Targeting DNA Repair Pathways in Hematological Malignancies
Abstract
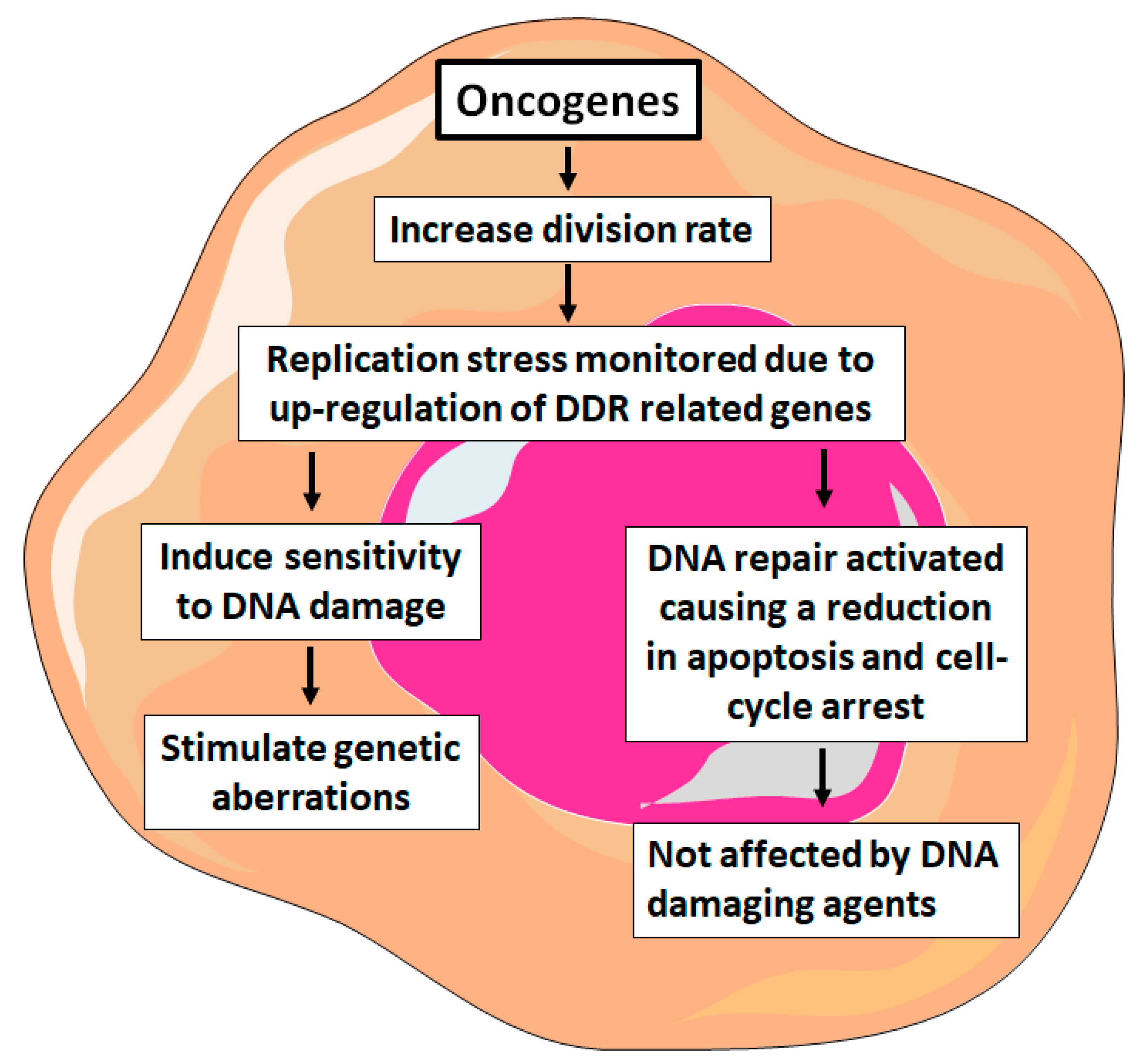
1. Introduction
2. Source of DNA Damage
3. The DNA Damage Response (DDR)
4. DNA Damage Regulators and Cell Cycle Checkpoints
5. DNA Repair
6. Defects of Ataxia-Telangiectasia in Hematological Malignancies
7. Gene Mutations of DNA Damage Response in Hematological Malignancies
8. Treatment of Hematological Malignancies and its Effect on DNA Damage and Repair
9. ATM-Deficient Cancer Therapies
9.1. Poly ADP ribose Polymerase (PARP) Inhibitors
9.2. Targeting ATR
9.3. CHK1 Inhibitors
9.4. Nucleoside Analogues
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AML: | Acute myeloid leukemia |
| ATM: | Ataxia telangiectasia mutated |
| BER: | Base excision repair |
| CIN: | Chromosomal instability |
| CLL: | Chronic lymphocytic leukemia |
| DDR: | DNA damage-response |
| DSB: | Double-strand breaks |
| GOF: | Gain-of-function |
| HDAC: | Histone deacetylase |
| HR: | Homologous recombination |
| MDS: | Myelodysplastic syndrome |
| MIN: | Microsatellite instability |
| MMR: | Mismatch repair |
| MMS: | Methyl-methane-sulfonate |
| NBS: | Nijmegen breakage syndrome |
| NCS: | Neocarzinostatin |
| NER: | Nucleotide excision repair |
| NHEJ: | Non-homologous end joining |
| ROS: | Reactive oxygen species |
| SDDC: | S phase DNA damage checkpoint |
| SSB: | Single-strand breaks |
References
- Chakarov, S.; Petkova, R.; Russev, G.C.; Zhelev, N. DNA damage and mutation. Types of DNA damage. BioDiscovery 2014, 11, 1. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Karaman, M.W.; Hacia, J.G. Genomes, 2nd edition. J. Hered. 2003, 94, 432–433. [Google Scholar] [CrossRef]
- Smith, A.G.; Howell, D.A.; Patmore, R.; Jack, A.; Roman, E. Incidence of haematological malignancy by sub-type: A report from the Haematological Malignancy Research Network. Br. J. Cancer 2011, 105, 1684–1692. [Google Scholar] [CrossRef]
- Thierry, S.; Jdey, W.; Alculumbre, S.; Soumelis, V.; Noguiez-Hellin, P.; Dutreix, M. The DNA Repair Inhibitor Dbait Is Specific for Malignant Hematologic Cells in Blood. Mol. Cancer Ther. 2017, 16, 2817–2827. [Google Scholar] [CrossRef]
- Davar, D.; Beumer, J.H.; Hamieh, L.; Tawbi, H. Role of PARP inhibitors in cancer biology and therapy. Curr. Med. Chem. 2012, 19, 3907–3921. [Google Scholar] [CrossRef]
- Vinayak, S.; Ford, J.M. PARP Inhibitors for the Treatment and Prevention of Breast Cancer. Curr. Breast Cancer Rep. 2010, 2, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Xiao, W.; Abdel-Wahab, O. Diagnosis and classification of hematologic malignancies on the basis of genetics. Blood 2017, 130, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Puiggros, A.; Blanco, G.; Espinet, B. Genetic Abnormalities in Chronic Lymphocytic Leukemia: Where We Are and Where We Go. BioMed Res. Int. 2014, 2014, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Haase, D.; Germing, U.; Schanz, J.; Pfeilstöcker, M.; Nösslinger, T.; Hildebrandt, B.; Kundgen, A.; Lübbert, M.; Kunzmann, R.; Giagounidis, A.A.N.; et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: Evidence from a core dataset of 2124 patients. Blood 2007, 110, 4385–4395. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, P.J.; Caldecott, K.W. DNA Strand Break Repair and Human Genetic Disease. Annu. Rev. Genom. Hum. Genet. 2007, 8, 37–55. [Google Scholar] [CrossRef] [PubMed]
- Weston, V.J.; Oldreive, C.E.; Skowronska, A.; Oscier, D.G.; Pratt, G.; Dyer, M.J.S.; Smith, G.; Powell, J.E.; Rudzki, Z.; Kearns, P.; et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood 2010, 116, 4578–4587. [Google Scholar] [CrossRef] [PubMed]
- Horton, T.M.; Jenkins, G.; Pati, D.; Zhang, L.; Dolan, M.E.; Ribes-Zamora, A.; Bertuch, A.A.; Blaney, S.M.; Delaney, S.L.; Hegde, M.; et al. Poly(ADP-ribose) polymerase inhibitor ABT-888 potentiates the cytotoxic activity of temozolomide in leukemia cells: Influence of mismatch repair status and O6-methylguanine-DNA methyltransferase activity. Mol. Cancer Ther. 2009, 8, 2232–2242. [Google Scholar] [CrossRef]
- Friedberg, E.C.; McDaniel, L.D.; Schultz, R.A. The role of endogenous and exogenous DNA damage and mutagenesis. Curr. Opin. Genet. Dev. 2004, 14, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, U.S.; Tan, B.W.; Vellayappan, B.; Jeyasekharan, A. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef]
- Cannan, W.J.; Pederson, D.S. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J. Cell. Physiol. 2016, 231, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nat. Cell Biol. 2009, 461, 1071–1078. [Google Scholar] [CrossRef]
- Basu, A.K. DNA Damage, Mutagenesis and Cancer. Int. J. Mol. Sci. 2018, 19, 970. [Google Scholar] [CrossRef]
- Vignard, J.; Mirey, G.; Salles, B. Ionizing-radiation induced DNA double-strand breaks: A direct and indirect lighting up. Radiother. Oncol. 2013, 108, 362–369. [Google Scholar] [CrossRef]
- Abrahamson, S. Adverse Reproductive Outcomes in Families of Atomic Veterans: The Feasibility of Epidemiologic Studies. Radiat. Res. 1995, 144, 248. [Google Scholar] [CrossRef]
- Principles of Ionizing Radiation. Industrial Hygiene Engineering; Elsevier: Amsterdam, The Netherlands, 1998; pp. 621–647. [Google Scholar]
- Lomax, M.E.; Folkes, L.K.; O’Neill, P. Biological Consequences of Radiation-induced DNA Damage: Relevance to Radiotherapy. Clin. Oncol. 2013, 25, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Baskar, R.; Dai, J.; Wenlong, N.; Yeo, R.; Yeoh, K.-W. Biological response of cancer cells to radiation treatment. Front. Mol. Biosci. 2014, 1, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Terradas, M.; Martin, M.; Tusell, L.; Genescà, A. Genetic activities in micronuclei: Is the DNA entrapped in micronuclei lost for the cell? Mutat. Res. Mutat. Res. 2010, 705, 60–67. [Google Scholar] [CrossRef]
- Caldecott, K.W.; Abrahams, B.S.; Geschwind, D.H. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008, 9, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J.H.J. DNA Damage, Aging, and Cancer. New Engl. J. Med. 2009, 361, 1475–1485. [Google Scholar] [CrossRef]
- Pucci, B.; Kasten, M.; Giordano, A. Cell Cycle and Apoptosis. Neoplasia 2000, 2, 291–299. [Google Scholar] [CrossRef]
- Cooper, G.M.; Hausman, R.E. The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2004. [Google Scholar]
- Weber, A.M.; Ryan, A.J. ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 2015, 149, 124–138. [Google Scholar] [CrossRef]
- Shiloh, Y. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer 2003, 3, 155–168. [Google Scholar] [CrossRef]
- Huen, M.S.; Chen, J. The DNA damage response pathways: At the crossroad of protein modifications. Cell Res. 2007, 18, 8–16. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R.; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef]
- Campisi, J.; Di Fagagna, F.D. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007, 8, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Bartkova, J.; Horejsi, Z.; Koed, K.; Krämer, A.; Tort, F.; Zieger, K.; Guldberg, P.; Sehested, M.; Nesland, J.M.; Lukas, C.; et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nat. Cell Biol. 2005, 434, 864–870. [Google Scholar] [CrossRef]
- Gorgoulis, V.G.; Vassiliou, L.-V.F.; Karakaidos, P.; Zacharatos, P.; Kotsinas, A.; Liloglou, T.; Venere, M.; DiTullio, R.A., Jr.; Kastrinakis, N.G.; Levy, B.; et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nat. Cell Biol. 2005, 434, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Riley, T.; Sontag, E.; Chen, P.A.; Levine, A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 2008, 9, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Misteli, T.; Soutoglou, E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 2009, 10, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Ziv, Y.; Bielopolski, D.; Galanty, Y.; Lukas, C.; Taya, Y.; Schultz, D.C.; Lukas, J.; Bekker-Jensen, S.; Blow, J.J.; Shiloh, Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006, 8, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Cook, P.J.; Ju, B.G.; Telese, F.; Wang, X.; Glass, C.K.; Rosenfeld, M.G. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nat. Cell Biol. 2009, 458, 591–596. [Google Scholar] [CrossRef]
- Vilenchik, M.M.; Knudson, A.G. Endogenous DNA double-strand breaks: Production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. USA 2003, 100, 12871–12876. [Google Scholar] [CrossRef]
- Shammas, M.A. Telomeres, lifestyle, cancer, and aging. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 28–34. [Google Scholar] [CrossRef]
- Castedo, M.; Vitale, I.; Kroemer, G. A novel source of tetraploid cancer cell precursors: Telomere insufficiency links aging to oncogenesis. Oncogene 2010, 29, 5869–5872. [Google Scholar] [CrossRef]
- Davoli, T.; De Lange, T. Telomere-Driven Tetraploidization Occurs in Human Cells Undergoing Crisis and Promotes Transformation of Mouse Cells. Cancer Cell 2012, 21, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Olaharski, A.J.; Sotelo, R.; Solorza-Luna, G.; Gonsebatt, M.E.; Guzman, P.; Mohar, A.; Eastmond, D.A. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinog. 2006, 27, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, J.; Li, G.; Tan, Y.; Chen, X.; Ren, F.; Guo, H.; Wang, H. A tetraploid minimally differentiated acute myeloblastic leukemia with extensive erythrophagocytosis: A case report and literature review. Int. J. Hematol. 2012, 96, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Kjeldsen, E. Telomere Shortening in Hematological Malignancies with Tetraploidization—A Mechanism for Chromosomal Instability? Cancers 2017, 9, 165. [Google Scholar] [CrossRef]
- Ganem, N.J.; Pellman, D. Limiting the Proliferation of Polyploid Cells. Cell 2007, 131, 437–440. [Google Scholar] [CrossRef]
- Maser, R.S.; Depinho, R.A. Connecting Chromosomes, Crisis, and Cancer. Science 2002, 297, 565–569. [Google Scholar] [CrossRef]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An Oncogene-Induced DNA Damage Model for Cancer Development. Science 2008, 319, 1352–1355. [Google Scholar] [CrossRef]
- Bristow, R.G.; Hill, R.P. Hypoxia, DNA repair and genetic instability. Nat. Rev. Cancer 2008, 8, 180–192. [Google Scholar] [CrossRef]
- Current topics in developmental biology. Mol. Reprod. Dev. 1998, 51, 477. [CrossRef]
- Bertoli, C.; Skotheim, J.M.; De Bruin, R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef]
- Willis, N.; Rhind, N. Regulation of DNA replication by the S-phase DNA damage checkpoint. Cell Div. 2009, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Löbrich, M.; Jeggo, P.A. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer 2007, 7, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Dasika, G.K.; Lin, S.-C.J.; Zhao, S.; Sung, P.; Tomkinson, A.; Lee, E.Y.-H.P. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene 1999, 18, 7883–7899. [Google Scholar] [CrossRef] [PubMed]
- Dulic, V.; Kaufmann, W.K.; Wilson, S.J.; Tisty, T.D.; Lees, E.; Harper, J.W.; Elledge, S.J.; Reed, S.I. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell 1994, 76, 1013–1023. [Google Scholar] [CrossRef]
- Kitagawa, K.; Kotake, Y.; Kitagawa, M. Ubiquitin-mediated control of oncogene and tumor suppressor gene products. Cancer Sci. 2009, 100, 1374–1381. [Google Scholar] [CrossRef]
- Hartwell, L.; Kastan, M. Cell cycle control and cancer. Science 1994, 266, 1821–1828. [Google Scholar] [CrossRef] [PubMed]
- Cottini, F.; Hideshima, T.; Suzuki, R.; Tai, Y.-T.; Bianchini, G.; Richardson, P.G.; Anderson, K.C.; Tonon, G. Synthetic Lethal Approaches Exploiting DNA Damage in Aggressive Myeloma. Cancer Discov. 2015, 5, 972–987. [Google Scholar] [CrossRef]
- Pantazopoulos, A.; Pappa, V.; Papageorgiou, S.; Dervenoulas, J.; Economopoulos, T. Abnormalities of DNA repair mechanisms in common hematological malignancies. Leuk. Lymphoma 2011, 52, 567–582. [Google Scholar] [CrossRef]
- Kim, K.H.; Roberts, C.W. Targeting EZH2 in cancer. Nat. Med. 2016, 22, 128–134. [Google Scholar] [CrossRef]
- Ezponda, T.; Licht, J.D. Molecular pathways: Deregulation of histone h3 lysine 27 methylation in cancer-different paths, same destination. Clin. Cancer Res. 2014, 20, 5001–5008. [Google Scholar] [CrossRef]
- Herviou, L.; Cavalli, G.; Cartron, G.; Klein, B.; Moreaux, J. EZH2 in normal hematopoiesis and hematological malignancies. Oncotarget 2015, 7, 2284–2296. [Google Scholar] [CrossRef] [PubMed]
- Popovic, R.; Shah, M.Y.; Licht, J.D. Epigenetic therapy of hematological malignancies: Where are we now? Ther. Adv. Hematol. 2013, 4, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Benyoucef, A.; Palii, C.G.; Wang, C.; Porter, C.J.; Chu, A.; Dai, F.; Tremblay, V.; Rakopoulos, P.; Singh, K.; Huang, S.; et al. UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev. 2016, 30, 508–521. [Google Scholar] [CrossRef]
- Campbell, S.; Ismail, I.H.; Young, L.C.; Poirier, G.G.; Hendzel, M.J. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell Cycle 2013, 12, 2675–2683. [Google Scholar] [CrossRef]
- Johnson, D.P.; Spitz, G.S.; Tharkar, S.; Quayle, S.N.; Shearstone, J.R.; Jones, S.; McDowell, M.E.; Wellman, H.; Tyler, J.K.; Cairns, B.R.; et al. HDAC1,2 inhibition impairs EZH2- and BBAP- mediated DNA repair to overcome chemoresistance in EZH2 gain-of-function mutant diffuse large B-cell lymphoma. Oncotarget 2015, 6, 4863–4887. [Google Scholar] [CrossRef] [PubMed]
- Alhmoud, J.F.; Woolley, J.F.; Al Moustafa, A.-E.; Malki, M.I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. [Google Scholar] [CrossRef]
- Li, G.-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2007, 18, 85–98. [Google Scholar] [CrossRef]
- Bader, A.S.; Hawley, B.R.; Wilczynska, A.; Bushell, M. The roles of RNA in DNA double-strand break repair. Br. J. Cancer 2020, 122, 613–623. [Google Scholar] [CrossRef]
- Altmann, T.; Gennery, A.R. DNA ligase IV syndrome; a review. Orphanet J. Rare Dis. 2016, 11, 137. [Google Scholar] [CrossRef]
- Prakash, R.; Zhang, Y.; Feng, W.; Jasin, M. Homologous Recombination and Human Health: The Roles of BRCA1, BRCA2, and Associated Proteins. Cold Spring Harb. Perspect. Biol. 2015, 7, a016600. [Google Scholar] [CrossRef]
- Johnson, D.P.; Spitz-Becker, G.S.; Chakraborti, K.; Bhaskara, S. Assessment of epigenetic mechanisms and DNA double-strand break repair using laser micro-irradiation technique developed for hematological cells. EBioMedicine 2019, 43, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Rothblum-Oviatt, C.; Wright, J.; Lefton-Greif, M.A.; McGrath-Morrow, S.A.; Crawford, T.O.; Lederman, H.M. Ataxia telangiectasia: A review. Orphanet J. Rare Dis. 2016, 11, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Suarez, F.; Mahlaoui, N.; Canioni, D.; Andriamanga, C.; D’Enghien, C.D.; Brousse, N.; Jais, J.-P.; Fischer, A.; Hermine, O.; Stoppa-Lyonnet, D. Incidence, Presentation, and Prognosis of Malignancies in Ataxia-Telangiectasia: A Report From the French National Registry of Primary Immune Deficiencies. J. Clin. Oncol. 2015, 33, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Reiman, A.; Srinivasan, V.; Barone, G.; Last, J.I.; Wootton, L.L.; Davies, E.G.; Verhagen, M.M.; Willemsen, M.A.; Weemaes, C.M.; Byrd, P.J.; et al. Lymphoid tumours and breast cancer in ataxia telangiectasia; substantial protective effect of residual ATM kinase activity against childhood tumours. Br. J. Cancer 2011, 105, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Yuille, M.A.R.; Coignet, L.J.A.; Abraham, S.M.; Yaqub, F.; Luo, L.; Matutes, E.; Brito-Babapulle, V.; Vorechovsky, I.; Dyer, M.J.S.; Catovsky, D. ATM is usually rearranged in T-cell prolymphocytic leukaemia. Oncogene 1998, 16, 789–796. [Google Scholar] [CrossRef]
- Camacho, E.; Hernández, L.; Hernández, S.; Tort, F.; Bellosillo, B.; Beà, S.; Bosch, F.; Montserrat, E.; Cardesa, A.; Fernández, P.L.; et al. ATM gene inactivation in mantle cell lymphoma mainly occurs by truncating mutations and missense mutations involving the phosphatidylinositol-3 kinase domain and is associated with increasing numbers of chromosomal imbalances. Blood 2002, 99, 238–244. [Google Scholar] [CrossRef]
- Lähdesmäki, A.; Kimby, E.; Duke, V.; Foroni, L.; Hammarström, L. ATM mutations in B-cell chronic lymphocytic leukemia. Haematology 2004, 89, 24–31. [Google Scholar] [CrossRef]
- Stankovic, T.; Stewart, G.S.; Fegan, C.; Biggs, P.; Last, J.; Byrd, P.J.; Keenan, R.D.; Moss, P.A.H.; Taylor, A.M.R. Ataxia telangiectasia mutated–deficient B-cell chronic lymphocytic leukemia occurs in pregerminal center cells and results in defective damage response and unrepaired chromosome damage. Blood 2002, 99, 300–309. [Google Scholar] [CrossRef]
- Wan, Y.; Wu, C.J. SF3B1 mutations in chronic lymphocytic leukemia. Blood 2013, 121, 4627–4634. [Google Scholar] [CrossRef]
- Bose, S.; Starczynski, J.; Chukwuma, M.; Baumforth, K.; Wei, W.; Morgan, S.; Byrd, P.; Ying, J.; Grundy, R.; Mann, J.; et al. Down-regulation of ATM protein in HRS cells of nodular sclerosis Hodgkin’s lymphoma in children occurs in the absence ofATMgene inactivation. J. Pathol. 2007, 213, 329–336. [Google Scholar] [CrossRef]
- Bartkova, J.; Hamerlik, P.; Stockhausen, M.-T.; Ehrmann, J.; Hlobilkova, A.; Laursen, H.; Kalita, O.; Kolar, Z.; Poulsen, H.S.; Broholm, H.; et al. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene 2010, 29, 5095–5102. [Google Scholar] [CrossRef] [PubMed]
- Di Rora’, A.G.L.; Iacobucci, I.; Martinelli, G. The cell cycle checkpoint inhibitors in the treatment of leukemias. J. Hematol. Oncol. 2017, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.J.; Baltimore, D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003, 17, 615–628. [Google Scholar] [CrossRef] [PubMed]
- De Klein, A.; Muijtjens, M.; Van Os, R.; Verhoeven, Y.; Smit, B.; Carr, A.M.; Lehmann, A.; Hoeijmakers, J. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr. Biol. 2000, 10, 479–482. [Google Scholar] [CrossRef]
- Sawyers, C.L. Molecular Consequences of the BCR-ABL Translocation in Chronic Myelogenous Leukemia. Leuk. Lymphoma 1993, 11, 101–103. [Google Scholar] [CrossRef]
- Kim, K.-T.; Baird, K.; Davis, S.; Piloto, O.; Levis, M.; Li, L.; Chen, P.; Meltzer, P.; Small, D. Constitutive Fms-like tyrosine kinase 3 activation results in specific changes in gene expression in myeloid leukaemic cells. Br. J. Haematol. 2007, 138, 603–615. [Google Scholar] [CrossRef]
- Faderl, S.; O’Brien, S.; Pui, C.-H.; Stock, W.; Wetzler, M.; Hoelzer, D.; Kantarjian, H.M. Adult acute lymphoblastic leukemia. Cancer 2010, 116, 1165–1176. [Google Scholar] [CrossRef]
- Muvarak, N.; Kelley, S.; Robert, C.; Baer, M.R.; Perrotti, D.; Gambacorti-Passerini, C.; Civin, C.; Scheibner, K.; Rassool, F.V. c-MYC Generates Repair Errors via Increased Transcription of Alternative-NHEJ Factors, LIG3 and PARP1, in Tyrosine Kinase-Activated Leukemias. Mol. Cancer Res. 2015, 13, 699–712. [Google Scholar] [CrossRef]
- Cavelier, C.; Didier, C.; Prade, N.; Mas, V.M.-D.; Manenti, S.; Recher, C.; Demur, C.; Ducommun, B. Constitutive Activation of the DNA Damage Signaling Pathway in Acute Myeloid Leukemia with Complex Karyotype: Potential Importance for Checkpoint Targeting Therapy. Cancer Res. 2009, 69, 8652–8661. [Google Scholar] [CrossRef]
- Iacobucci, I.; Di Rora’, A.G.L.; Falzacappa, M.V.V.; Agostinelli, C.; Derenzini, E.; Ferrari, A.; Papayannidis, C.; Lonetti, A.; Righi, S.; Imbrogno, E.; et al. In vitro and in vivo single-agent efficacy of checkpoint kinase inhibition in acute lymphoblastic leukemia. J. Hematol. Oncol. 2015, 8, 125. [Google Scholar] [CrossRef]
- Sarmento, L.M.; Póvoa, V.; Nascimento, R.; Real, G.; Antunes, I.; Martins, L.R.; Moita, C.F.; Alves, P.M.; Abecasis, M.; Moita, L.F.; et al. CHK1 overexpression in T-cell acute lymphoblastic leukemia is essential for proliferation and survival by preventing excessive replication stress. Oncogene 2014, 34, 2978–2990. [Google Scholar] [CrossRef]
- Nieborowska-Skorska, M.; Stoklosa, T.; Datta, M.; Czechowska, A.; Rink, L.; Slupianek, A.; Koptyra, M.; Seferynska, I.; Krszyna, K.; Blasiak, J.; et al. ATR-Chk1 Axis Protects BCR/ABL Leukemia Cells from the Lethal Effect of DNA Double-Strand Breaks. Cell Cycle 2006, 5, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Delhommeau, F.; Dupont, S.; Della Valle, V.; James, C.; Trannoy, S.; Masse, A.; Kosmider, O.; Le Couédic, J.-P.; Robert, F.; Alberdi, A.; et al. Mutation inTET2in Myeloid Cancers. New Engl. J. Med. 2009, 360, 2289–2301. [Google Scholar] [CrossRef] [PubMed]
- Moran-Crusio, K.; Reavie, L.; Shih, A.; Abdel-Wahab, O.; Ndiaye-Lobry, D.; Lobry, C.; Figueroa, M.E.; VasanthaKumar, A.; Patel, J.; Zhao, X.; et al. Tet2 Loss Leads to Increased Hematopoietic Stem Cell Self-Renewal and Myeloid Transformation. Cancer Cell 2011, 20, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Coulter, J.; Lopez-Bertoni, H.; Kuhns, K.J.; Lee, R.S.; Laterra, J.; Bressler, J.P. TET1 deficiency attenuates the DNA damage response and promotes resistance to DNA damaging agents. Epigenetics 2017, 12, 854–864. [Google Scholar] [CrossRef]
- Zhong, J.; Li, X.; Cai, W.; Wang, Y.; Dong, S.; Yang, J.; Zhang, J.; Wu, N.; Li, Y.; Mao, F.; et al. TET1 modulates H4K16 acetylation by controlling auto-acetylation of hMOF to affect gene regulation and DNA repair function. Nucleic Acids Res. 2016, 45, 672–684. [Google Scholar] [CrossRef]
- Cimmino, L.; Dawlaty, M.M.; Ndiaye-Lobry, D.; Yap, Y.S.; Bakogianni, S.; Yu, Y.; Bhattacharyya, S.; Shaknovich, R.; Geng, H.; Lobry, C.; et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat. Immunol. 2015, 16, 653–662. [Google Scholar] [CrossRef]
- An, J.; González-Avalos, E.; Chawla, A.; Jeong, M.; López-Moyado, I.F.; Li, W.; Goodell, M.A.; Chavez, L.; Ko, M.; Rao, A. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat. Commun. 2015, 6, 10071. [Google Scholar] [CrossRef]
- Jiang, D.; Wei, S.; Chen, F.; Zhang, Y.; Li, J. TET3-mediated DNA oxidation promotes ATR-dependent DNA damage response. EMBO Rep. 2017, 18, 781–796. [Google Scholar] [CrossRef]
- Kafer, G.; Li, X.; Horii, T.; Suetake, I.; Tajima, S.; Hatada, I.; Carlton, P.M. 5-Hydroxymethylcytosine Marks Sites of DNA Damage and Promotes Genome Stability. Cell Rep. 2016, 14, 1283–1292. [Google Scholar] [CrossRef]
- Mahfoudhi, E.; Talhaoui, I.; Cabagnols, X.; Della Valle, V.; Secardin, L.; Rameau, P.; Bernard, A.O.; Ishchenko, A.A.; Abbes, S.; Vainchenker, W.; et al. TET2-mediated 5-hydroxymethylcytosine induces genetic instability and mutagenesis. DNA Repair 2016, 43, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Shanbhag, S.; Ambinder, R.F. Hodgkin lymphoma: A review and update on recent progress. CA A Cancer J. Clin. 2017, 68, 116–132. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-T.; Huang, Y.-W.; Yang, C.-H.; Huang, K.-S. Drug Delivery Systems and Combination Therapy by Using Vinca Alkaloids. Curr. Top. Med. Chem. 2015, 15, 1491–1500. [Google Scholar] [CrossRef] [PubMed]
- Cook, A.M.; McDonnell, A.M.; Lake, R.A.; Nowak, A.K. Dexamethasone co-medication in cancer patients undergoing chemotherapy causes substantial immunomodulatory effects with implications for chemo-immunotherapy strategies. OncoImmunology 2015, 5, e1066062. [Google Scholar] [CrossRef]
- Ventola, C.L. Cancer Immunotherapy, Part 2: Efficacy, Safety, and Other Clinical Considerations. PT 2017, 42, 452–463. [Google Scholar]
- Zhao, P.; Li, L.; Jiang, X.; Li, Q. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J. Hematol. Oncol. 2019, 12, 54. [Google Scholar] [CrossRef]
- Cramer-Morales, K.; Nieborowska-Skorska, M.; Scheibner, K.; Padget, M.; Irvine, D.A.; Sliwinski, T.; Haas, K.; Lee, J.; Geng, H.; Roy, D.; et al. Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood 2013, 122, 1293–1304. [Google Scholar] [CrossRef]
- Kubota, E.; Williamson, C.T.; Ye, R.; Elegbede, A.; Peterson, L.; Lees-Miller, S.P.; Bebb, D.G. Low ATM protein expression and depletion of p53 correlates with olaparib sensitivity in gastric cancer cell lines. Cell Cycle 2014, 13, 2129–2137. [Google Scholar] [CrossRef]
- Williamson, C.T.; Kubota, E.; Hamill, J.D.; Klimowicz, A.; Ye, R.; Muzik, H.; Dean, M.; Tu, L.; Gilley, D.; Magliocco, A.M.; et al. Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53. EMBO Mol. Med. 2012, 4, 515–527. [Google Scholar] [CrossRef]
- Menezes, D.L.; Holt, J.; Tang, Y.; Feng, J.; Barsanti, P.; Ghoddusi, M.; Holash, J.; Lees, E.; Taricani, L.; Pan, Y.; et al. A Synthetic Lethal Screen Reveals Enhanced Sensitivity to ATR Inhibitor Treatment in Mantle Cell Lymphoma with ATM Loss-of-Function. Mol. Cancer Res. 2015, 13, 120–129. [Google Scholar] [CrossRef]
- Fokas, E.; Prevo, R.; Pollard, J.R.; Reaper, P.M.; Charlton, A.P.; Cornelissen, B.; Vallis, K.A.; Hammond, E.M.; Olcina, M.M.; McKenna, W.G.; et al. Targeting ATR in vivo using the novel inhibitor VE-822 results in selective sensitization of pancreatic tumors to radiation. Cell Death Dis. 2012, 3, e441. [Google Scholar] [CrossRef] [PubMed]
- Prevo, R.; Fokas, E.; Reaper, P.M.; Charlton, P.A.; Pollard, J.R.; McKenna, W.G.; Muschel, R.J.; Brunner, T.B. The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol. Ther. 2012, 13, 1072–1081. [Google Scholar] [CrossRef] [PubMed]
- Foote, K.M.; Nissink, J.W.M.; McGuire, T.M.; Turner, P.; Guichard, S.; Yates, J.W.T.; Lau, A.; Blades, K.; Heathcote, D.; Odedra, R.; et al. Discovery and Characterization of AZD6738, a Potent Inhibitor of Ataxia Telangiectasia Mutated and Rad3 Related (ATR) Kinase with Application as an Anticancer Agent. J. Med. Chem. 2018, 61, 9889–9907. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.Y.; Fecteau, J.-F.; Brown, J.; Lau, A.; Kipps, T.J. Abstract 5485: Induction of proliferation sensitizes chronic lymphocytic leukemia cells to apoptosis mediated by the ATR inhibitor AZD6738. Cancer Res. 2014, 74, 5485. [Google Scholar] [CrossRef]
- Kwok, M.; Davies, N.; Agathanggelou, A.; Smith, E.; Oldreive, C.; Petermann, E.; Stewart, G.; Brown, J.; Lau, A.; Pratt, G.; et al. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood 2016, 127, 582–595. [Google Scholar] [CrossRef]
- Prudhomme, M. Novel checkpoint 1 inhibitors. Recent Pat. Anti-Cancer Drug Discov. 2006, 1, 55–68. [Google Scholar] [CrossRef]
- Wayne, J.; Brooks, T.; Massey, A.J. Inhibition of Chk1 with the small molecule inhibitor V158411 induces DNA damage and cell death in an unperturbed S-phase. Oncotarget 2016, 7, 85033–85048. [Google Scholar] [CrossRef]
- Daud, A.I.; Ashworth, M.T.; Strosberg, J.; Goldman, J.W.; Mendelson, D.; Springett, G.; Venook, A.P.; Loechner, S.; Rosen, L.S.; Shanahan, F.; et al. Phase I Dose-Escalation Trial of Checkpoint Kinase 1 Inhibitor MK-8776 As Monotherapy and in Combination With Gemcitabine in Patients With Advanced Solid Tumors. J. Clin. Oncol. 2015, 33, 1060–1066. [Google Scholar] [CrossRef]
- Dai, Y.; Chen, S.; Kmieciak, M.; Zhou, L.; Lin, H.; Pei, X.-Y.; Grant, S. The novel Chk1 inhibitor MK-8776 sensitizes human leukemia cells to HDAC inhibitors by targeting the intra-S checkpoint and DNA replication and repair. Mol. Cancer Ther. 2013, 12, 878–889. [Google Scholar] [CrossRef]
- Zemanova, J.; Hylse, O.; Collakova, J.; Veselý, P.; Oltova, A.; Borsky, M.; Zaprazna, K.; Kasparkova, M.; Janovska, P.; Verner, J.; et al. Chk1 inhibition significantly potentiates activity of nucleoside analogs in TP53-mutated B-lymphoid cells. Oncotarget 2016, 7, 62091–62106. [Google Scholar] [CrossRef]
- Morgan, M.A.; Parsels, L.A.; Zhao, L.; Parsels, J.D.; Davis, M.A.; Hassan, M.C.; Arumugarajah, S.; Hylander-Gans, L.; Morosini, D.; Simeone, D.M.; et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 2010, 70, 4972–4981. [Google Scholar] [CrossRef] [PubMed]
- Landau, H.; McNeely, S.C.; Nair, J.S.; Comenzo, R.; Asai, T.; Friedman, H.; Jhanwar, S.C.; Nimer, S.D.; Schwartz, G.K. The Checkpoint Kinase Inhibitor AZD7762 Potentiates Chemotherapy-Induced Apoptosis of p53-Mutated Multiple Myeloma Cells. Mol. Cancer Ther. 2012, 11, 1781–1788. [Google Scholar] [CrossRef] [PubMed]
- Didier, C.; Demur, C.; Grimal, F.; Jullien, D.; Manenti, S.; Ducommun, B. Evaluation of checkpoint kinase targeting therapy in Acute Myeloid Leukemia with complex karyotype. Cancer Biol. Ther. 2012, 13, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Hawkins, E.; Kolluri, A.; Kmieciak, M.; Park, H.; Lin, H.; Grant, S. Synergism between bosutinib (SKI-606) and the Chk1 inhibitor (PF-00477736) in highly imatinib-resistant BCR/ABL⁺ leukemia cells. Leuk. Res. 2014, 39, 65–71. [Google Scholar] [CrossRef][Green Version]
- King, C.; Diaz, H.B.; McNeely, S.; Barnard, D.; Dempsey, J.; Blosser, W.; Beckmann, R.; Barda, D.; Marshall, M. LY2606368 Causes Replication Catastrophe and Antitumor Effects through CHK1-Dependent Mechanisms. Mol. Cancer Ther. 2015, 14, 2004–2013. [Google Scholar] [CrossRef]
- Di Rora’, A.G.L.; Iacobucci, I.; Imbrogno, E.; Papayannidis, C.; Derenzini, E.; Ferrari, A.; Guadagnuolo, V.; Robustelli, V.; Parisi, S.; Sartor, C.; et al. Prexasertib, a Chk1/Chk2 inhibitor, increases the effectiveness of conventional therapy in B-/T- cell progenitor acute lymphoblastic leukemia. Oncotarget 2016, 7, 53377–53391. [Google Scholar] [CrossRef]
- Calvo, E.; Chen, V.J.; Marshall, M.; Ohnmacht, U.; Hynes, S.M.; Kumm, E.; Diaz, H.B.; Barnard, D.; Merzoug, F.F.; Huber, L.; et al. Preclinical analyses and phase I evaluation of LY2603618 administered in combination with Pemetrexed and cisplatin in patients with advanced cancer. Investig. New Drugs 2014, 32, 955–968. [Google Scholar] [CrossRef]
- Zhao, J.; Niu, X.; Li, X.; Edwards, H.; Wang, G.; Wang, Y.; Taub, J.W.; Lin, H.; Ge, Y. Inhibition of CHK1 enhances cell death induced by the Bcl-2-selective inhibitor ABT-199 in acute myeloid leukemia cells. Oncotarget 2016, 7, 34785–34799. [Google Scholar] [CrossRef]
- Luedtke, A.D.; Niu, X.; Pan, Y.; Zhao, J.; Liu, S.; Edwards, H.; Chen, K.; Lin, H.; Taub, J.W.; Ge, Y. Inhibition of Mcl-1 enhances cell death induced by the Bcl-2-selective inhibitor ABT-199 in acute myeloid leukemia cells. Signal Transduct. Target. Ther. 2017, 2, 17012. [Google Scholar] [CrossRef]
- Galmarini, C.M.; Mackey, J.R.; Dumontet, C. Nucleoside analogues: Mechanisms of drug resistance and reversal strategies. Leukemia 2001, 15, 875–890. [Google Scholar] [CrossRef]
- Liu, X.-J.; Nowak, B.; Wang, Y.-Q.; Plunkett, W. Sapacitabine, the prodrug of CNDAC, is a nucleoside analog with a unique action mechanism of inducing DNA strand breaks. Chin. J. Cancer 2012, 31, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.; Faderl, S.; Garcia-Manero, G.; Luger, S.; Venugopal, P.; Maness, L.; Wetzler, M.; Coutre, S.; Stock, W.; Claxton, D.; et al. Oral sapacitabine for the treatment of acute myeloid leukaemia in elderly patients: A randomised phase 2 study. Lancet Oncol. 2012, 13, 1096–1104. [Google Scholar] [CrossRef]
- Choi, M.; Kipps, T.; Kurzrock, R. ATM Mutations in Cancer: Therapeutic Implications. Mol. Cancer Ther. 2016, 15, 1781–1791. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhmoud, J.F.; Mustafa, A.G.; Malki, M.I. Targeting DNA Repair Pathways in Hematological Malignancies. Int. J. Mol. Sci. 2020, 21, 7365. https://doi.org/10.3390/ijms21197365
Alhmoud JF, Mustafa AG, Malki MI. Targeting DNA Repair Pathways in Hematological Malignancies. International Journal of Molecular Sciences. 2020; 21(19):7365. https://doi.org/10.3390/ijms21197365
Chicago/Turabian StyleAlhmoud, Jehad F., Ayman G. Mustafa, and Mohammed Imad Malki. 2020. "Targeting DNA Repair Pathways in Hematological Malignancies" International Journal of Molecular Sciences 21, no. 19: 7365. https://doi.org/10.3390/ijms21197365
APA StyleAlhmoud, J. F., Mustafa, A. G., & Malki, M. I. (2020). Targeting DNA Repair Pathways in Hematological Malignancies. International Journal of Molecular Sciences, 21(19), 7365. https://doi.org/10.3390/ijms21197365




