Roles of ASYMMETRIC LEAVES2 (AS2) and Nucleolar Proteins in the Adaxial–Abaxial Polarity Specification at the Perinucleolar Region in Arabidopsis
Abstract
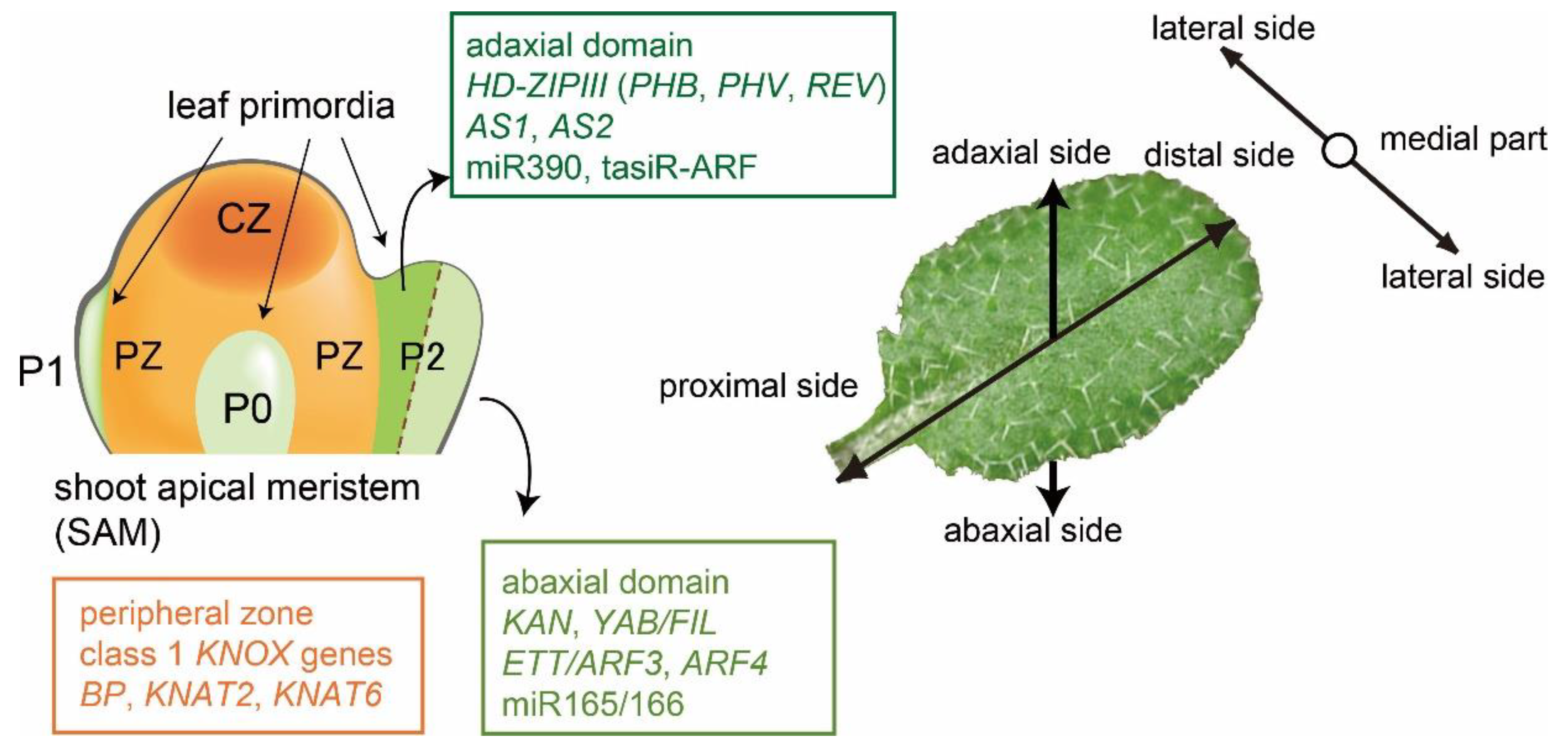
1. Leaf Developments in Arabidopsis
2. Roles of AS2–AS1 in the Development of Leaf Polarity
3. Modifier Mutations That Enhance Defects of AS2 and AS1 in Leaf Adaxial–Abaxial Polarity
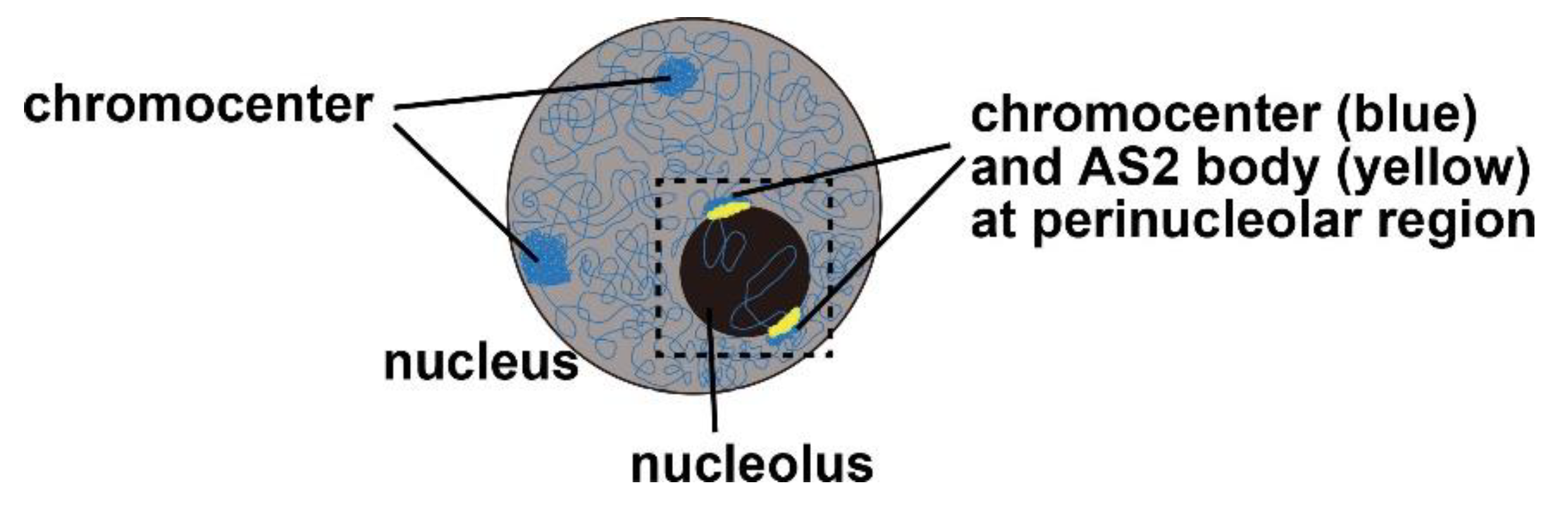
4. AS2 Bodies: Perinucleolar Granules Co-Localized Partially with the Chromocenter
5. AS2–AS1 Binds to Exon 1 of the Target Gene ETT/ARF3, and Is Involved in Maintaining CpG Methylation in Exon 6
6. Subcellular localization of AS2
7. Possible Roles of AS2 Bodies in Epigenetic Repression of ETT/ARF3
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| FISH | Fluorescence in situ Hybridization |
| GFP | Green Fluorescent Protein |
| BY-2 | Bright Yellow-2 |
| tasiR | trans-acting small interfering RNA |
References
- Bar, M.; Ori, N. Compound leaf development in model plant species. Curr. Opin. Plant. Biol. 2015, 23, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Machida, C.; Nakagawa, A.; Kojima, S.; Takahashi, H.; Machida, Y. The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in Arabidopsis. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 655–671. [Google Scholar] [CrossRef] [PubMed]
- Fouracre, J.P.; Poethig, R.S. The role of small RNAs in vegetative shoot development. Curr. Opin. Plant. Biol. 2016, 29, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Guan, C.; Jiao, Y. Molecular Mechanisms of Leaf Morphogenesis. Mol. Plant 2018, 11, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.P.; Furumizu, C.; Efroni, I.; Eshed, Y.; Bowman, J.L. Active suppression of a leaf meristem orchestrates determinate leaf growth. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Conklin, P.A.; Strable, J.; Li, S.; Scanlon, M.J. On the mechanisms of development in monocot and eudicot leaves. New Phytol. 2019, 221, 706–724. [Google Scholar] [CrossRef]
- Kuhlemeier, C.; Timmermans, M.C. The Sussex signal: Insights into leaf dorsiventrality. Development 2016, 143, 3230–3237. [Google Scholar] [CrossRef]
- Satterlee, J.W.; Scanlon, M.J. Coordination of Leaf Development Across Developmental Axes. Plants 2019, 8, 433. [Google Scholar] [CrossRef]
- Nakata, M.T.; Tameshige, T.; Takahara, M.; Mitsuda, N.; Okada, K. The functional balance between the WUSCHEL-RELATED HOMEOBOX1 gene and the phytohormone auxin is a key factor for cell proliferation in Arabidopsis seedlings. Plant. Biotechnol. 2018, 35, 141–154. [Google Scholar] [CrossRef]
- Byrne, M.E.; Barley, R.; Curtis, M.; Arroyo, J.M.; Dunham, M.; Hudson, A.; Martienssen, R.A. Asymmetric leaves 1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 2000, 408, 967–971. [Google Scholar] [CrossRef]
- Semiarti, E.; Ueno, Y.; Tsukaya, H.; Iwakawa, H.; Machida, C.; Machida, Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 2001, 128, 1771–1783. [Google Scholar] [PubMed]
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C.; et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.C.; Shuai, B.; Springer, P.S. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 2003, 15, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Timmermans, M.C.; Hudson, A.; Becraft, P.W.; Nelson, T. ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 1999, 284, 151–153. [Google Scholar] [CrossRef]
- Tsiantis, M.; Schneeberger, R.; Golz, J.F.; Freeling, M.; Langdale, J.A. The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 1999, 284, 154–156. [Google Scholar] [CrossRef]
- Waites, R.; Selvadurai, H.R.; Oliver, I.R.; Hudson, A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 1998, 93, 779–789. [Google Scholar] [CrossRef]
- Waites, R.; Hudson, A. The Handlebars gene is required with Phantastica for dorsoventral asymmetry of organs and for stem cell activity in Antirrhinum. Development 2001, 128, 1923–1931. [Google Scholar]
- Ikezaki, M.; Kojima, M.; Sakakibara, H.; Kojima, S.; Ueno, Y.; Machida, C.; Machida, Y. Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J. 2010, 61, 70–82. [Google Scholar] [CrossRef]
- Guo, M.; Thomas, J.; Collins, G.; Timmermans, M.C. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 2008, 20, 48–58. [Google Scholar] [CrossRef]
- McConnell, J.R.; Emery, J.; Eshed, Y.; Bao, N.; Bowman, J.; Barton, M.K. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001, 411, 709–713. [Google Scholar] [CrossRef]
- Emery, J.F.; Floyd, S.K.; Alvarez, J.; Eshed, Y.; Hawker, N.P.; Izhaki, A.; Baum, S.F.; Bowman, J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003, 13, 1768–1774. [Google Scholar] [CrossRef] [PubMed]
- Merelo, P.; Paredes, E.B.; Heisler, M.G.; Wenkel, S. The shady side of leaf development: The role of the REVOLUTA/KANADI1 module in leaf patterning and auxin-mediated growth promotion. Curr. Opin. Plant. Biol. 2017, 35, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Tatematsu, K.; Toyokura, K.; Miyashima, S.; Nakajima, K.; Okada, K. A molecular mechanism that confines the activity pattern of miR165 in Arabidopsis leaf primordia. Plant J. 2015, 82, 596–608. [Google Scholar] [CrossRef]
- Kerstetter, R.A.; Bollman, K.; Taylor, R.A.; Bomblies, K.; Poethig, R.S. KANADI regulates organ polarity in Arabidopsis. Nature 2001, 411, 706–709. [Google Scholar] [CrossRef]
- Sawa, S.; Ito, T.; Shimura, Y.; Okada, K. FILAMENTOUS FLOWER controls the formation and development of arabidopsis inflorescences and floral meristems. Plant Cell 1999, 11, 69–86. [Google Scholar] [CrossRef]
- Siegfried, K.R.; Eshed, Y.; Baum, S.F.; Otsuga, D.; Drews, G.N.; Bowman, J.L. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 1999, 126, 4117–4128. [Google Scholar]
- Watanabe, K.; Okada, K. Two discrete cis elements control the Abaxial side-specific expression of the FILAMENTOUS FLOWER gene in Arabidopsis. Plant Cell 2003, 15, 2592–2602. [Google Scholar] [CrossRef]
- Golz, J.F.; Roccaro, M.; Kuzoff, R.; Hudson, A. GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development 2004, 131, 3661–3670. [Google Scholar] [CrossRef]
- Sarojam, R.; Sappl, P.G.; Goldshmidt, A.; Efroni, I.; Floyd, S.K.; Eshed, Y.; Bowman, J.L. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 2010, 22, 2113–2130. [Google Scholar] [CrossRef]
- Sessions, A.; Nemhauser, J.L.; McColl, A.; Roe, J.L.; Feldmann, K.A.; Zambryski, P.C. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 1997, 124, 4481–4491. [Google Scholar] [PubMed]
- Shi, J.; Dong, J.; Xue, J.; Wang, H.; Yang, Z.; Jiao, Y.; Xu, L.; Huang, H. Model for the role of auxin polar transport in patterning of the leaf adaxial-abaxial axis. Plant J. 2017, 92, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Phelps-Durr, T.L.; Thomas, J.; Vahab, P.; Timmermans, M.C. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 2005, 17, 2886–2898. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Iwasaki, M.; Machida, C.; Machida, Y.; Zhou, X. Chua NH: BetaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 2008, 22, 2564–2577. [Google Scholar] [CrossRef]
- Ueno, Y.; Ishikawa, T.; Watanabe, K.; Terakura, S.; Iwakawa, H.; Okada, K.; Machida, C.; Machida, Y. Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 2007, 19, 445–457. [Google Scholar] [CrossRef]
- Luo, M.; Yu, C.W.; Chen, F.F.; Zhao, L.; Tian, G.; Liu, X.; Cui, Y.; Yang, J.Y.; Wu, K. Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in arabidopsis. PLoS Genet 2012, 8, e1003114. [Google Scholar] [CrossRef]
- Iwakawa, H.; Iwasaki, M.; Kojima, S.; Ueno, Y.; Soma, T.; Tanaka, H.; Semiarti, E.; Machida, Y.; Machida, C. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 2007, 51, 173–184. [Google Scholar] [CrossRef]
- Shuai, B.; Reynaga-Peña, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant. Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef]
- Matsumura, Y.; Iwakawa, H.; Machida, Y.; Machida, C. Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009, 58, 525–537. [Google Scholar] [CrossRef]
- Song, B.; Tang, Z.; Li, X.; Li, J.; Zhang, M.; Zhao, K.; Liu, H.; Zhang, S.; Wu, J. Mining and evolution analysis of lateral organ boundaries domain (LBD) genes in Chinese white pear (Pyrus bretschneideri). BMC Genom. 2020, 21, 644. [Google Scholar] [CrossRef]
- Guo, B.J.; Wang, J.; Lin, S.; Tian, Z.; Zhou, K.; Luan, H.Y.; Lyu, C.; Zhang, X.Z.; Xu, R.G. A genome-wide analysis of the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) gene family in barley (Hordeum vulgare L.). J. Zhejiang Univ. Sci. B 2016, 17, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Xu, H.; Lei, Q.; Du, J.; Li, C.; Wang, C.; Yang, Y.; Sun, X. The Arabidopsis transcription factor LBD15 mediates ABA signaling and tolerance of water-deficit stress by regulating ABI4 expression. Plant J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Z.; Ma, B.; Hou, Q.; Wan, X. Phylogeny and Functions of LOB Domain Proteins in Plants. Int. J. Mol. Sci. 2020, 21, 2278. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Xie, Q.; Li, C.; Dong, Y.; Zhu, S.; Chen, J. Comprehensive characterization and gene expression patterns of LBD gene family in Gossypium. Planta 2020, 251, 81. [Google Scholar] [CrossRef]
- Liu, H.; Cao, M.; Chen, X.; Ye, M.; Zhao, P.; Nan, Y.; Li, W.; Zhang, C.; Kong, L.; Kong, N.; et al. Genome-Wide Analysis of the Lateral Organ Boundaries Domain (LBD) Gene Family in. Int. J. Mol. Sci. 2019, 20, 5360. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, H.; Li, W.; Wu, J.; Zhou, Z.; Zhou, F.; Chen, H.; Lin, Y. Genome-wide association study of callus induction variation to explore the callus formation mechanism of rice. J. Integr. Plant. Biol. 2019, 61, 1134–1150. [Google Scholar] [CrossRef]
- Chen, W.F.; Wei, X.B.; Rety, S.; Huang, L.Y.; Liu, N.N.; Dou, S.X.; Xi, X.G. Structural analysis reveals a “molecular calipers” mechanism for a LATERAL ORGAN BOUNDARIES DOMAIN transcription factor protein from wheat. J. Biol. Chem. 2019, 294, 142–156. [Google Scholar] [CrossRef]
- Takahashi, H.; Iwakawa, H.; Nakao, S.; Ojio, T.; Morishita, R.; Morikawa, S.; Machida, Y.; Machida, C.; Kobayashi, T. Knowledge-based fuzzy adaptive resonance theory and its application to the analysis of gene expression in plants. J. Biosci. Bioeng. 2008, 106, 587–593. [Google Scholar] [CrossRef]
- Iwasaki, M.; Takahashi, H.; Iwakawa, H.; Nakagawa, A.; Ishikawa, T.; Tanaka, H.; Matsumura, Y.; Pekker, I.; Eshed, Y.; Vial-Pradel, S.; et al. Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development 2013, 140, 1958–1969. [Google Scholar] [CrossRef]
- Husbands, A.Y.; Benkovics, A.H.; Nogueira, F.T.; Lodha, M.; Timmermans, M.C. The ASYMMETRIC LEAVES Complex Employs Multiple Modes of Regulation to Affect Adaxial-Abaxial Patterning and Leaf Complexity. Plant Cell 2015, 27, 3321–3335. [Google Scholar] [CrossRef]
- Hunter, C.; Willmann, M.R.; Wu, G.; Yoshikawa, M.; de la Luz Gutiérrez-Nava, M.; Poethig, S.R. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 2006, 133, 2973–2981. [Google Scholar] [CrossRef] [PubMed]
- Luong, T.Q.; Keta, S.; Asai, T.; Kojima, S.; Nakagawa, A.; Micol, J.L.; Xia, S.; Machida, Y.; Machida, C. A genetic link between epigenetic repressor AS1-AS2 and DNA replication factors in establishment of adaxial-abaxial leaf polarity of Arabidopsis. Plant. Biotechnol. 2018, 35, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Pinon, V.; Etchells, J.P.; Rossignol, P.; Collier, S.A.; Arroyo, J.M.; Martienssen, R.A.; Byrne, M.E. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 2008, 135, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Ling, Q.; Wang, H.; Huang, H. Ribosomal proteins promote leaf adaxial identity. Development 2008, 135, 1325–1334. [Google Scholar] [CrossRef]
- Szakonyi, D.; Byrne, M.E. Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J. 2011, 65, 269–281. [Google Scholar] [CrossRef]
- Horiguchi, G.; Mollá-Morales, A.; Pérez-Pérez, J.M.; Kojima, K.; Robles, P.; Ponce, M.R.; Micol, J.L.; Tsukaya, H. Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J. 2011, 65, 724–736. [Google Scholar] [CrossRef]
- Casanova-Sáez, R.; Candela, H.; Micol, J.L. Combined haploinsufficiency and purifying selection drive retention of RPL36a paralogs in Arabidopsis. Sci. Rep. 2014, 4, 4122. [Google Scholar] [CrossRef]
- Kojima, K.; Tamura, J.; Chiba, H.; Fukada, K.; Tsukaya, H.; Horiguchi, G. Two Nucleolar Proteins, GDP1 and OLI2, Function as Ribosome Biogenesis Factors and Are Preferentially Involved in Promotion of Leaf Cell Proliferation without Strongly Affecting Leaf Adaxial-Abaxial Patterning in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 2240. [Google Scholar] [CrossRef]
- Matsumura, Y.; Ohbayashi, I.; Takahashi, H.; Kojima, S.; Ishibashi, N.; Keta, S.; Nakagawa, A.; Hayashi, R.; Saéz-Vásquez, J.; Echeverria, M.; et al. A genetic link between epigenetic repressor AS1-AS2 and a putative small subunit processome in leaf polarity establishment of Arabidopsis. Biol. Open 2016, 5, 942–954. [Google Scholar] [CrossRef]
- Ohbayashi, I.; Konishi, M.; Ebine, K.; Sugiyama, M. Genetic identification of Arabidopsis RID2 as an essential factor involved in pre-rRNA processing. Plant J. 2011, 67, 49–60. [Google Scholar] [CrossRef]
- Pontvianne, F.; Matía, I.; Douet, J.; Tourmente, S.; Medina, F.J.; Echeverria, M.; Sáez-Vásquez, J. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol. Biol. Cell 2007, 18, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Sáez-Vásquez, J.; Delseny, M. Ribosome Biogenesis in Plants: From Functional 45S Ribosomal DNA Organization to Ribosome Assembly Factors. Plant Cell 2019, 31, 1945–1967. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, N.; Kim, H.B.; Park, N.I.; Kim, H.S.; Kim, Y.K.; Park, Y.I.; Choi, S.B. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010, 64, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Iwakawa, H.; Ishibashi, N.; Kojima, S.; Matsumura, Y.; Prananingrum, P.; Iwasaki, M.; Takahashi, A.; Ikezaki, M.; Luo, L.; et al. Meta-analyses of microarrays of Arabidopsis asymmetric leaves1 (as1), as2 and their modifying mutants reveal a critical role for the ETT pathway in stabilization of adaxial-abaxial patterning and cell division during leaf development. Plant Cell Physiol. 2013, 54, 418–431. [Google Scholar] [CrossRef] [PubMed]
- Vial-Pradel, S.; Keta, S.; Nomoto, M.; Luo, L.; Takahashi, H.; Suzuki, M.; Yokoyama, Y.; Sasabe, M.; Kojima, S.; Tada, Y.; et al. Arabidopsis Zinc-Finger-Like Protein ASYMMETRIC LEAVES2 (AS2) and Two Nucleolar Proteins Maintain Gene Body DNA Methylation in the Leaf Polarity Gene ETTIN (ARF3). Plant Cell Physiol. 2018, 59, 1385–1397. [Google Scholar] [CrossRef]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef]
- Pontvianne, F.; Carpentier, M.C.; Durut, N.; Pavlištová, V.; Jaške, K.; Schořová, Š.; Parrinello, H.; Rohmer, M.; Pikaard, C.S.; Fojtová, M.; et al. Identification of Nucleolus-Associated Chromatin Domains Reveals a Role for the Nucleolus in 3D Organization of the A. thaliana Genome. Cell Rep. 2016, 16, 1574–1587. [Google Scholar] [CrossRef]
- Bersaglieri, C.; Santoro, R. Genome Organization in and around the Nucleolus. Cells 2019, 8, 579. [Google Scholar] [CrossRef]
- Ginisty, H.; Sicard, H.; Roger, B.; Bouvet, P. Structure and functions of nucleolin. J. Cell Sci. 1999, 112 Pt 6, 761–772. [Google Scholar]
- Kojima, H.; Suzuki, T.; Kato, T.; Enomoto, K.; Sato, S.; Tabata, S.; Sáez-Vasquez, J.; Echeverría, M.; Nakagawa, T.; Ishiguro, S.; et al. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 2007, 49, 1053–1063. [Google Scholar] [CrossRef]
- Petricka, J.J.; Nelson, T.M. Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 2007, 144, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Micol-Ponce, R.; Sarmiento-Mañús, R.; Ruiz-Bayón, A.; Montacié, C.; Sáez-Vasquez, J.; Ponce, M.R. Arabidopsis RIBOSOMAL RNA PROCESSING7 Is Required for 18S rRNA Maturation. Plant Cell 2018, 30, 2855–2872. [Google Scholar] [CrossRef] [PubMed]
- Durut, N.; Sáez-Vásquez, J. Nucleolin: Dual roles in rDNA chromatin transcription. Gene 2015, 556, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Montacié, C.; Durut, N.; Opsomer, A.; Palm, D.; Comella, P.; Picart, C.; Carpentier, M.C.; Pontvianne, F.; Carapito, C.; Schleiff, E.; et al. Nucleolar Proteome Analysis and Proteasomal Activity Assays Reveal a Link between Nucleolus and 26S Proteasome in. Front. Plant Sci. 2017, 8, 1815. [Google Scholar] [CrossRef]
- Picart, C.; Pontvianne, F. Plant nucleolar DNA: Green light shed on the role of Nucleolin in genome organization. Nucleus 2017, 8, 11–16. [Google Scholar] [CrossRef]
- Turner, A.J.; Knox, A.A.; Prieto, J.L.; McStay, B.; Watkins, N.J. A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol. Cell Biol. 2009, 29, 3007–3017. [Google Scholar] [CrossRef]
- Phipps, K.R.; Charette, J.; Baserga, S.J. The small subunit processome in ribosome biogenesis—Progress and prospects. Wiley Interdiscip Rev. RNA 2011, 2, 1–21. [Google Scholar] [CrossRef]
- Granneman, S.; Bernstein, K.A.; Bleichert, F.; Baserga, S.J. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis. Mol. Cell Biol. 2006, 26, 1183–1194. [Google Scholar] [CrossRef]
- Dragon, F.; Gallagher, J.E.; Compagnone-Post, P.A.; Mitchell, B.M.; Porwancher, K.A.; Wehner, K.A.; Wormsley, S.; Settlage, R.E.; Shabanowitz, J.; Osheim, Y.; et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 2002, 417, 967–970. [Google Scholar] [CrossRef]
- Feng, J.M.; Tian, H.F.; Wen, J.F. Origin and evolution of the eukaryotic SSU processome revealed by a comprehensive genomic analysis and implications for the origin of the nucleolus. Genome Biol. Evol. 2013, 5, 2255–2267. [Google Scholar] [CrossRef]
- You, K.T.; Park, J.; Kim, V.N. Role of the small subunit processome in the maintenance of pluripotent stem cells. Genes Dev. 2015, 29, 2004–2009. [Google Scholar] [CrossRef]
- Sardana, R.; White, J.P.; Johnson, A.W. The rRNA methyltransferase Bud23 shows functional interaction with components of the SSU processome and RNase MRP. RNA 2013, 19, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Sardana, R.; Zhu, J.; Gill, M.; Johnson, A.W. Physical and functional interaction between the methyltransferase Bud23 and the essential DEAH-box RNA helicase Ecm16. Mol. Cell Biol. 2014, 34, 2208–2220. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Liu, X.; Anjos, M.; Correll, C.C.; Johnson, A.W. Utp14 Recruits and Activates the RNA Helicase Dhr1 to Undock U3 snoRNA from the Preribosome. Mol. Cell Biol. 2016, 36, 965–978. [Google Scholar] [CrossRef] [PubMed]
- Ohbayashi, I.; Lin, C.Y.; Shinohara, N.; Matsumura, Y.; Machida, Y.; Horiguchi, G.; Tsukaya, H.; Sugiyama, M. Evidence for a Role of ANAC082 as a Ribosomal Stress Response Mediator Leading to Growth Defects and Developmental Alterations in Arabidopsis. Plant Cell 2017, 29, 2644–2660. [Google Scholar] [CrossRef]
- Ohbayashi, I.; Sugiyama, M. Plant Nucleolar Stress Response, a New Face in the NAC-Dependent Cellular Stress Responses. Front. Plant Sci. 2017, 8, 2247. [Google Scholar] [CrossRef]
- Huang, K.C.; Lin, W.C.; Cheng, W.H. Salt hypersensitive mutant 9, a nucleolar APUM23 protein, is essential for salt sensitivity in association with the ABA signaling pathway in Arabidopsis. BMC Plant Biol. 2018, 18, 40. [Google Scholar] [CrossRef]
- Bernstein, K.A.; Gallagher, J.E.; Mitchell, B.M.; Granneman, S.; Baserga, S.J. The small-subunit processome is a ribosome assembly intermediate. Eukaryot. Cell 2004, 3, 1619–1626. [Google Scholar] [CrossRef]
- McCann, K.L.; Charette, J.M.; Vincent, N.G.; Baserga, S.J. A protein interaction map of the LSU processome. Genes Dev. 2015, 29, 862–875. [Google Scholar] [CrossRef]
- Lawrence, R.J.; Earley, K.; Pontes, O.; Silva, M.; Chen, Z.J.; Neves, N.; Viegas, W.; Pikaard, C.S. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 2004, 13, 599–609. [Google Scholar] [CrossRef]
- Pontes, O.; Lawrence, R.J.; Silva, M.; Preuss, S.; Costa-Nunes, P.; Earley, K.; Neves, N.; Viegas, W.; Pikaard, C.S. Postembryonic establishment of megabase-scale gene silencing in nucleolar dominance. PLoS ONE 2007, 2, e1157. [Google Scholar] [CrossRef]
- Zhou, C.; Labbe, H.; Sridha, S.; Wang, L.; Tian, L.; Latoszek-Green, M.; Yang, Z.; Brown, D.; Miki, B.; Wu, K. Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J. 2004, 38, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Ando, S.; Sasabe, M.; Machida, C.; Kurihara, D.; Higashiyama, T.; Machida, Y. Arabidopsis ASYMMETRIC LEAVES2 protein required for leaf morphogenesis consistently forms speckles during mitosis of tobacco BY-2 cells via signals in its specific sequence. J. Plant Res. 2012, 125, 661–668. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luo, L.; Ando, S.; Sakamoto, Y.; Suzuki, T.; Takahashi, H.; Ishibashi, N.; Kojima, S.; Kurihara, D.; Higashiyama, T.; Yamamoto, K.T.; et al. The formation of perinucleolar bodies is important for normal leaf development and requires the zinc-finger DNA-binding motif in Arabidopsis ASYMMETRIC LEAVES2. Plant J. 2020, 101, 1118–1134. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Wu, M. Identification of a novel nucleolar localization signal and a degradation signal in Survivin-deltaEx3: A potential link between nucleolus and protein degradation. Oncogene 2005, 24, 2723–2734. [Google Scholar] [CrossRef] [PubMed]
- Musinova, Y.R.; Lisitsyna, O.M.; Golyshev, S.A.; Tuzhikov, A.I.; Polyakov, V.Y.; Sheval, E.V. Nucleolar localization/retention signal is responsible for transient accumulation of histone H2B in the nucleolus through electrostatic interactions. Biochim. Biophys. Acta 2011, 1813, 27–38. [Google Scholar] [CrossRef] [PubMed]
- de Melo, I.S.; Jimenez-Nuñez, M.D.; Iglesias, C.; Campos-Caro, A.; Moreno-Sanchez, D.; Ruiz, F.A.; Bolívar, J. NOA36 protein contains a highly conserved nucleolar localization signal capable of directing functional proteins to the nucleolus, in mammalian cells. PLoS ONE 2013, 8, e59065. [Google Scholar] [CrossRef]
- Earley, L.F.; Kawano, Y.; Adachi, K.; Sun, X.X.; Dai, M.S.; Nakai, H. Identification and characterization of nuclear and nucleolar localization signals in the adeno-associated virus serotype 2 assembly-activating protein. J. Virol. 2015, 89, 3038–3048. [Google Scholar] [CrossRef]
- Ye, J.; Yang, J.; Sun, Y.; Zhao, P.; Gao, S.; Jung, C.; Qu, J.; Fang, R.; Chua, N.H. Geminivirus Activates ASYMMETRIC LEAVES 2 to Accelerate Cytoplasmic DCP2-Mediated mRNA Turnover and Weakens RNA Silencing in Arabidopsis. PLoS Pathog. 2015, 11, e1005196. [Google Scholar] [CrossRef]
- Vial-Pradel, S.; Hasegawa, Y.; Nakagawa, A.; Miyaki, S.; Machida, Y.; Kojima, S.; Machida, C.; Takahashi, H. SIMON: Simple methods for analyzing DNA methylation by targeted bisulfite next-generation sequencing. Plant. Biotechnol. 2019, 36, 213–222. [Google Scholar] [CrossRef]
- Husbands, A.; Bell, E.M.; Shuai, B.; Smith, H.M.; Springer, P.S. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007, 35, 6663–6671. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Iwamoto, K.; Fukuda, H. LOB DOMAIN-CONTAINING PROTEIN 15 Positively Regulates Expression of VND7, a Master Regulator of Tracheary Elements. Plant Cell Physiol. 2018, 59, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T. Gene Body Methylation Involved in Leaf Development. Plant Cell Physiol. 2018, 59, 1288–1289. [Google Scholar] [CrossRef] [PubMed]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Song, J.; Rechkoblit, O.; Bestor, T.H.; Patel, D.J. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 2011, 331, 1036–1040. [Google Scholar] [CrossRef]
- Long, H.K.; Blackledge, N.P.; Klose, R.J. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem. Soc. Trans. 2013, 41, 727–740. [Google Scholar] [CrossRef]
- Nishiyama, A.; Yamaguchi, L.; Sharif, J.; Johmura, Y.; Kawamura, T.; Nakanishi, K.; Shimamura, S.; Arita, K.; Kodama, T.; Ishikawa, F.; et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 2013, 502, 249–253. [Google Scholar] [CrossRef]
- Song, J.; Du, Z.; Ravasz, M.; Dong, B.; Wang, Z.; Ewing, R.M. A Protein Interaction between β-Catenin and Dnmt1 Regulates Wnt Signaling and DNA Methylation in Colorectal Cancer Cells. Mol. Cancer Res. 2015, 13, 969–981. [Google Scholar] [CrossRef]
- Zhang, Z.M.; Liu, S.; Lin, K.; Luo, Y.; Perry, J.J.; Wang, Y.; Song, J. Crystal Structure of Human DNA Methyltransferase 1. J. Mol. Biol. 2015, 427, 2520–2531. [Google Scholar] [CrossRef]
- Du, J. Structure and Mechanism of Plant DNA Methyltransferases. Adv. Exp. Med. Biol. 2016, 945, 173–192. [Google Scholar] [CrossRef]
- Ferry, L.; Fournier, A.; Tsusaka, T.; Adelmant, G.; Shimazu, T.; Matano, S.; Kirsh, O.; Amouroux, R.; Dohmae, N.; Suzuki, T.; et al. Methylation of DNA Ligase 1 by G9a/GLP Recruits UHRF1 to Replicating DNA and Regulates DNA Methylation. Mol. Cell 2017, 67, 550–565.e555. [Google Scholar] [CrossRef] [PubMed]
- Ryazanova, A.Y.; Abrosimova, L.A.; Oretskaya, T.S.; Kubareva, E.A. Diverse Domains of (Cytosine-5)-DNA Methyltransferases: Structural and Functional Characterization, Methylation-From DNA, RNA and Histones to Diseases and Treatment; Dricu, A., Ed.; InTech: Rijeka, Croatia, 2012; pp. 29–69. [Google Scholar]
- Pontvianne, F.; Abou-Ellail, M.; Douet, J.; Comella, P.; Matia, I.; Chandrasekhara, C.; Debures, A.; Blevins, T.; Cooke, R.; Medina, F.J.; et al. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet 2010, 6, e1001225. [Google Scholar] [CrossRef] [PubMed]
- Pontvianne, F.; Blevins, T.; Chandrasekhara, C.; Mozgová, I.; Hassel, C.; Pontes, O.M.; Tucker, S.; Mokros, P.; Muchová, V.; Fajkus, J.; et al. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev. 2013, 27, 1545–1550. [Google Scholar] [CrossRef] [PubMed]
- To, T.K.; Kim, J.M.; Matsui, A.; Kurihara, Y.; Morosawa, T.; Ishida, J.; Tanaka, M.; Endo, T.; Kakutani, T.; Toyoda, T.; et al. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet 2011, 7, e1002055. [Google Scholar] [CrossRef]
- Liu, X.; Yu, C.W.; Duan, J.; Luo, M.; Wang, K.; Tian, G.; Cui, Y.; Wu, K. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 2012, 158, 119–129. [Google Scholar] [CrossRef]
- Lodha, M.; Marco, C.F.; Timmermans, M.C. The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev. 2013, 27, 596–601. [Google Scholar] [CrossRef]
- Li, Z.; Li, B.; Liu, J.; Guo, Z.; Liu, Y.; Li, Y.; Shen, W.H.; Huang, Y.; Huang, H.; Zhang, Y.; et al. Transcription factors AS1 and AS2 interact with LHP1 to repress KNOX genes in Arabidopsis. J. Integr. Plant Biol. 2016, 58, 959–970. [Google Scholar] [CrossRef]
- Lin, X.; Gu, D.; Zhao, H.; Peng, Y.; Zhang, G.; Yuan, T.; Li, M.; Wang, Z.; Wang, X.; Cui, S. LFR is functionally associated with AS2 to mediate leaf development in Arabidopsis. Plant J. 2018, 95, 598–612. [Google Scholar] [CrossRef]
- Xu, C.; Luo, F.; Hochholdinger, F. LOB Domain Proteins: Beyond Lateral Organ Boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, J. Identification of nuclear localization signal in ASYMMETRIC LEAVES2-LIKE18/LATERAL ORGAN BOUNDARIES DOMAIN16 (ASL18/LBD16) from Arabidopsis. J. Plant Physiol. 2012, 169, 1221–1226. [Google Scholar] [CrossRef]
- Correll, C.C.; Bartek, J.; Dundr, M. The Nucleolus: A Multiphase Condensate Balancing Ribosome Synthesis and Translational Capacity in Health, Aging and Ribosomopathies. Cells 2019, 8, 869. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Kumazawa, T.; Kuroda, T.; Katagiri, N.; Tsuchiya, M.; Goto, N.; Furumai, R.; Murayama, A.; Yanagisawa, J.; Kimura, K. Perturbation of ribosome biogenesis drives cells into senescence through 5S RNP-mediated p53 activation. Cell Rep. 2015, 10, 1310–1323. [Google Scholar] [CrossRef] [PubMed]
- Padeken, J.; Heun, P. Nucleolus and nuclear periphery: Velcro for heterochromatin. Curr. Opin. Cell Biol. 2014, 28, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, M.C.; Picart-Picolo, A.; Pontvianne, F. A Method to Identify Nucleolus-Associated Chromatin Domains (NADs). Methods Mol. Biol. 2018, 1675, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekhara, C.; Mohannath, G.; Blevins, T.; Pontvianne, F.; Pikaard, C.S. Chromosome-specific NOR inactivation explains selective rRNA gene silencing and dosage control in Arabidopsis. Genes Dev. 2016, 30, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Mohannath, G.; Pontvianne, F.; Pikaard, C.S. Selective nucleolus organizer inactivation in Arabidopsis is a chromosome position-effect phenomenon. Proc. Natl. Acad. Sci. USA 2016, 113, 13426–13431. [Google Scholar] [CrossRef] [PubMed]
- Pavlištová, V.; Dvořáčková, M.; Jež, M.; Mozgová, I.; Mokroš, P.; Fajkus, J. Phenotypic reversion in fas mutants of Arabidopsis thaliana by reintroduction of FAS genes: Variable recovery of telomeres with major spatial rearrangements and transcriptional reprogramming of 45S rDNA genes. Plant J. 2016, 88, 411–424. [Google Scholar] [CrossRef]
- O’Malley, R.C.; Huang, S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef]




| 1. Gene Name (Mutant Name) | 2. AGI Code | 3. Protein | 4. Cellular Process and Status | 5.References |
|---|---|---|---|---|
| I. Genes involved in rRNA processing | ||||
| NUCLEOLIN1 (nuc1) | AT1G48920 | NUCLEOLIN | rRNA processing and ribosome biogenesis Components of SSUP-like complex | [59,61,70,71] |
| RNA HELICASE10 (rh10) | AT5G60990 | DEAD-box RNA helicase family protein | pre-rRNA processing Components of SSUP-like complex | [59] |
| ROOT INITIATION DEFECTIVE2 (rid2) | AT5G57280 | RNA methyltransferase-like protein | pre-rRNA processing | [59,60] |
| APUM23 (apum23) | AT1G72320 | Pumillio protein containing PUF domain | pre-rRNA processing and rRNA maturation | [63] |
| II. Genes for ribosomal proteins | ||||
| RPL4D (rpl4d) | AT5G02870 | Ribosomal proteins | Subunits of ribosome; components of pre-rRNA-protein complex | [53,54,55,56,57] |
| RPL5A (pgy3/ae6/oli5/rpl5a) | AT3G25520 | |||
| RPL5B (rpl5b/oli7) | AT5G39740 | |||
| RPL7B (rpl7b) | AT2G01250 | |||
| RPL9c (rpl9c/pgy2) | AT1G33140 | |||
| RPL10aB (rpl10ab/pgy1) | AT2G27530 | |||
| RPL18C (rpl18c) | AT5G27850 | |||
| RPL24b (stv1) | AT3G53020 | |||
| RPL27ac (rpl27ac) | AT1G70600 | |||
| RPL28A (ae5/rpl28a) | AT2G19730 | |||
| PRL36aB (api2) | AT4G14320 | |||
| RPL36aA (rpl36aa) | AT3G59540 | |||
| RPL38B (rpl38b) | AT4G31985 | |||
| RPL39C (rpl39c) | AT3G23390 | |||
| RPS6A * (rps6a) | AT4G31700 | |||
| RPS21B (rps21b) | AT3G53890 | |||
| RPS24B (rps24b) | AT5G28060 | |||
| RPS28B (rps28b) | AT5G03850 | |||
| III. Genes involved in histone modification | ||||
| HDT1 (hdt1/hd2a/hda3) | AT3G44750 | Histone deacetylase (plant-specific class) | Deacetylation of nucleosomal histone H3, transcription of rDNAs | [35,90,92] |
| HDT2 (hdt2/hd2b) | AT5G22650 | Histone deacetylase (plant-specific class) | Deacetylation of nucleosomal histone H3, transcription of rDNAs | [35,90,92] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwakawa, H.; Takahashi, H.; Machida, Y.; Machida, C. Roles of ASYMMETRIC LEAVES2 (AS2) and Nucleolar Proteins in the Adaxial–Abaxial Polarity Specification at the Perinucleolar Region in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 7314. https://doi.org/10.3390/ijms21197314
Iwakawa H, Takahashi H, Machida Y, Machida C. Roles of ASYMMETRIC LEAVES2 (AS2) and Nucleolar Proteins in the Adaxial–Abaxial Polarity Specification at the Perinucleolar Region in Arabidopsis. International Journal of Molecular Sciences. 2020; 21(19):7314. https://doi.org/10.3390/ijms21197314
Chicago/Turabian StyleIwakawa, Hidekazu, Hiro Takahashi, Yasunori Machida, and Chiyoko Machida. 2020. "Roles of ASYMMETRIC LEAVES2 (AS2) and Nucleolar Proteins in the Adaxial–Abaxial Polarity Specification at the Perinucleolar Region in Arabidopsis" International Journal of Molecular Sciences 21, no. 19: 7314. https://doi.org/10.3390/ijms21197314
APA StyleIwakawa, H., Takahashi, H., Machida, Y., & Machida, C. (2020). Roles of ASYMMETRIC LEAVES2 (AS2) and Nucleolar Proteins in the Adaxial–Abaxial Polarity Specification at the Perinucleolar Region in Arabidopsis. International Journal of Molecular Sciences, 21(19), 7314. https://doi.org/10.3390/ijms21197314





