Inhibitory Effect of a Human MicroRNA, miR-6133-5p, on the Fibrotic Activity of Hepatic Stellate Cells in Culture
Abstract
1. Introduction
2. Results
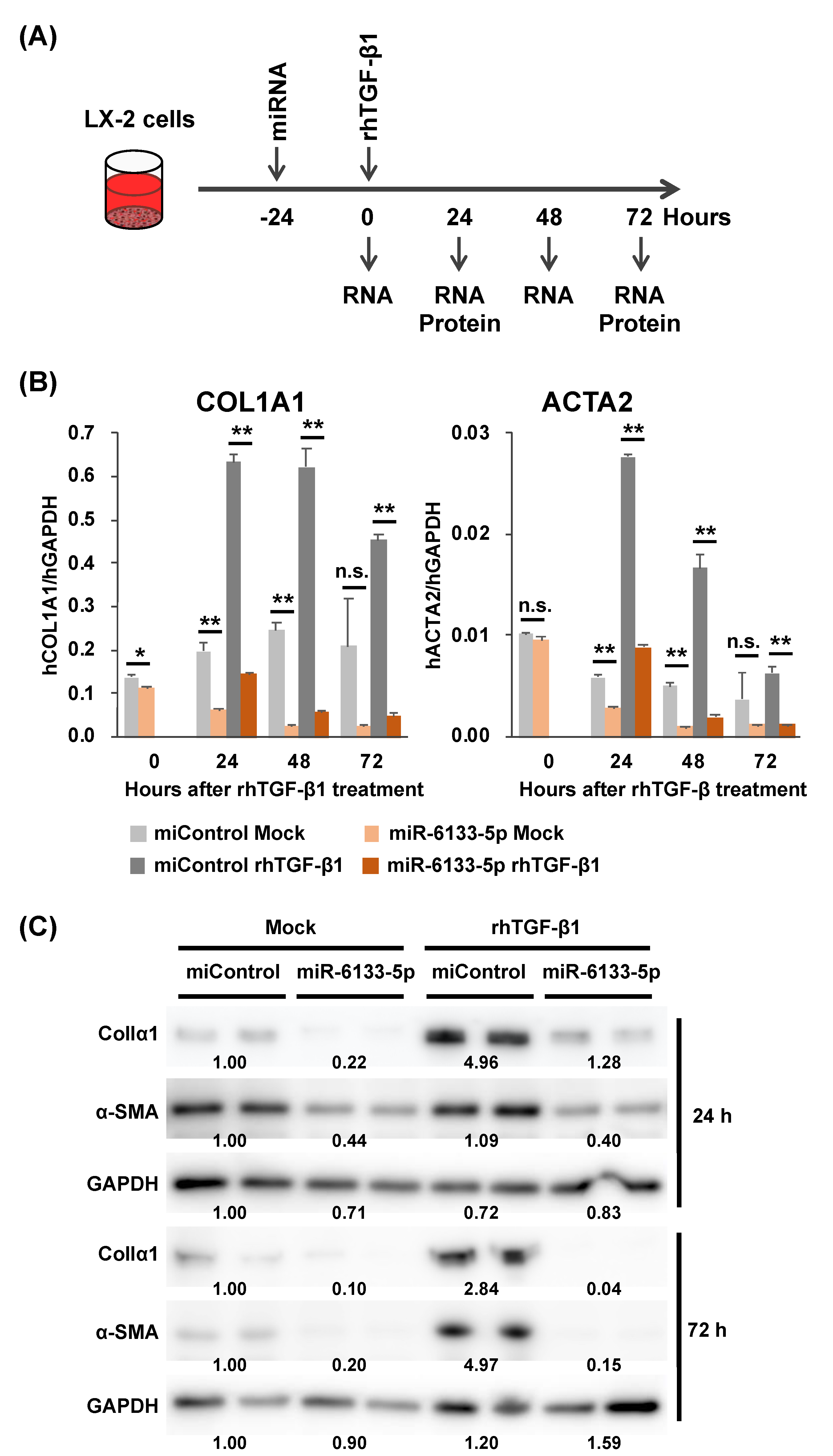
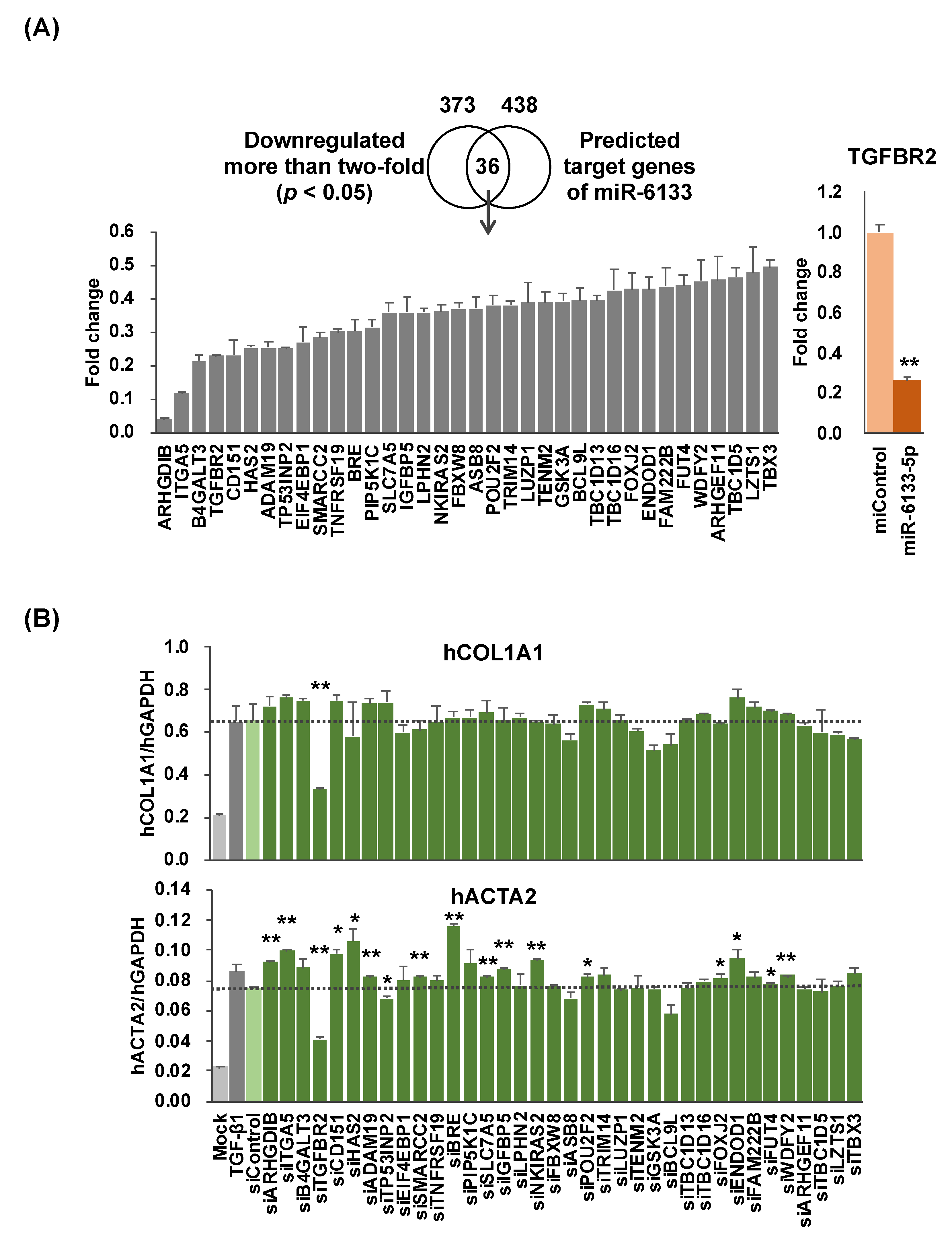
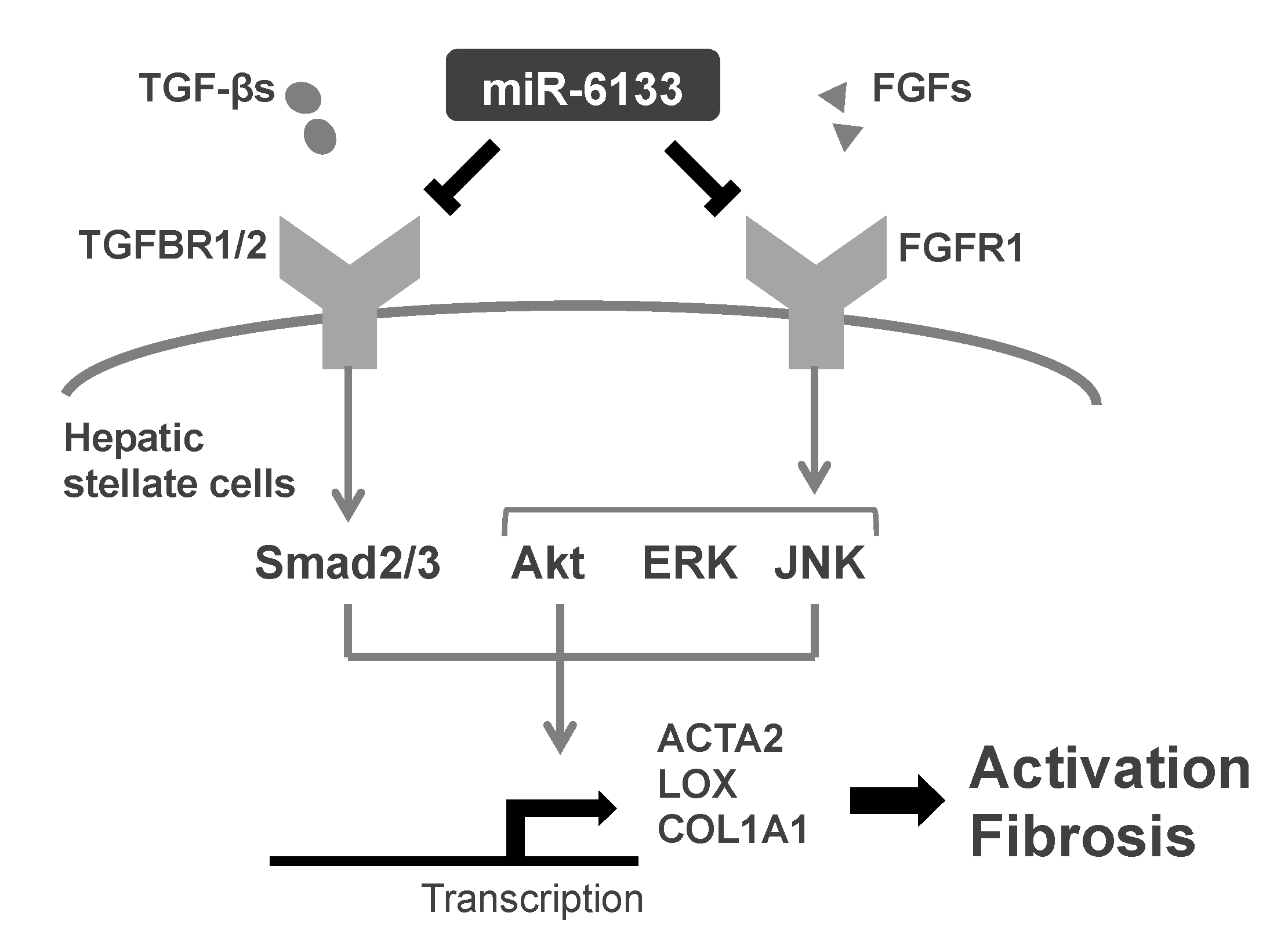
2.1. MiR-6133-5p Suppresses the Synthesis of α-Chain of Collagen Type I and α-Smooth Muscle Actin in LX-2 Cells
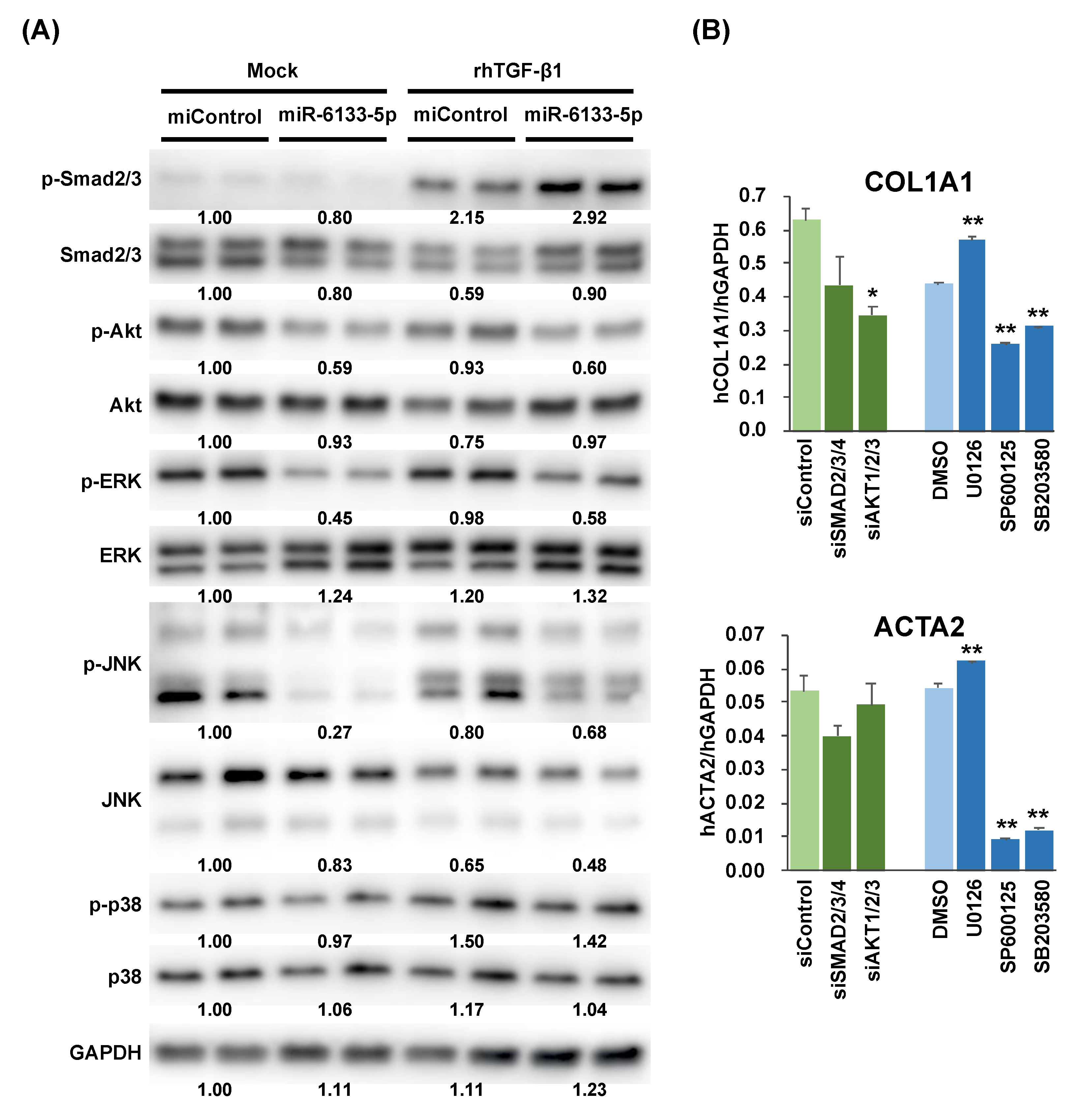
2.2. MiR-6133 Decreased Phosphorylation of Akt, ERK, and JNK
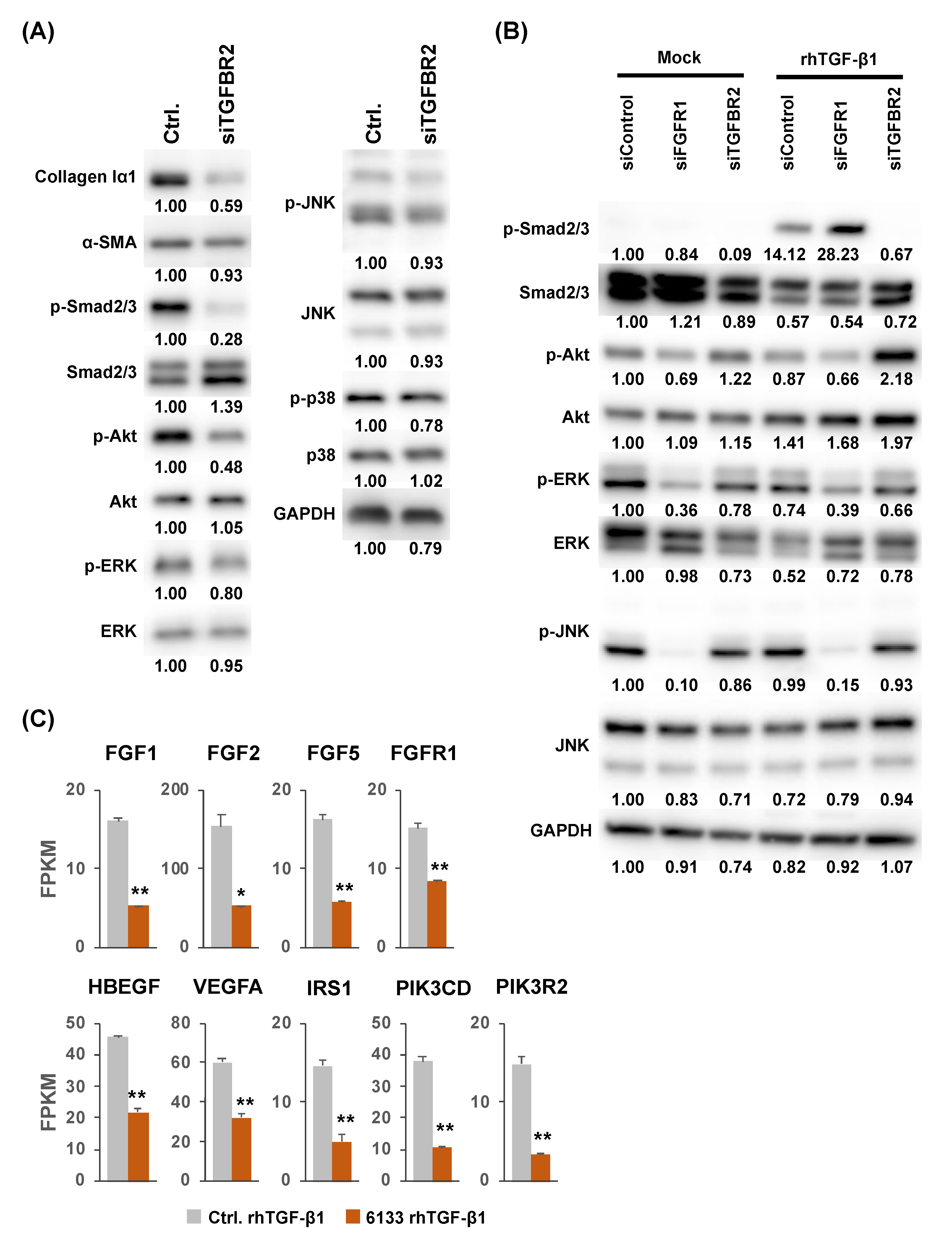
2.3. Possible Involvement of the Fibroblast Growth Factor Receptor 1 (FGFR1) Gene in the Suppression of JNK Phosphorylation by MiR-6133-5p
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Transfection
4.2. Gene Expression Analysis by RT-qPCR
4.3. Quantification of Protein
4.4. RNAseq Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HBV | hepatitis B virus |
| HSCs | hepatic stellate cells |
| TGF-β | transforming growth factor β |
| α-SMA | α-smooth muscle actin |
| miRNA | microRNA |
| 3′-UTRs | 3′-untranslated regions |
| ERK | extracellular signal-regulated kinase |
| JNK | c-Jun N-terminal kinase |
| FGFR1 | fibroblast growth factor 1 |
| siRNA | small interfering RNA |
| GSEA | gene set enrichment analysis |
| GO | gene ontology |
References
- Sepanlou, S.G.; Safiri, S.; Bisignano, C.; Ikuta, K.S.; Merat, S.; Saberifiroozi, M.; Poustchi, H.; Tsoi, D.; Colombara, D.V.; Abdoli, A.; et al. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 245–266. [Google Scholar] [CrossRef]
- Graber-Stiehl, I. The silent epidemic killing more people than HIV, malaria or TB. Nature 2018, 564, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Bataller, R.; Brenner, D.A. Liver fibrosis. J. Clin. Investig. 2005, 115, 209–218. [Google Scholar] [CrossRef]
- Cai, C.X.; Buddha, H.; Castelino-Prabhu, S.; Zhang, Z.; Britton, R.S.; Bacon, B.R.; Neuschwander-Tetri, B.A. Activation of Insulin-PI3K/Akt-p70S6K Pathway in Hepatic Stellate Cells Contributes to Fibrosis in Nonalcoholic Steatohepatitis. Dig. Dis. Sci. 2017, 62, 968–978. [Google Scholar] [CrossRef] [PubMed]
- Reif, S.; Scanga, A.E.; Brenner, D.A.; Rippe, R.A. The role of AKT in hepatic stellate cell activation. YGAST 2001, 120, A27. [Google Scholar]
- Schnabl, B.; Bradham, C.A.; Bennett, B.L.; Manning, A.M.; Stefanovic, B.; Brenner, D.A. TAK1/JNK and p38 have opposite effects on rat hepatic stellate cells. Hepatology 2001, 34, 953–963. [Google Scholar] [CrossRef]
- Anania, F. Aldehydes potentiate α2(I) collagen gene activity by JNK in hepatic stellate cells. Free Radic. Biol. Med. 2001, 30, 846–857. [Google Scholar] [CrossRef]
- Kluwe, J.; Pradere, J.P.; Gwak, G.Y.; Mencin, A.; De Minicis, S.; Österreicher, C.H.; Colmenero, J.; Bataller, R.; Schwabe, R.F. Modulation of Hepatic Fibrosis by c-Jun-N-Terminal Kinase Inhibition. YGAST 2010, 138, 347–359. [Google Scholar] [CrossRef]
- Higashi, T.; Friedman, S.L.; Hoshida, Y. Hepatic stellate cells as key target in liver fibrosis. Adv. Drug Deliv. Rev. 2017, 121, 27–42. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, 101. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Garcia, I.; Miska, E.A. MicroRNA functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef] [PubMed]
- Ivey, K.N.; Srivastava, D. microRNAs as Developmental Regulators. Cold Spring Harb. Perspect. Biol. 2015, 7, a008144. [Google Scholar] [CrossRef] [PubMed]
- Baltimore, D.; Boldin, M.P.; O’Connell, R.M.; Rao, D.S.; Taganov, K.D. MicroRNAs: New regulators of immune cell development and function. Nat. Immunol. 2008, 9, 839–845. [Google Scholar] [CrossRef]
- Gailhouste, L.; Histopathol, T.O.H. Cancer-related microRNAs and their role as tumor suppressors and oncogenes in hepatocellular carcinoma. Histol. Histopathol. 2013, 28, 437–451. [Google Scholar]
- Chuma, M.; Toyoda, H.; Matsuzaki, J.; Saito, Y.; Kumada, T.; Tada, T.; Kaneoka, Y.; Maeda, A.; Yokoo, H.; Ogawa, K.; et al. Circulating microRNA-1246 as a possible biomarker for early tumor recurrence of hepatocellular carcinoma. Hepatol. Res. 2019, 49, 810–822. [Google Scholar] [CrossRef]
- Zheng, Z.; Wen, Y.; Nie, K.; Tang, S.; Chen, X.; Lan, S.; Pan, J.; Jiang, K.; Jiang, X.; Liu, P.; et al. Construction of a 13-microRNA-based signature and prognostic nomogram for predicting overall survival in patients with hepatocellular carcinoma. Hepatol. Res. 2020, 50, hepr.13538. [Google Scholar] [CrossRef]
- Roderburg, C.; Urban, G.W.; Bettermann, K.; Vucur, M.; Zimmermann, H.; Schmidt, S.; Janssen, J.; Koppe, C.; Knolle, P.; Castoldi, M.; et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 2011, 53, 209–218. [Google Scholar] [CrossRef]
- Ogawa, T.; Iizuka, M.; Sekiya, Y.; Yoshizato, K.; Ikeda, K.; Kawada, N. Suppression of type I collagen production by microRNA-29b in cultured human stellate cells. Biochem. Biophys. Res. Commun. 2010, 391, 316–321. [Google Scholar] [CrossRef]
- Oba, S.; Kumano, S.; Suzuki, E.; Nishimatsu, H.; Takahashi, M.; Takamori, H.; Kasuya, M.; Ogawa, Y.; Sato, K.; Kimura, K.; et al. miR-200b Precursor Can Ameliorate Renal Tubulointerstitial Fibrosis. PLoS ONE 2010, 5, e13614. [Google Scholar] [CrossRef]
- Hyun, J.; Wang, S.; Kim, J.; Rao, K.M.; Park, S.Y.; Chung, I.; Ha, C.-S.; Kim, S.-W.; Yun, Y.H.; Jung, Y. MicroRNA-378 limits activation of hepatic stellate cells and liver fibrosis by suppressing Gli3 expression. Nat. Commun. 2016, 7, 10993. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; De Giorgi, V.; Schechterly, C.; Wang, R.Y.; Farci, P.; Tanaka, Y.; Alter, H.J. Circulating let-7 levels in plasma and extracellular vesicles correlate with hepatic fibrosis progression in chronic hepatitis C. Hepatology 2016, 64, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Hamada-Tsutsumi, S.; Yamamoto, Y.; Kogure, A.; Yoshioka, Y.; Watashi, K.; Ochiya, T.; Tanaka, Y. Screening of microRNAs for a repressor of hepatitis B virus replication. Oncotarget 2018, 9, 29857–29868. [Google Scholar] [CrossRef]
- McMahon, B.J. The natural history of chronic hepatitis B virus infection. Hepatology 2009, 49, S45–S55. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-Y.; Lin, Y.-C.-D.; Li, J.; Huang, K.-Y.; Shrestha, S.; Hong, H.-C.; Tang, Y.; Chen, Y.-G.; Jin, C.-N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef]
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shi, S.; Srivastava, S.P.; Kitada, M.; Nagai, T.; Nitta, K.; Kohno, M.; Kanasaki, K.; Koya, D. FGFR1 is critical for the anti-endothelial mesenchymal transition effect of N-acetyl-seryl-aspartyl-lysyl-proline via induction of the MAP4K4 pathway. Cell Death Dis. 2017, 8, e2965. [Google Scholar] [CrossRef]
- Schuster-Gaul, S.; Geisler, L.J.; McGeough, M.D.; Johnson, C.D.; Zagorska, A.; Li, L.; Wree, A.; Barry, V.; Mikaelian, I.; Jih, L.J.; et al. ASK1 inhibition reduces cell death and hepatic fibrosis in an Nlrp3 mutant liver injury model. JCI Insight 2020, 5, 3030. [Google Scholar] [CrossRef]
- Delić, D.; Eisele, C.; Schmid, R.; Baum, P.; Wiech, F.; Gerl, M.; Zimdahl, H.; Pullen, S.S.; Urquhart, R. Urinary Exosomal miRNA Signature in Type II Diabetic Nephropathy Patients. PLoS ONE 2016, 11, e0150154. [Google Scholar] [CrossRef]
- Dandri, M.; Burda, M.R.; Török, E.; Pollok, J.M.; Iwanska, A.; Sommer, G.; Rogiers, X.; Rogler, C.E.; Gupta, S.; Will, H.; et al. Repopulation of mouse liver with human hepatocytes and in vivo infection with hepatitis B virus. Hepatology 2001, 33, 981–988. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamada-Tsutsumi, S.; Onishi, M.; Matsuura, K.; Isogawa, M.; Kawashima, K.; Sato, Y.; Tanaka, Y. Inhibitory Effect of a Human MicroRNA, miR-6133-5p, on the Fibrotic Activity of Hepatic Stellate Cells in Culture. Int. J. Mol. Sci. 2020, 21, 7251. https://doi.org/10.3390/ijms21197251
Hamada-Tsutsumi S, Onishi M, Matsuura K, Isogawa M, Kawashima K, Sato Y, Tanaka Y. Inhibitory Effect of a Human MicroRNA, miR-6133-5p, on the Fibrotic Activity of Hepatic Stellate Cells in Culture. International Journal of Molecular Sciences. 2020; 21(19):7251. https://doi.org/10.3390/ijms21197251
Chicago/Turabian StyleHamada-Tsutsumi, Susumu, Masaya Onishi, Kentaro Matsuura, Masanori Isogawa, Keigo Kawashima, Yusuke Sato, and Yasuhito Tanaka. 2020. "Inhibitory Effect of a Human MicroRNA, miR-6133-5p, on the Fibrotic Activity of Hepatic Stellate Cells in Culture" International Journal of Molecular Sciences 21, no. 19: 7251. https://doi.org/10.3390/ijms21197251
APA StyleHamada-Tsutsumi, S., Onishi, M., Matsuura, K., Isogawa, M., Kawashima, K., Sato, Y., & Tanaka, Y. (2020). Inhibitory Effect of a Human MicroRNA, miR-6133-5p, on the Fibrotic Activity of Hepatic Stellate Cells in Culture. International Journal of Molecular Sciences, 21(19), 7251. https://doi.org/10.3390/ijms21197251





