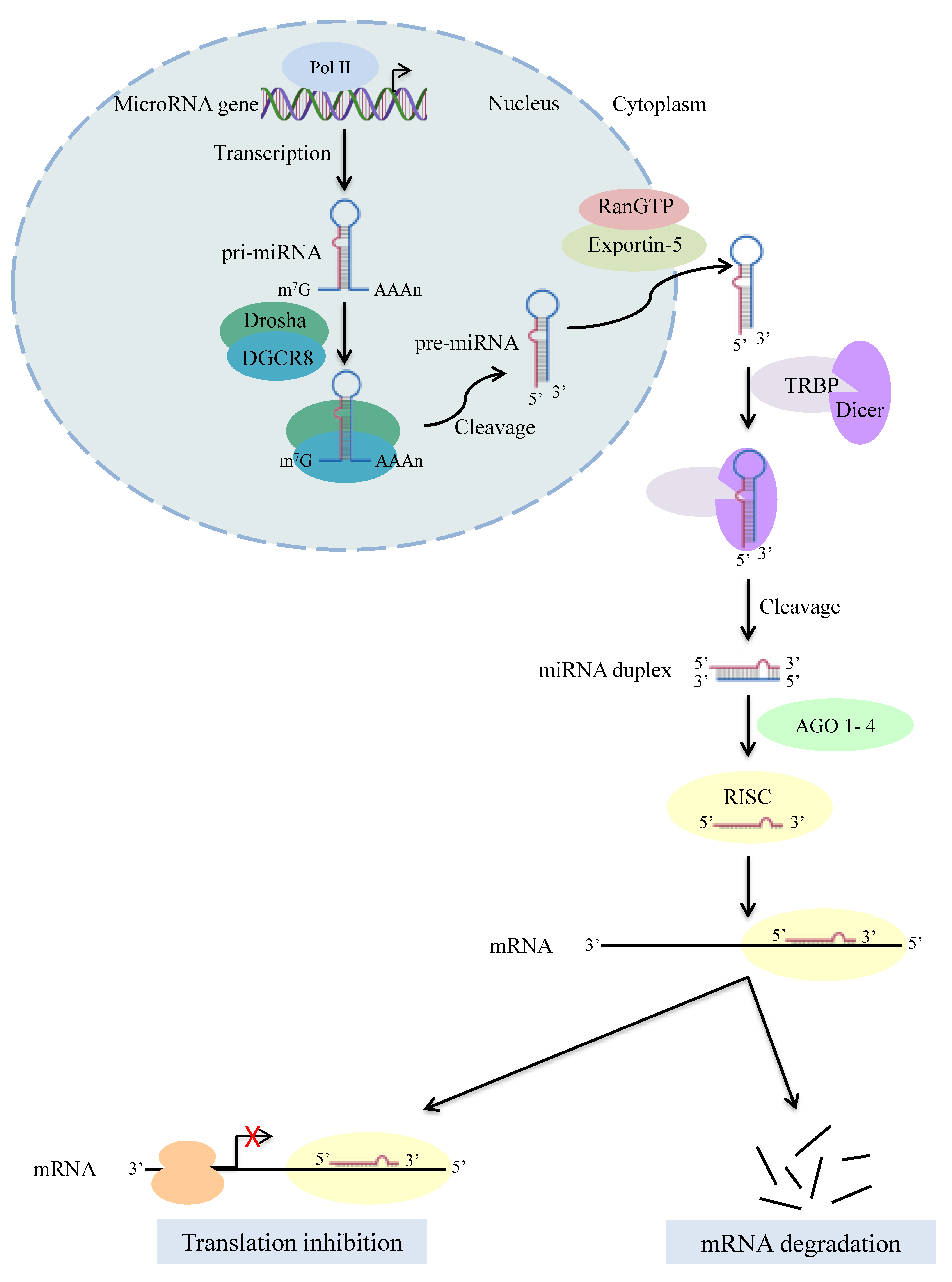
:1. Introduction
2. Dicer in Human Diseases
2.1. Dicer1 Syndrome
2.2. Dicer and Other Cancer Types
2.3. Dicer in Geographic Atrophy
2.4. Dicer in Psychiatric and Neurological Diseases
2.4.1. Chronic Stress and Depression
2.4.2. Parkinson’s Disease
2.4.3. Triplet Repeat Expansion Diseases
2.5. Dicer in Autoimmune Disorders
2.5.1. Psoriasis
2.5.2. Ankylosing Spondylitis
2.5.3. Rheumatoid Arthritis
2.5.4. Multiple Sclerosis
2.5.5. Autoimmune Thyroid Diseases
2.6. Dicer in Cardiovascular Diseases
2.7. Dicer in Female and Male Fertility
2.8. Dicer-Dependent Antiviral Defense Mechanism
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Svobodova, E.; Kubikova, J.; Svoboda, P. Production of small RNAs by mammalian Dicer. Pflugers Arch. 2016, 468, 1089–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, D.; Dancis, B.; Brown, J.R. The evolution of core proteins involved in microRNA biogenesis. BMC Evol. Biol. 2008, 8, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- de Jong, D.; Eitel, M.; Jakob, W.; Osigus, H.J.; Hadrys, H.; Desalle, R.; Schierwater, B. Multiple dicer genes in the early-diverging metazoa. Mol. Biol. Evol. 2009, 26, 1333–1340. [Google Scholar] [CrossRef]
- Ji, X. The mechanism of RNase III action: How dicer dices. Curr. Top. MicroBiol. Immunol. 2008, 320, 99–116. [Google Scholar] [CrossRef]
- Robertson, H.D.; Webster, R.E.; Zinder, N.D. Purification and properties of ribonuclease III from Escherichia coli. J. Biol. Chem. 1968, 243, 82–91. [Google Scholar]
- Nicholson, A.W. Structure, reactivity, and biology of double-stranded RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996, 52, 1–65. [Google Scholar] [CrossRef]
- Filippov, V.; Solovyev, V.; Filippova, M.; Gill, S.S. A novel type of RNase III family proteins in eukaryotes. Gene 2000, 245, 213–221. [Google Scholar] [CrossRef]
- Lau, P.W.; Guiley, K.Z.; De, N.; Potter, C.S.; Carragher, B.; MacRae, I.J. The molecular architecture of human Dicer. Nat. Struct. Mol. Biol. 2012, 19, 436–440. [Google Scholar] [CrossRef] [Green Version]
- Song, M.S.; Rossi, J.J. Molecular mechanisms of Dicer: Endonuclease and enzymatic activity. BioChem. J. 2017, 474, 1603–1618. [Google Scholar] [CrossRef] [Green Version]
- Sinkkonen, L.; Hugenschmidt, T.; Filipowicz, W.; Svoboda, P. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PLoS ONE 2010, 5, e12175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, R.C.; Tambe, A.; Kidwell, M.A.; Noland, C.L.; Schneider, C.P.; Doudna, J.A. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell 2015, 57, 397–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.W.; Noland, C.; Siridechadilok, B.; Taylor, D.W.; Ma, E.; Felderer, K.; Doudna, J.A.; Nogales, E. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 2009, 16, 1148–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveto, S.; Mancino, M.; Manfrini, N.; Biffo, S. Role of microRNAs in translation regulation and cancer. World J. Biol. Chem. 2017, 8, 45–56. [Google Scholar] [CrossRef]
- Li, Z.; Rana, T.M. Molecular mechanisms of RNA-triggered gene silencing machineries. Acc. Chem. Res. 2012, 45, 1122–1131. [Google Scholar] [CrossRef]
- Pong, S.K.; Gullerova, M. Noncanonical functions of microRNA pathway enzymes-Drosha, DGCR8, Dicer and Ago proteins. FEBS Lett. 2018, 592, 2973–2986. [Google Scholar] [CrossRef] [Green Version]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Maniataki, E.; Mourelatos, Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005, 19, 2979–2990. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Hur, I.; Park, S.Y.; Kim, Y.K.; Suh, M.R.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006, 25, 522–532. [Google Scholar] [CrossRef]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef] [Green Version]
- Tahbaz, N.; Kolb, F.A.; Zhang, H.; Jaronczyk, K.; Filipowicz, W.; Hobman, T.C. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004, 5, 189–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langenberger, D.; Cakir, M.V.; Hoffmann, S.; Stadler, P.F. Dicer-processed small RNAs: Rules and exceptions. J. Exp. Zool. B Mol. Dev. Evol. 2013, 320, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009, 15, 2147–2160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsui, M.; Li, L.; Janowski, B.A.; Corey, D.R. Reduced Expression of Argonaute 1, Argonaute 2, and TRBP Changes Levels and Intracellular Distribution of RNAi Factors. Sci. Rep. 2015, 5, 12855. [Google Scholar] [CrossRef] [Green Version]
- Doyle, M.; Badertscher, L.; Jaskiewicz, L.; Guttinger, S.; Jurado, S.; Hugenschmidt, T.; Kutay, U.; Filipowicz, W. The double-stranded RNA binding domain of human Dicer functions as a nuclear localization signal. RNA 2013, 19, 1238–1252. [Google Scholar] [CrossRef] [Green Version]
- Roche, B.; Arcangioli, B.; Martienssen, R. New roles for Dicer in the nucleolus and its relevance to cancer. Cell Cycle 2017, 16, 1643–1653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fragkos, M.; Barra, V.; Egger, T.; Bordignon, B.; Lemacon, D.; Naim, V.; Coquelle, A. Dicer prevents genome instability in response to replication stress. Oncotarget 2019, 10, 4407–4423. [Google Scholar] [CrossRef]
- Francia, S.; Michelini, F.; Saxena, A.; Tang, D.; de Hoon, M.; Anelli, V.; Mione, M.; Carninci, P.; d’Adda di Fagagna, F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012, 488, 231–235. [Google Scholar] [CrossRef]
- Gullerova, M.; Proudfoot, N.J. Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells. Nat. Struct. Mol. Biol. 2012, 19, 1193–1201. [Google Scholar] [CrossRef] [Green Version]
- White, E.; Schlackow, M.; Kamieniarz-Gdula, K.; Proudfoot, N.J.; Gullerova, M. Human nuclear Dicer restricts the deleterious accumulation of endogenous double-stranded RNA. Nat. Struct. Mol. Biol. 2014, 21, 552–559. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Zhang, H. Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep. 2013, 14, 568–576. [Google Scholar] [CrossRef] [Green Version]
- Gibbings, D.; Mostowy, S.; Jay, F.; Schwab, Y.; Cossart, P.; Voinnet, O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat. Cell Biol. 2012, 14, 1314–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rybak-Wolf, A.; Jens, M.; Murakawa, Y.; Herzog, M.; Landthaler, M.; Rajewsky, N. A variety of dicer substrates in human and C. elegans. Cell 2014, 159, 1153–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machitani, M.; Sakurai, F.; Wakabayashi, K.; Tomita, K.; Tachibana, M.; Mizuguchi, H. Dicer functions as an antiviral system against human adenoviruses via cleavage of adenovirus-encoded noncoding RNA. Sci. Rep. 2016, 6, 27598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machitani, M.; Yamaguchi, T.; Shimizu, K.; Sakurai, F.; Katayama, K.; Kawabata, K.; Mizuguchi, H. Adenovirus Vector-Derived VA-RNA-Mediated Innate Immune Responses. Pharmaceutics 2011, 3, 338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathews, M.B.; Shenk, T. Adenovirus virus-associated RNA and translation control. J. Virol. 1991, 65, 5657–5662. [Google Scholar] [CrossRef] [Green Version]
- Robertson, J.C.; Jorcyk, C.L.; Oxford, J.T. DICER1 Syndrome: DICER1 Mutations in Rare Cancers. Cancers 2018, 10, 143. [Google Scholar] [CrossRef] [Green Version]
- Matskevich, A.A.; Moelling, K. Stimuli-dependent cleavage of Dicer during apoptosis. BioChem. J. 2008, 412, 527–534. [Google Scholar] [CrossRef] [Green Version]
- Schultz, K.A.P.; Stewart, D.R.; Kamihara, J.; Bauer, A.J.; Merideth, M.A.; Stratton, P.; Huryn, L.A.; Harris, A.K.; Doros, L.; Field, A.; et al. DICER1 Tumor Predisposition. In GeneReviews; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Hill, D.A.; Ivanovich, J.; Priest, J.R.; Gurnett, C.A.; Dehner, L.P.; Desruisseau, D.; Jarzembowski, J.A.; Wikenheiser-Brokamp, K.A.; Suarez, B.K.; Whelan, A.J.; et al. DICER1 mutations in familial pleuropulmonary blastoma. Science 2009, 325, 965. [Google Scholar] [CrossRef] [Green Version]
- de Kock, L.; Wang, Y.C.; Revil, T.; Badescu, D.; Rivera, B.; Sabbaghian, N.; Wu, M.; Weber, E.; Sandoval, C.; Hopman, S.M.; et al. High-sensitivity sequencing reveals multi-organ somatic mosaicism causing DICER1 syndrome. J. Med. Genet. 2016, 53, 43–52. [Google Scholar] [CrossRef] [Green Version]
- Slade, I.; Bacchelli, C.; Davies, H.; Murray, A.; Abbaszadeh, F.; Hanks, S.; Barfoot, R.; Burke, A.; Chisholm, J.; Hewitt, M.; et al. DICER1 syndrome: Clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J. Med. Genet. 2011, 48, 273–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Kock, L.; Rivera, B.; Revil, T.; Thorner, P.; Goudie, C.; Bouron-Dal Soglio, D.; Choong, C.S.; Priest, J.R.; van Diest, P.J.; Tanboon, J.; et al. Sequencing of DICER1 in sarcomas identifies biallelic somatic DICER1 mutations in an adult-onset embryonal rhabdomyosarcoma. Br. J. Cancer 2017, 116, 1621–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melendez-Zajgla, J.; Mercado-Celis, G.E.; Gaytan-Cervantes, J.; Torres, A.; Gabino, N.B.; Zapata-Tarres, M.; Juarez-Villegas, L.E.; Lezama, P.; Maldonado, V.; Ruiz-Monroy, K.; et al. Genomics of a pediatric ovarian fibrosarcoma. Association with the DICER1 syndrome. Sci. Rep. 2018, 8, 3252. [Google Scholar] [CrossRef]
- Stewart, C.J.; Charles, A.; Foulkes, W.D. Gynecologic Manifestations of the DICER1 Syndrome. Surg. Pathol. Clin. 2016, 9, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.K.; Sabbaghian, N.; Xu, B.; Addidou-Kalucki, S.; Bernard, C.; Zou, D.; Reeve, A.E.; Eccles, M.R.; Cole, C.; Choong, C.S.; et al. Biallelic DICER1 mutations occur in Wilms tumours. J. Pathol. 2013, 230, 154–164. [Google Scholar] [CrossRef] [Green Version]
- de Kock, L.; Bah, I.; Revil, T.; Berube, P.; Wu, M.K.; Sabbaghian, N.; Priest, J.R.; Ragoussis, J.; Foulkes, W.D. Deep Sequencing Reveals Spatially Distributed Distinct Hot Spot Mutations in DICER1-Related Multinodular Goiter. J. Clin. Endocrinol. Metab. 2016, 101, 3637–3645. [Google Scholar] [CrossRef]
- Anglesio, M.S.; Wang, Y.; Yang, W.; Senz, J.; Wan, A.; Heravi-Moussavi, A.; Salamanca, C.; Maines-Bandiera, S.; Huntsman, D.G.; Morin, G.B. Cancer-associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse-strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J. Pathol. 2013, 229, 400–409. [Google Scholar] [CrossRef] [Green Version]
- Noren Hooten, N.; Martin-Montalvo, A.; Dluzen, D.F.; Zhang, Y.; Bernier, M.; Zonderman, A.B.; Becker, K.G.; Gorospe, M.; de Cabo, R.; Evans, M.K. Metformin-mediated increase in DICER1 regulates microRNA expression and cellular senescence. Aging Cell 2016, 15, 572–581. [Google Scholar] [CrossRef]
- Wu, X.; Yang, Y.; Huang, Y.; Chen, Y.; Wang, T.; Wu, S.; Tong, L.; Wang, Y.; Lin, L.; Hao, M.; et al. RNA-binding protein AUF1 suppresses miR-122 biogenesis by down-regulating Dicer1 in hepatocellular carcinoma. Oncotarget 2018, 9, 14815–14827. [Google Scholar] [CrossRef] [Green Version]
- Dedes, K.J.; Natrajan, R.; Lambros, M.B.; Geyer, F.C.; Lopez-Garcia, M.A.; Savage, K.; Jones, R.L.; Reis-Filho, J.S. Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. Eur. J. Cancer 2011, 47, 138–150. [Google Scholar] [CrossRef]
- Karube, Y.; Tanaka, H.; Osada, H.; Tomida, S.; Tatematsu, Y.; Yanagisawa, K.; Yatabe, Y.; Takamizawa, J.; Miyoshi, S.; Mitsudomi, T.; et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005, 96, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Prodromaki, E.; Korpetinou, A.; Giannopoulou, E.; Vlotinou, E.; Chatziathanasiadou, M.; Papachristou, N.I.; Scopa, C.D.; Papadaki, H.; Kalofonos, H.P.; Papachristou, D.J. Expression of the microRNA regulators Drosha, Dicer and Ago2 in non-small cell lung carcinomas. Cell Oncol. 2015, 38, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Chiosea, S.; Jelezcova, E.; Chandran, U.; Luo, J.; Mantha, G.; Sobol, R.W.; Dacic, S. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007, 67, 2345–2350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faber, C.; Horst, D.; Hlubek, F.; Kirchner, T. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur. J. Cancer 2011, 47, 1414–1419. [Google Scholar] [CrossRef]
- Piroozian, F.; Bagheri Varkiyani, H.; Koolivand, M.; Ansari, M.; Afsa, M.; AtashAbParvar, A.; MalekZadeh, K. The impact of variations in transcription of DICER and AGO2 on exacerbation of childhood B-cell lineage acute lymphoblastic leukaemia. Int. J. Exp. Pathol. 2019, 100, 184–191. [Google Scholar] [CrossRef]
- Chiosea, S.; Jelezcova, E.; Chandran, U.; Acquafondata, M.; McHale, T.; Sobol, R.W.; Dhir, R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am. J. Pathol. 2006, 169, 1812–1820. [Google Scholar] [CrossRef] [Green Version]
- Flavin, R.J.; Smyth, P.C.; Finn, S.P.; Laios, A.; O’Toole, S.A.; Barrett, C.; Ring, M.; Denning, K.M.; Li, J.; Aherne, S.T.; et al. Altered eIF6 and Dicer expression is associated with clinicopathological features in ovarian serous carcinoma patients. Mod. Pathol. 2008, 21, 676–684. [Google Scholar] [CrossRef] [Green Version]
- Pampalakis, G.; Diamandis, E.P.; Katsaros, D.; Sotiropoulou, G. Down-regulation of dicer expression in ovarian cancer tissues. Clin. BioChem. 2010, 43, 324–327. [Google Scholar] [CrossRef]
- Kuang, Y.; Cai, J.; Li, D.; Han, Q.; Cao, J.; Wang, Z. Repression of Dicer is associated with invasive phenotype and chemoresistance in ovarian cancer. Oncol. Lett. 2013, 5, 1149–1154. [Google Scholar] [CrossRef] [Green Version]
- Puppin, C.; Durante, C.; Sponziello, M.; Verrienti, A.; Pecce, V.; Lavarone, E.; Baldan, F.; Campese, A.F.; Boichard, A.; Lacroix, L.; et al. Overexpression of genes involved in miRNA biogenesis in medullary thyroid carcinomas with RET mutation. Endocrine 2014, 47, 528–536. [Google Scholar] [CrossRef]
- Erler, P.; Keutgen, X.M.; Crowley, M.J.; Zetoune, T.; Kundel, A.; Kleiman, D.; Beninato, T.; Scognamiglio, T.; Elemento, O.; Zarnegar, R.; et al. Dicer expression and microRNA dysregulation associate with aggressive features in thyroid cancer. Surgery 2014, 156, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.; Au, G.K.; et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008, 299, 425–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, H.J.; Baek, G.O.; Seo, C.W.; Ahn, H.R.; Sung, S.; Son, J.A.; Kim, S.S.; Cho, S.W.; Jang, J.W.; Nam, S.W.; et al. Exosomal microRNA-4661-5p-based serum panel as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. Cancer Med. 2020, 9, 5459–5472. [Google Scholar] [CrossRef] [PubMed]
- Jinushi, T.; Shibayama, Y.; Kinoshita, I.; Oizumi, S.; Jinushi, M.; Aota, T.; Takahashi, T.; Horita, S.; Dosaka-Akita, H.; Iseki, K. Low expression levels of microRNA-124-5p correlated with poor prognosis in colorectal cancer via targeting of SMC4. Cancer Med. 2014, 3, 1544–1552. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Chen, G.; Cheng, X.; Zhang, C.; Xu, H.; Qi, M.; Chen, X.; Wang, T.; Li, L. Inhibitory effects of miR26b5p on thyroid cancer. Mol. Med. Rep. 2019, 20, 1196–1202. [Google Scholar] [CrossRef] [Green Version]
- Michalik, K.M.; Bottcher, R.; Forstemann, K. A small RNA response at DNA ends in Drosophila. Nucleic Acids Res. 2012, 40, 9596–9603. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Rendtlew Danielsen, J.M.; Yang, Y.G.; Qi, Y. A role for small RNAs in DNA double-strand break repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef] [Green Version]
- Meister, G. Vision: Dicer leaps into view. Nature 2011, 471, 308–309. [Google Scholar] [CrossRef]
- Ambati, J.; Ambati, B.K.; Yoo, S.H.; Ianchulev, S.; Adamis, A.P. Age-related macular degeneration: Etiology, pathogenesis, and therapeutic strategies. Surv. OphthalMol. 2003, 48, 257–293. [Google Scholar] [CrossRef]
- Kaneko, H.; Dridi, S.; Tarallo, V.; Gelfand, B.D.; Fowler, B.J.; Cho, W.G.; Kleinman, M.E.; Ponicsan, S.L.; Hauswirth, W.W.; Chiodo, V.A.; et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 2011, 471, 325–330. [Google Scholar] [CrossRef] [Green Version]
- Takeda, A.; Baffi, J.Z.; Kleinman, M.E.; Cho, W.G.; Nozaki, M.; Yamada, K.; Kaneko, H.; Albuquerque, R.J.; Dridi, S.; Saito, K.; et al. CCR3 is a target for age-related macular degeneration diagnosis and therapy. Nature 2009, 460, 225–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarallo, V.; Hirano, Y.; Gelfand, B.D.; Dridi, S.; Kerur, N.; Kim, Y.; Cho, W.G.; Kaneko, H.; Fowler, B.J.; Bogdanovich, S.; et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 2012, 149, 847–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batzer, M.A.; Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Teo, C.H.; Soga, T.; Parhar, I.S. Brain Beta-Catenin Signalling During Stress and Depression. Neurosignals 2018, 26, 31–42. [Google Scholar] [CrossRef]
- Tsigos, C.; Kyrou, I.; Kassi, E.; Chrousos, G.P. Stress, Endocrine Physiology and Pathophysiology. In Endtext; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Tafet, G.E.; Bernardini, R. Psychoneuroendocrinological links between chronic stress and depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 2003, 27, 893–903. [Google Scholar] [CrossRef]
- McEwen, B.S. Glucocorticoids, depression, and mood disorders: Structural remodeling in the brain. Metabolism 2005, 54, 20–23. [Google Scholar] [CrossRef]
- Wieschaus, E.; Riggleman, R. Autonomous requirements for the segment polarity gene armadillo during Drosophila embryogenesis. Cell 1987, 49, 177–184. [Google Scholar] [CrossRef]
- Chen, Y.C.; Tan, Q.R.; Dang, W.; Wang, H.N.; Zhang, R.B.; Li, Z.Y.; Lin, H.; Liu, R. The effect of citalopram on chronic stress-induced depressive-like behavior in rats through GSK3beta/beta-catenin activation in the medial prefrontal cortex. Brain Res. Bull. 2012, 88, 338–344. [Google Scholar] [CrossRef]
- Dias, C.; Feng, J.; Sun, H.; Shao, N.Y.; Mazei-Robison, M.S.; Damez-Werno, D.; Scobie, K.; Bagot, R.; LaBonte, B.; Ribeiro, E.; et al. beta-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature 2014, 516, 51–55. [Google Scholar] [CrossRef] [Green Version]
- Miller, G.E.; Chen, E.; Sze, J.; Marin, T.; Arevalo, J.M.; Doll, R.; Ma, R.; Cole, S.W. A functional genomic fingerprInt. of chronic stress in humans: Blunted glucocorticoid and increased NF-kappaB signaling. Biol. Psychiatry 2008, 64, 266–272. [Google Scholar] [CrossRef] [Green Version]
- Wingo, A.P.; Almli, L.M.; Stevens, J.S.; Klengel, T.; Uddin, M.; Li, Y.; Bustamante, A.C.; Lori, A.; Koen, N.; Stein, D.J.; et al. DICER1 and microRNA regulation in post-traumatic stress disorder with comorbid depression. Nat. Commun. 2015, 6, 10106. [Google Scholar] [CrossRef]
- Zar, H.J.; Barnett, W.; Myer, L.; Stein, D.J.; Nicol, M.P. Investigating the early-life determinants of illness in Africa: The Drakenstein Child Health Study. Thorax 2015, 70, 592–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higuchi, F.; Uchida, S.; Yamagata, H.; Abe-Higuchi, N.; Hobara, T.; Hara, K.; Kobayashi, A.; Shintaku, T.; Itoh, Y.; Suzuki, T.; et al. Hippocampal MicroRNA-124 Enhances Chronic Stress Resilience in Mice. J. NeuroSci. 2016, 36, 7253–7267. [Google Scholar] [CrossRef] [PubMed]
- Volk, N.; Pape, J.C.; Engel, M.; Zannas, A.S.; Cattane, N.; Cattaneo, A.; Binder, E.B.; Chen, A. Amygdalar MicroRNA-15a Is Essential for Coping with Chronic Stress. Cell Rep. 2016, 17, 1882–1891. [Google Scholar] [CrossRef] [Green Version]
- Issler, O.; Haramati, S.; Paul, E.D.; Maeno, H.; Navon, I.; Zwang, R.; Gil, S.; Mayberg, H.S.; Dunlop, B.W.; Menke, A.; et al. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron 2014, 83, 344–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjorklund, A.; Dunnett, S.B. Dopamine neuron systems in the brain: An update. Trends NeuroSci. 2007, 30, 194–202. [Google Scholar] [CrossRef]
- Moore, D.J.; West, A.B.; Dawson, V.L.; Dawson, T.M. Molecular pathophysiology of Parkinson’s disease. Annu. Rev. Neurosci. 2005, 28, 57–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, M.A.; Raghavan, P.; Thomou, T.; Boucher, J.; Robida-Stubbs, S.; Macotela, Y.; Russell, S.J.; Kirkland, J.L.; Blackwell, T.K.; Kahn, C.R. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012, 16, 336–347. [Google Scholar] [CrossRef] [Green Version]
- Boon, R.A.; Iekushi, K.; Lechner, S.; Seeger, T.; Fischer, A.; Heydt, S.; Kaluza, D.; Treguer, K.; Carmona, G.; Bonauer, A.; et al. MicroRNA-34a regulates cardiac ageing and function. Nature 2013, 495, 107–110. [Google Scholar] [CrossRef]
- Simunovic, F.; Yi, M.; Wang, Y.; Stephens, R.; Sonntag, K.C. Evidence for gender-specific transcriptional profiles of nigral dopamine neurons in Parkinson disease. PLoS ONE 2010, 5, e8856. [Google Scholar] [CrossRef]
- Chmielarz, P.; Konovalova, J.; Najam, S.S.; Alter, H.; Piepponen, T.P.; Erfle, H.; Sonntag, K.C.; Schutz, G.; Vinnikov, I.A.; Domanskyi, A. Dicer and microRNAs protect adult dopamine neurons. Cell Death Dis. 2017, 8, e2813. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.H.; Cao, T.; Zheng, L.T.; Waddington, J.L.; Zhen, X.C. Development and characterization of an inducible Dicer conditional knockout mouse model of Parkinson’s disease: Validation of the antiparkinsonian effects of a sigma-1 receptor agonist and dihydromyricetin. Acta Pharmacol. Sin. 2020, 41, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; He, Q.; Chen, Y.; Shao, W.; Yuan, C.; Wang, Y. JNK-mediated microglial DICER degradation potentiates inflammatory responses to induce dopaminergic neuron loss. J. Neuroinflamm. 2018, 15, 184. [Google Scholar] [CrossRef] [PubMed]
- Colonna, M.; Butovsky, O. Microglia Function in the Central Nervous System during Health and Neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441–468. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Y.; Zhou, J. Neuroinflammation in Parkinson’s disease and its potential as therapeutic target. Transl. Neurodegener 2015, 4, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatchel, J.R.; Zoghbi, H.Y. Diseases of unstable repeat expansion: Mechanisms and common principles. Nat. Rev. Genet. 2005, 6, 743–755. [Google Scholar] [CrossRef]
- Krol, J.; Fiszer, A.; Mykowska, A.; Sobczak, K.; de Mezer, M.; Krzyzosiak, W.J. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol. Cell 2007, 25, 575–586. [Google Scholar] [CrossRef]
- Napierala, M.; Krzyzosiak, W.J. CUG repeats present in myotonin kinase RNA form metastable “slippery” hairpins. J. Biol. Chem. 1997, 272, 31079–31085. [Google Scholar] [CrossRef] [Green Version]
- Tian, B.; White, R.J.; Xia, T.; Welle, S.; Turner, D.H.; Mathews, M.B.; Thornton, C.A. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA 2000, 6, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Neueder, A.; Bates, G.P. RNA Related Pathology in Huntington’s Disease. Adv. Exp. Med. Biol. 2018, 1049, 85–101. [Google Scholar] [CrossRef]
- Banez-Coronel, M.; Porta, S.; Kagerbauer, B.; Mateu-Huertas, E.; Pantano, L.; Ferrer, I.; Guzman, M.; Estivill, X.; Marti, E. A pathogenic mechanism in Huntington’s disease involves small CAG-repeated RNAs with neurotoxic activity. PLoS Genet. 2012, 8, e1002481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llamusi, B.; Artero, R. Molecular Effects of the CTG Repeats in Mutant Dystrophia Myotonica Protein Kinase Gene. Curr. Genom. 2008, 9, 509–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malinina, L. Possible involvement of the RNAi pathway in trinucleotide repeat expansion diseases. J. BioMol. Struct. Dyn. 2005, 23, 233–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rostami Mogaddam, M.; Safavi Ardabili, N.; Shafaeei, Y.; Maleki, N.; Jafari, N.; Jafari, A. Overexpression of Drosha, DiGeorge syndrome critical region gene 8 (DGCR8), and Dicer mRNAs in the pathogenesis of psoriasis. J. Cosmet. Dermatol. 2017, 16, e48–e53. [Google Scholar] [CrossRef]
- Sonkoly, E. The expanding microRNA world in psoriasis. Exp. Dermatol. 2017, 26, 375–376. [Google Scholar] [CrossRef] [Green Version]
- Di Meglio, P.; Villanova, F.; Nestle, F.O. Psoriasis. Cold Spring Harb. Perspect. Med. 2014, 4. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, C.E.; Barker, J.N. Pathogenesis and clinical features of psoriasis. Lancet 2007, 370, 263–271. [Google Scholar] [CrossRef]
- Gaspari, A.A. Innate and adaptive immunity and the pathophysiology of psoriasis. J. Am. Acad. Dermatol. 2006, 54, S67–S80. [Google Scholar] [CrossRef]
- Zhou, X.; Krueger, J.G.; Kao, M.C.; Lee, E.; Du, F.; Menter, A.; Wong, W.H.; Bowcock, A.M. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol. Genom. 2003, 13, 69–78. [Google Scholar] [CrossRef] [Green Version]
- Sonkoly, E.; Wei, T.; Janson, P.C.; Saaf, A.; Lundeberg, L.; Tengvall-Linder, M.; Norstedt, G.; Alenius, H.; Homey, B.; Scheynius, A.; et al. MicroRNAs: Novel regulators involved in the pathogenesis of psoriasis? PLoS ONE 2007, 2, e610. [Google Scholar] [CrossRef] [Green Version]
- Yi, R.; O’Carroll, D.; Pasolli, H.A.; Zhang, Z.; Dietrich, F.S.; Tarakhovsky, A.; Fuchs, E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 2006, 38, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Ning, M.S.; Andl, T. Control by a hair’s breadth: The role of microRNAs in the skin. Cell Mol. Life Sci. 2013, 70, 1149–1169. [Google Scholar] [CrossRef] [Green Version]
- Tabrizi, Z.; Mansouri, R.; Aslani, S.; Jamshidi, A.R.; Mahmoudi, M. Expression levels of the microRNA maturing microprocessor complex components; Drosha, Dicer, and DGCR8 in PBMCs from ankylosing spondylitis patients. Mediterr. J. Rheumatol. 2017, 28, 80–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goie The, H.S.; Steven, M.M.; van der Linden, S.M.; Cats, A. Evaluation of diagnostic criteria for ankylosing spondylitis: A comparison of the Rome, New York and modified New York criteria in patients with a positive clinical history screening test for ankylosing spondylitis. Br. J. Rheumatol. 1985, 24, 242–249. [Google Scholar] [CrossRef] [Green Version]
- Jamshidi, A.R.; Shahlaee, A.; Farhadi, E.; Fallahi, S.; Nicknam, M.H.; Bidad, K.; Barghamadi, M.; Mahmoudi, M. Clinical characteristics and medical management of Iranian patients with ankylosing spondylitis. Mod. Rheumatol. 2014, 24, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.S.; Burgos-Vargas, R.; Louthrenoo, W.; Mok, M.Y.; Wordsworth, P.; Zeng, Q.Y. Features of spondyloarthritis around the world. Rheum Dis. Clin. N. Am. 1998, 24, 753–770. [Google Scholar] [CrossRef]
- Reveille, J.D. Recent studies on the genetic basis of ankylosing spondylitis. Curr. Rheumatol. Rep. 2009, 11, 340–348. [Google Scholar] [CrossRef]
- Lai, N.S.; Yu, H.C.; Chen, H.C.; Yu, C.L.; Huang, H.B.; Lu, M.C. Aberrant expression of microRNAs in T cells from patients with ankylosing spondylitis contributes to the immunopathogenesis. Clin. Exp. Immunol. 2013, 173, 47–57. [Google Scholar] [CrossRef]
- Liston, A.; Lu, L.F.; O’Carroll, D.; Tarakhovsky, A.; Rudensky, A.Y. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J. Exp. Med. 2008, 205, 1993–2004. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Jeker, L.T.; Fife, B.T.; Zhu, S.; Anderson, M.S.; McManus, M.T.; Bluestone, J.A. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J. Exp. Med. 2008, 205, 1983–1991. [Google Scholar] [CrossRef]
- Kovaleva, V.; Mora, R.; Park, Y.J.; Plass, C.; Chiramel, A.I.; Bartenschlager, R.; Dohner, H.; Stilgenbauer, S.; Pscherer, A.; Lichter, P.; et al. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012, 72, 1763–1772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciccia, F.; Haroon, N. Autophagy in the pathogenesis of ankylosing spondylitis. Clin. Rheumatol. 2016, 35, 1433–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asadipour, M.; Hassan-Zadeh, V.; Aryaeian, N.; Shahram, F.; Mahmoudi, M. Histone variants expression in peripheral blood mononuclear cells of patients with rheumatoid arthritis. Int. J. Rheum. Dis. 2018, 21, 1831–1837. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.L. Biologics-based therapy for the treatment of rheumatoid arthritis. Clin. Pharmacol. Ther. 2012, 91, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.D.; Zhang, M.; Zhang, Y.J.; Ye, D.Q. IL-33 in rheumatoid arthritis: Potential role in pathogenesis and therapy. Hum. Immunol. 2013, 74, 1057–1060. [Google Scholar] [CrossRef]
- Suzuki, A.; Yamada, R.; Chang, X.; Tokuhiro, S.; Sawada, T.; Suzuki, M.; Nagasaki, M.; Nakayama-Hamada, M.; Kawaida, R.; Ono, M.; et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat. Genet. 2003, 34, 395–402. [Google Scholar] [CrossRef]
- Brooks, W.H.; Le Dantec, C.; Pers, J.O.; Youinou, P.; Renaudineau, Y. Epigenetics and autoimmunity. J. Autoimmun. 2010, 34, J207–J219. [Google Scholar] [CrossRef]
- Alsaleh, G.; Nehmar, R.; Bluml, S.; Schleiss, C.; Ostermann, E.; Dillenseger, J.P.; Sayeh, A.; Choquet, P.; Dembele, D.; Francois, A.; et al. Reduced DICER1 Expression Bestows Rheumatoid Arthritis Synoviocytes Proinflammatory Properties and Resistance to Apoptotic Stimuli. Arthritis Rheumatol. 2016, 68, 1839–1848. [Google Scholar] [CrossRef] [Green Version]
- Akhlaghi, M.; Soltani, S.; Jamshidi, F.; Faezi, S.T.; Aslani, S.; Poursani, S.; Jamshidi, A.; Mahmoudi, M. Downregulation of Drosha, Dicer, and DGCR8 mRNAs in Peripheral Blood Mononuclear Cells of Patients with Rheumatoid Arthritis. Rheumatol. Res. 2018, 3, 135–143. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [Green Version]
- Dooley, J.; Linterman, M.A.; Liston, A. MicroRNA regulation of T-cell development. Immunol. Rev. 2013, 253, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Jafari, N.; Shaghaghi, H.; Mahmoodi, D.; Shirzad, Z.; Alibeiki, F.; Bohlooli, S.; Dogaheh, H.P. Overexpression of microRNA biogenesis machinery: Drosha, DGCR8 and Dicer in multiple sclerosis patients. J. Clin. Neurosci. 2015, 22, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.; Barkhof, F.; Montalban, X.; Thompson, A.; Filippi, M. Clinically isolated syndromes suggestive of multiple sclerosis, part I: Natural history, pathogenesis, diagnosis, and prognosis. Lancet Neurol. 2005, 4, 281–288. [Google Scholar] [CrossRef]
- Bremer, J.; O’Connor, T.; Tiberi, C.; Rehrauer, H.; Weis, J.; Aguzzi, A. Ablation of Dicer from murine Schwann cells increases their proliferation while blocking myelination. PLoS ONE 2010, 5, e12450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, D.; Shin, J.Y.; McManus, M.T.; Ptacek, L.J.; Fu, Y.H. Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann. Neurol. 2009, 66, 843–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asada, S.; Takahashi, T.; Isodono, K.; Adachi, A.; Imoto, H.; Ogata, T.; Ueyama, T.; Matsubara, H.; Oh, H. Downregulation of Dicer expression by serum withdrawal sensitizes human endothelial cells to apoptosis. Am. J. Physiol. Heart Circ. Physiol. 2008, 295, H2512–H2521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Gonzalez, E.; Lopez-Casas, P.P.; del Mazo, J. The expression patterns of genes involved in the RNAi pathways are tissue-dependent and differ in the germ and somatic cells of mouse testis. Biochim. Biophys. Acta 2008, 1779, 306–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiesen, J.L.; Tomasi, T.B. Dicer is regulated by cellular stresses and interferons. Mol. Immunol. 2009, 46, 1222–1228. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Lopez, J.; del Mazo, J. Expression dynamics of microRNA biogenesis during preimplantation mouse development. Biochim. Biophys. Acta 2012, 1819, 847–854. [Google Scholar] [CrossRef]
- Magner, W.J.; Weinstock-Guttman, B.; Rho, M.; Hojnacki, D.; Ghazi, R.; Ramanathan, M.; Tomasi, T.B. Dicer and microRNA expression in multiple sclerosis and response to interferon therapy. J. NeuroImmunol. 2016, 292, 68–78. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Aung, L.L.; Balashov, K.E. Decreased Dicer expression is linked to increased expression of co-stimulatory molecule CD80 on B cells in multiple sclerosis. Mult. Scler. J. 2015, 21, 1131–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koralov, S.B.; Muljo, S.A.; Galler, G.R.; Krek, A.; Chakraborty, T.; Kanellopoulou, C.; Jensen, K.; Cobb, B.S.; Merkenschlager, M.; Rajewsky, N.; et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 2008, 132, 860–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belver, L.; de Yebenes, V.G.; Ramiro, A.R. MicroRNAs prevent the generation of autoreactive antibodies. Immunity 2010, 33, 713–722. [Google Scholar] [CrossRef] [Green Version]
- Jacobson, D.L.; Gange, S.J.; Rose, N.R.; Graham, N.M. Epidemiology and estimated population burden of selected autoimmune diseases in the United States. Clin. Immunol. ImmunoPathol. 1997, 84, 223–243. [Google Scholar] [CrossRef] [Green Version]
- Antonelli, A.; Ferrari, S.M.; Corrado, A.; Di Domenicantonio, A.; Fallahi, P. Autoimmune thyroid disorders. Autoimmun. Rev. 2015, 14, 174–180. [Google Scholar] [CrossRef]
- Saeki, M.; Watanabe, M.; Inoue, N.; Tokiyoshi, E.; Takuse, Y.; Arakawa, Y.; Hidaka, Y.; Iwatani, Y. DICER and DROSHA gene expression and polymorphisms in autoimmune thyroid diseases. Autoimmunity 2016, 49, 514–522. [Google Scholar] [CrossRef]
- Ma, H.; Yuan, H.; Yuan, Z.; Yu, C.; Wang, R.; Jiang, Y.; Hu, Z.; Shen, H.; Chen, N. Genetic variations in key microRNA processing genes and risk of head and neck cancer: A case-control study in Chinese population. PLoS ONE 2012, 7, e47544. [Google Scholar] [CrossRef] [Green Version]
- Suarez, Y.; Fernandez-Hernando, C.; Pober, J.S.; Sessa, W.C. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 2007, 100, 1164–1173. [Google Scholar] [CrossRef] [Green Version]
- Kuehbacher, A.; Urbich, C.; Zeiher, A.M.; Dimmeler, S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ. Res. 2007, 101, 59–68. [Google Scholar] [CrossRef] [Green Version]
- Mishra, P.K.; Tyagi, N.; Kumar, M.; Tyagi, S.C. MicroRNAs as a therapeutic target for cardiovascular diseases. J. Cell Mol. Med. 2009, 13, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.J.; Yang, D.D.; Na, S.; Sandusky, G.E.; Zhang, Q.; Zhao, G. Dicer is required for embryonic angiogenesis during mouse development. J. Biol. Chem. 2005, 280, 9330–9335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Costa Martins, P.A.; Bourajjaj, M.; Gladka, M.; Kortland, M.; van Oort, R.J.; Pinto, Y.M.; Molkentin, J.D.; De Windt, L.J. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation 2008, 118, 1567–1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.F.; Murchison, E.P.; Tang, R.; Callis, T.E.; Tatsuguchi, M.; Deng, Z.; Rojas, M.; Hammond, S.M.; Schneider, M.D.; Selzman, C.H.; et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc. Natl. Acad. Sci. USA 2008, 105, 2111–2116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Corbalan-Campos, J.; Gurung, R.; Natarelli, L.; Zhu, M.; Exner, N.; Erhard, F.; Greulich, F.; Geissler, C.; Uhlenhaut, N.H.; et al. Dicer in Macrophages Prevents Atherosclerosis by Promoting Mitochondrial Oxidative Metabolism. Circulation 2018, 138, 2007–2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Q.; Bhatt, K.; He, H.Z.; Mi, Q.S.; Haase, V.H.; Dong, Z. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J. Am. Soc. Nephrol. 2010, 21, 756–761. [Google Scholar] [CrossRef] [Green Version]
- Borghini, A.; Pulignani, S.; Mercuri, A.; Vecoli, C.; Turchi, S.; Carpeggiani, C.; Andreassi, M.G. Influence of genetic polymorphisms in DICER and XPO5 genes on the risk of coronary artery disease and circulating levels of vascular miRNAs. Thromb. Res. 2019, 180, 32–36. [Google Scholar] [CrossRef]
- Carletti, M.Z.; Christenson, L.K. MicroRNA in the ovary and female reproductive tract. J. Anim. Sci. 2009, 87, E29–E38. [Google Scholar] [CrossRef] [Green Version]
- Kimmins, S.; Kotaja, N.; Davidson, I.; Sassone-Corsi, P. Testis-specific transcription mechanisms promoting male germ-cell differentiation. Reproduction 2004, 128, 5–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimmins, S.; Sassone-Corsi, P. Chromatin remodelling and epigenetic features of germ cells. Nature 2005, 434, 583–589. [Google Scholar] [CrossRef]
- Murchison, E.P.; Stein, P.; Xuan, Z.; Pan, H.; Zhang, M.Q.; Schultz, R.M.; Hannon, G.J. Critical roles for Dicer in the female germline. Genes Dev. 2007, 21, 682–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maatouk, D.M.; Loveland, K.L.; McManus, M.T.; Moore, K.; Harfe, B.D. Dicer1 is required for differentiation of the mouse male germline. Biol. Reprod. 2008, 79, 696–703. [Google Scholar] [CrossRef] [Green Version]
- Korhonen, H.M.; Meikar, O.; Yadav, R.P.; Papaioannou, M.D.; Romero, Y.; Da Ros, M.; Herrera, P.L.; Toppari, J.; Nef, S.; Kotaja, N. Dicer is required for haploid male germ cell differentiation in mice. PLoS ONE 2011, 6, e24821. [Google Scholar] [CrossRef] [Green Version]
- Yuan, S.; Ortogero, N.; Wu, Q.; Zheng, H.; Yan, W. Murine follicular development requires oocyte DICER, but not DROSHA. Biol. Reprod. 2014, 91, 39. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Graham, I.; Hastings, R.; Gunewardena, S.; Brinkmeier, M.L.; Conn, P.M.; Camper, S.A.; Kumar, T.R. Gonadotrope-specific deletion of Dicer results in severely suppressed gonadotropins and fertility defects. J. Biol. Chem. 2015, 290, 2699–2714. [Google Scholar] [CrossRef] [PubMed]
- Luense, L.J.; Carletti, M.Z.; Christenson, L.K. Role of Dicer in female fertility. Trends Endocrinol. Metab. 2009, 20, 265–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattiske, D.M.; Han, L.; Mann, J.R. Meiotic maturation failure induced by DICER1 deficiency is derived from primary oocyte ooplasm. Reproduction 2009, 137, 625–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otsuka, M.; Zheng, M.; Hayashi, M.; Lee, J.D.; Yoshino, O.; Lin, S.; Han, J. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J. Clin. Investig. 2008, 118, 1944–1954. [Google Scholar] [CrossRef] [Green Version]
- Hong, X.; Luense, L.J.; McGinnis, L.K.; Nothnick, W.B.; Christenson, L.K. Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology 2008, 149, 6207–6212. [Google Scholar] [CrossRef] [Green Version]
- Nagaraja, A.K.; Andreu-Vieyra, C.; Franco, H.L.; Ma, L.; Chen, R.; Han, D.Y.; Zhu, H.; Agno, J.E.; Gunaratne, P.H.; DeMayo, F.J.; et al. Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol. Endocrinol. 2008, 22, 2336–2352. [Google Scholar] [CrossRef] [Green Version]
- Pastorelli, L.M.; Wells, S.; Fray, M.; Smith, A.; Hough, T.; Harfe, B.D.; McManus, M.T.; Smith, L.; Woolf, A.S.; Cheeseman, M.; et al. Genetic analyses reveal a requirement for Dicer1 in the mouse urogenital tract. Mamm. Genome 2009, 20, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, G.; Behringer, R.R. Dicer is required for female reproductive tract development and fertility in the mouse. Mol. Reprod. Dev. 2009, 76, 678–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arur, S. Context-dependent regulation of Dicer activity and small RNA production: Implications to oocyte-to-embryo transition. Worm 2015, 4, e1086062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, S.W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef]
- Aliyari, R.; Ding, S.W. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol. Rev. 2009, 227, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Li, W.X.; Ding, S.W. Induction and suppression of RNA silencing by an animal virus. Science 2002, 296, 1319–1321. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.W.; Han, Q.; Wang, J.; Li, W.X. Antiviral RNA interference in mammals. Curr. Opin. Immunol. 2018, 54, 109–114. [Google Scholar] [CrossRef]
- Cullen, B.R. Viral and cellular messenger RNA targets of viral microRNAs. Nature 2009, 457, 421–425. [Google Scholar] [CrossRef] [Green Version]
- Maillard, P.V.; Ciaudo, C.; Marchais, A.; Li, Y.; Jay, F.; Ding, S.W.; Voinnet, O. Antiviral RNA interference in mammalian cells. Science 2013, 342, 235–238. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Basavappa, M.; Lu, J.; Dong, S.; Cronkite, D.A.; Prior, J.T.; Reinecker, H.C.; Hertzog, P.; Han, Y.; Li, W.X.; et al. Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat. Microbiol. 2016, 2, 16250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, Y.; Xu, Y.; Zhang, Y.; Zhou, H.; Deng, Y.Q.; Li, X.F.; Miao, M.; Zhang, Q.; Zhong, B.; Hu, Y.; et al. Human Virus-Derived Small RNAs Can Confer Antiviral Immunity in Mammals. Immunity 2017, 46, 992–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coley, W.; Van Duyne, R.; Carpio, L.; Guendel, I.; Kehn-Hall, K.; Chevalier, S.; Narayanan, A.; Luu, T.; Lee, N.; Klase, Z.; et al. Absence of DICER in monocytes and its regulation by HIV-1. J. Biol. Chem. 2010, 285, 31930–31943. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Disease | Association | |
|---|---|---|
| Dicer1 syndrome | germline mutations | |
| Cancer | downregulation/upregulation | |
| Geographic atrophy (GA) | downregulation | |
| Psychiatric and Neurological Diseases | Chronic stress and depression | downregulation |
| Parkinson’s disease (PD) | downregulation | |
| Triplet repeat expansion diseases (TREDs) | normal expression/overexpression | |
| Autoimmune Disorders | Psoriasis | upregulation |
| Ankylosing spondylitis (AS) | downregulation | |
| Rheumatoid arthritis (RA) | downregulation | |
| Multiple sclerosis (MS) | downregulation | |
| Autoimmune thyroid diseases (AITDs) | downregulation | |
| Cardiovascular diseases | downregulation/polymorphism | |
| Infertility | downregulation | |
| Viral infections | vsiRNAs generation | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Theotoki, E.I.; Pantazopoulou, V.I.; Georgiou, S.; Kakoulidis, P.; Filippa, V.; Stravopodis, D.J.; Anastasiadou, E. Dicing the Disease with Dicer: The Implications of Dicer Ribonuclease in Human Pathologies. Int. J. Mol. Sci. 2020, 21, 7223. https://doi.org/10.3390/ijms21197223
Theotoki EI, Pantazopoulou VI, Georgiou S, Kakoulidis P, Filippa V, Stravopodis DJ, Anastasiadou E. Dicing the Disease with Dicer: The Implications of Dicer Ribonuclease in Human Pathologies. International Journal of Molecular Sciences. 2020; 21(19):7223. https://doi.org/10.3390/ijms21197223
Chicago/Turabian StyleTheotoki, Eleni I., Vasiliki I. Pantazopoulou, Stella Georgiou, Panos Kakoulidis, Vicky Filippa, Dimitrios J. Stravopodis, and Ema Anastasiadou. 2020. "Dicing the Disease with Dicer: The Implications of Dicer Ribonuclease in Human Pathologies" International Journal of Molecular Sciences 21, no. 19: 7223. https://doi.org/10.3390/ijms21197223
APA StyleTheotoki, E. I., Pantazopoulou, V. I., Georgiou, S., Kakoulidis, P., Filippa, V., Stravopodis, D. J., & Anastasiadou, E. (2020). Dicing the Disease with Dicer: The Implications of Dicer Ribonuclease in Human Pathologies. International Journal of Molecular Sciences, 21(19), 7223. https://doi.org/10.3390/ijms21197223






