Maternal Recognition of Pregnancy in the Horse: Are MicroRNAs the Secret Messengers?
Abstract
1. Introduction
2. Results
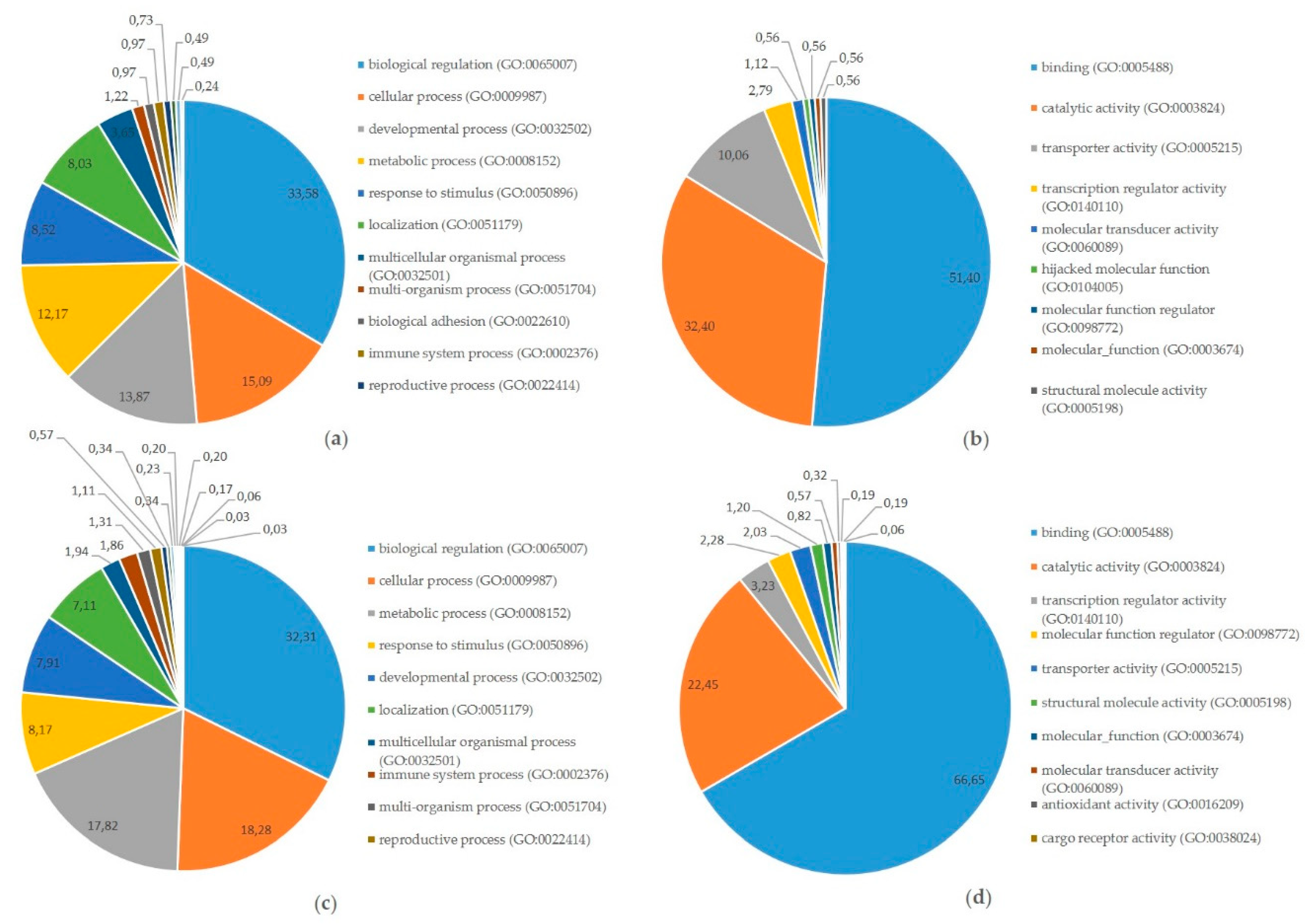
2.1. Messenger-RNA Sequencing
2.2. Micro-RNA Sequencing
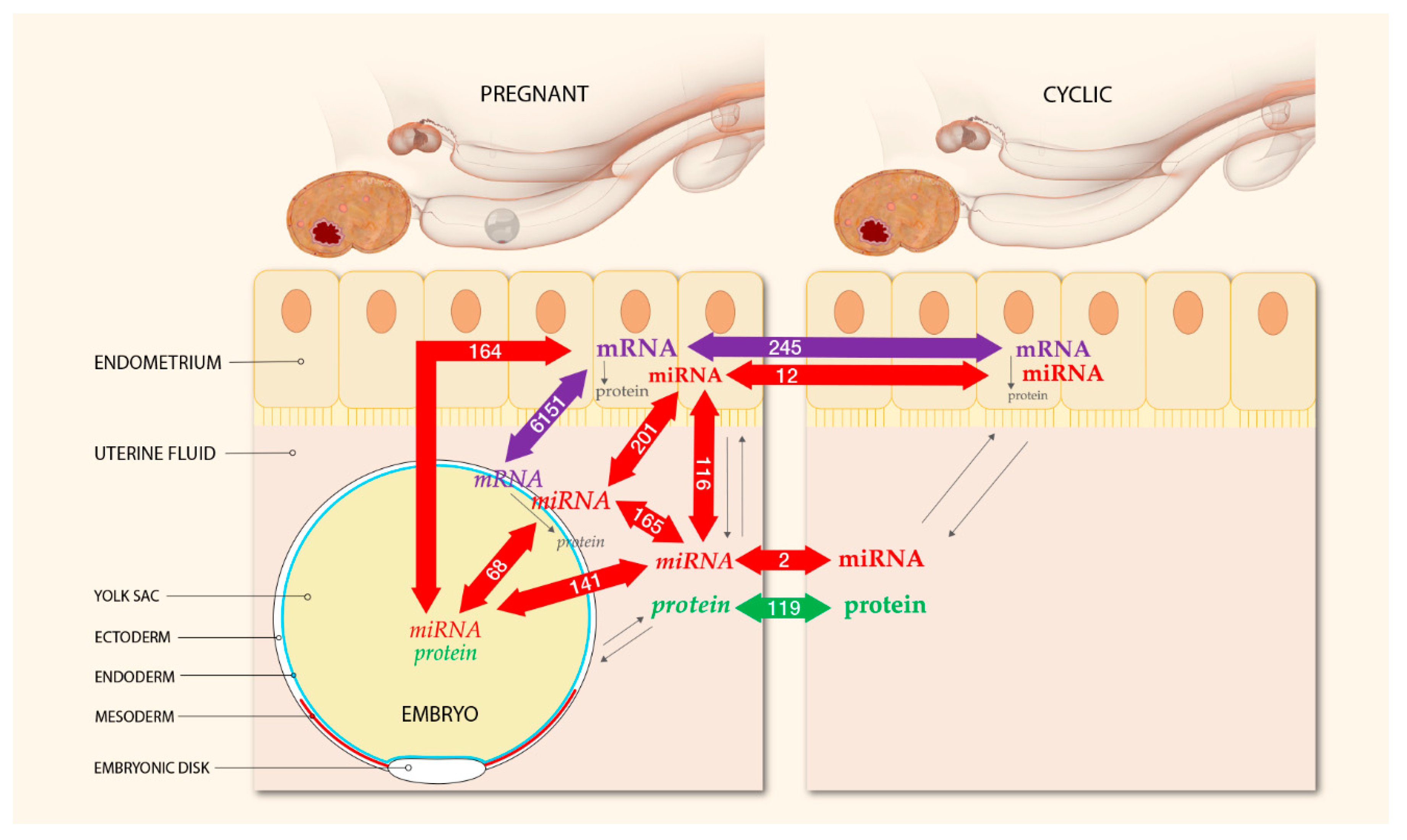
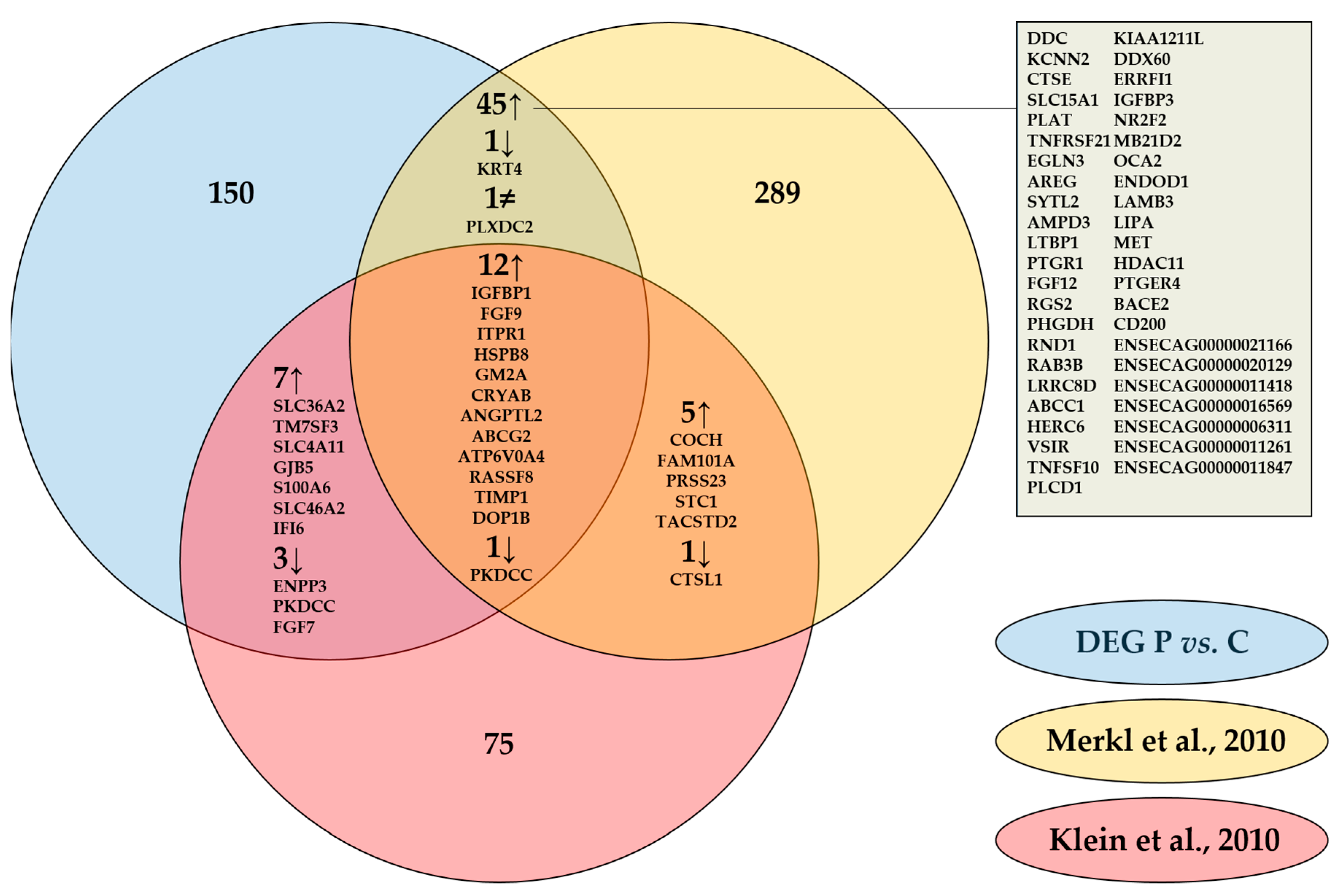
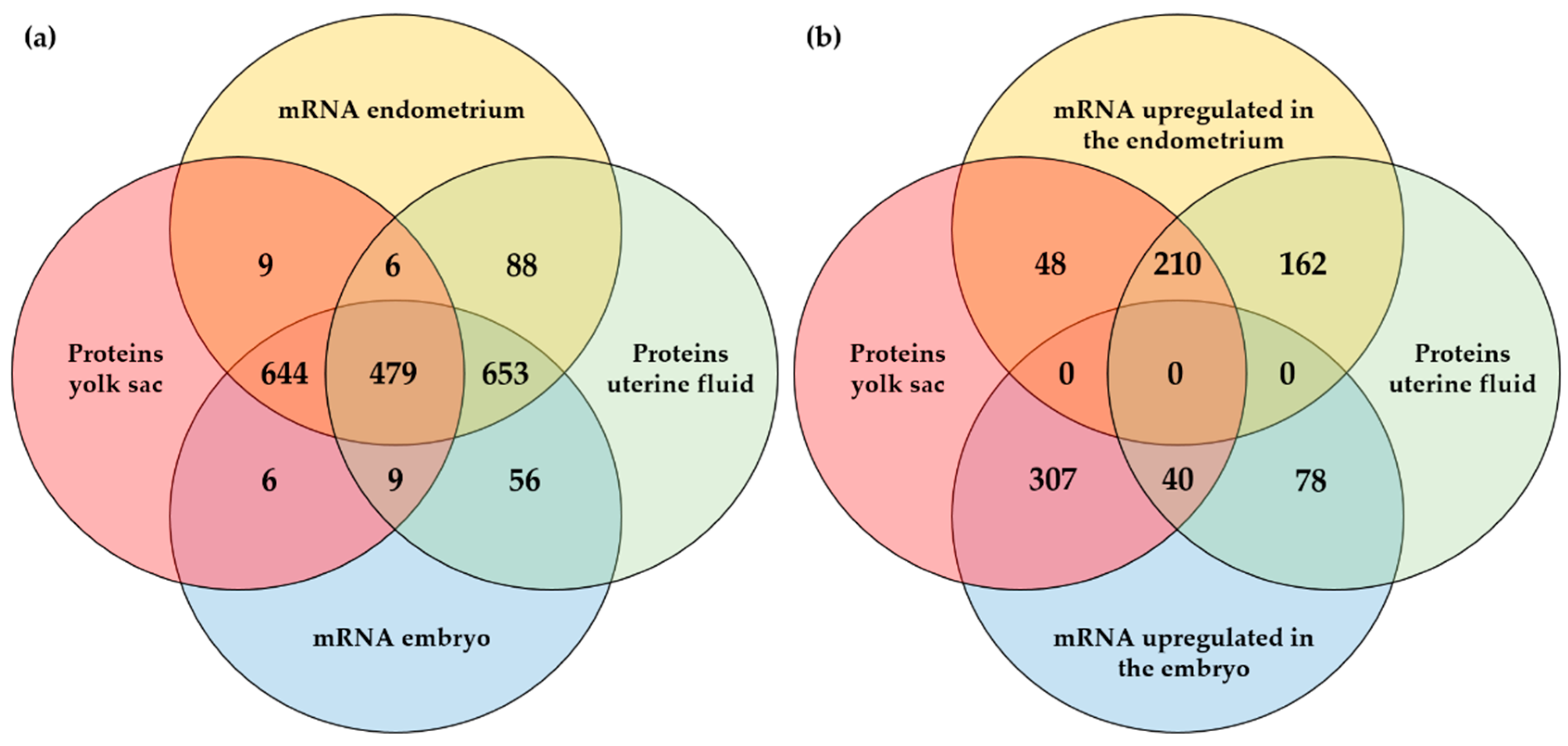
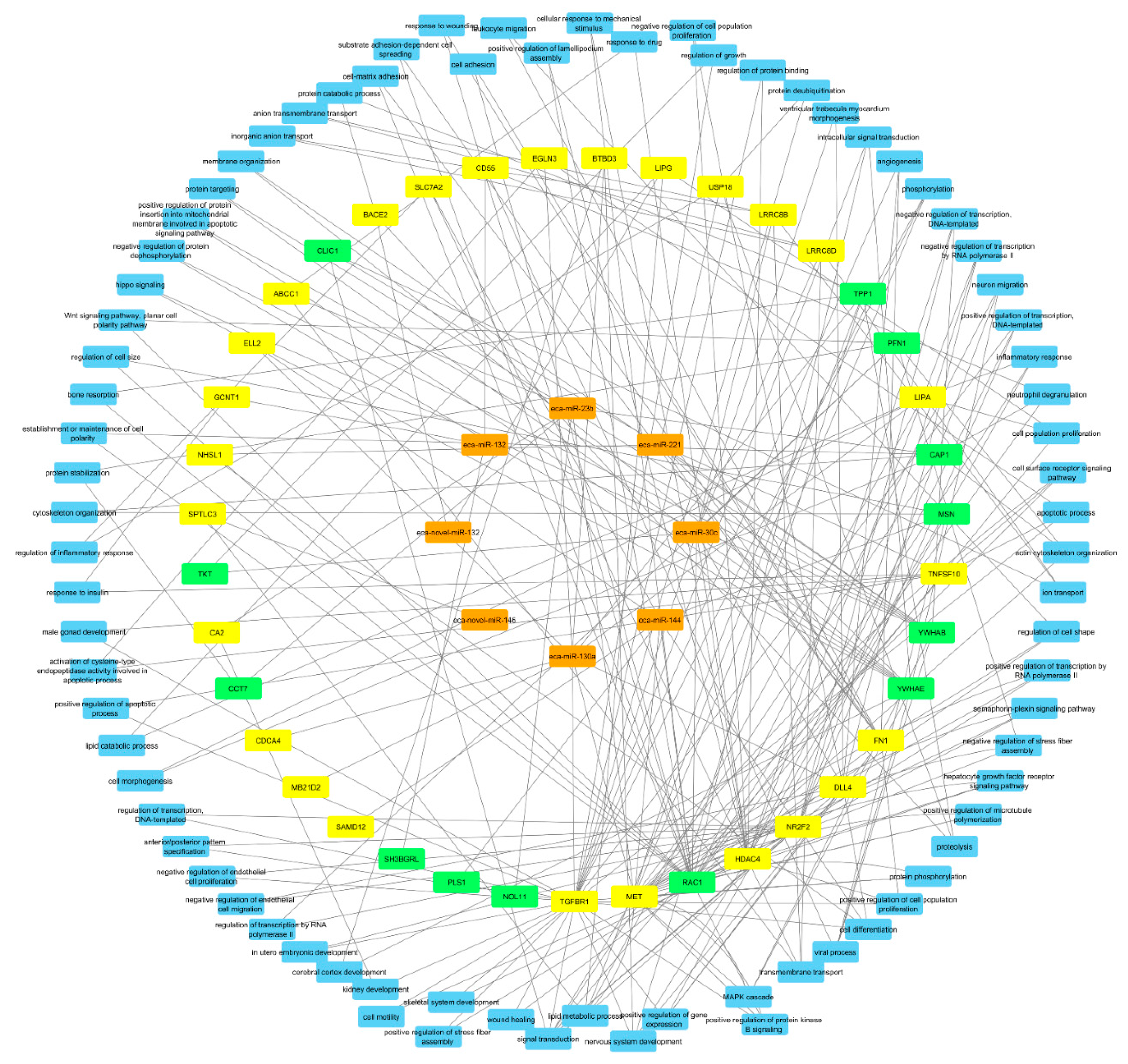
2.3. Integrated Analysis of Proteomics, Transcriptomics, and Micro-RNA Expression
3. Discussion
4. Materials and Methods
4.1. Sampling
4.2. RNA Extraction
4.3. Messenger-RNA-Sequencing and Data Analysis
4.4. MicroRNA-Sequencing and Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MRP | Maternal recognition of pregnancy |
| UF | Uterine fluid |
| YS | Yolk sac |
| P | Pregnant |
| C | Cyclic |
| DEGs | Differentially expressed genes |
| DE | Differential expression |
References
- Van Niekerk, C.H.; Gerneke, W.H. Persistence and parthenogentic cleavage of tubal ova in the mare. Onderstepoort J. Vet. Res. 1966, 33, 195. [Google Scholar]
- Oriol, J.G.; Betteridge, K.J.; Clarke, A.J.; Sharom, F.J. Mucin-like glycoproteins in the equine embryonic capsule. Mol. Reprod. Dev. 1993, 34, 255–265. [Google Scholar] [CrossRef]
- Allen, W.R. Fetomaternal interactions and influences during equine pregnancy. Reproduction 2001, 121, 513–527. [Google Scholar] [CrossRef]
- Betteridge, K.J. Comparative aspects of equine embryonic development. Anim. Reprod. Sci. 2000, 60, 691–702. [Google Scholar] [CrossRef]
- Short, R. Implantation and the maternal recognition of pregnancy. Foetal Auton. 1969, 2, 31. [Google Scholar]
- Boerboom, D.; Brown, K.A.; Vaillancourt, D.; Poitras, P.; Goff, A.K.; Watanabe, K.; Dore, M.; Sirois, J. Expression of key prostaglandin synthases in equine endometrium during late diestrus and early pregnancy. Biol. Reprod. 2004, 70, 391–399. [Google Scholar] [CrossRef]
- Vanderwall, D.K.; Silvia, W.J.; Fitzgerald, B.P. Concentrations of oxytocin in the intercavernous sinus of mares during luteolysis: Temporal relationship with concentrations of 13,14-dihydro-15-keto-prostaglandin F2 alpha. J. Reprod. Fertil. 1998, 112, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Sharp, D.C.; Thatcher, M.J.; Salute, M.E.; Fuchs, A.R. Relationship between endometrial oxytocin receptors and oxytocin-induced prostaglandin F2 alpha release during the oestrous cycle and early pregnancy in pony mares. J. Reprod. Fertil. 1997, 109, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Kindahl, H.; Knudsen, O.; Madej, A.; Edqvist, L.E. Progesterone, prostaglandin F-2 alpha, PMSG and oestrone sulphate during early pregnancy in the mare. J. Reprod. Fertil. Suppl. 1982, 32, 353–359. [Google Scholar] [PubMed]
- Goff, A.K.; Pontbriand, D.; Sirois, J. Oxytocin stimulation of plasma 15-keto-13,14-dihydro prostaglandin F-2 alpha during the oestrous cycle and early pregnancy in the mare. J. Reprod. Fertil. Suppl. 1987, 35, 253–260. [Google Scholar]
- Starbuck, G.R.; Stout, T.A.; Lamming, G.E.; Allen, W.R.; Flint, A.P. Endometrial oxytocin receptor and uterine prostaglandin secretion in mares during the oestrous cycle and early pregnancy. J. Reprod. Fertil. 1998, 113, 173–179. [Google Scholar] [CrossRef][Green Version]
- Ealy, A.D.; Eroh, M.L.; Sharp, D.C., 3rd. Prostaglandin H synthase Type 2 is differentially expressed in endometrium based on pregnancy status in pony mares and responds to oxytocin and conceptus secretions in explant culture. Anim. Reprod. Sci. 2010, 117, 99–105. [Google Scholar] [CrossRef] [PubMed]
- de Ruijter-Villani, M.; van Tol, H.T.; Stout, T.A. Effect of pregnancy on endometrial expression of luteolytic pathway components in the mare. Reprod. Fertil. Dev. 2015, 27, 834–845. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Troedsson, M.H. Maternal recognition of pregnancy in the horse: A mystery still to be solved. Reprod. Fertil. Dev. 2011, 23, 952–963. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Thatcher, W. Theory of maternal recognition of pregnancy in swine based on estrogen controlled endocrine versus exocrine secretion of prostaglandin F 2α by the uterine endometrium. Prostaglandins 1977, 14, 397–401. [Google Scholar] [CrossRef]
- Lamming, G.E.; Wathes, D.C.; Flint, A.P.; Payne, J.H.; Stevenson, K.R.; Vallet, J.L. Local action of trophoblast interferons in suppression of the development of oxytocin and oestradiol receptors in ovine endometrium. J. Reprod. Fertil. 1995, 105, 165–175. [Google Scholar] [CrossRef]
- Stout, T.A.; Allen, W.R. Prostaglandin E (2) and F (2 alpha) production by equine conceptuses and concentrations in conceptus fluids and uterine flushings recovered from early pregnant and dioestrous mares. Reproduction 2002, 123, 261–268. [Google Scholar] [CrossRef]
- McDowell, K.J.; Sharp, D.C.; Grubaugh, W.; Thatcher, W.W.; Wilcox, C.J. Restricted conceptus mobility results in failure of pregnancy maintenance in mares. Biol. Reprod. 1988, 39, 340–348. [Google Scholar] [CrossRef]
- Klein, C.; Scoggin, K.E.; Ealy, A.D.; Troedsson, M.H. Transcriptional profiling of equine endometrium during the time of maternal recognition of pregnancy. Biol. Reprod. 2010, 83, 102–113. [Google Scholar] [CrossRef]
- Klein, C.; Troedsson, M.H. Transcriptional profiling of equine conceptuses reveals new aspects of embryo-maternal communication in the horse. Biol. Reprod. 2011, 84, 872–885. [Google Scholar] [CrossRef]
- Klein, C. Novel equine conceptus? Endometrial interactions on Day 16 of pregnancy based on RNA sequencing. Reprod. Fertil. Dev. 2015, 2016, 1712–1720. [Google Scholar] [CrossRef] [PubMed]
- Merkl, M.; Ulbrich, S.E.; Otzdorff, C.; Herbach, N.; Wanke, R.; Wolf, E.; Handler, J.; Bauersachs, S. Microarray analysis of equine endometrium at days 8 and 12 of pregnancy. Biol. Reprod. 2010, 83, 874–886. [Google Scholar] [CrossRef] [PubMed]
- Bauersachs, S.; Wolf, E. Transcriptome analyses of bovine, porcine and equine endometrium during the pre-implantation phase. Anim. Reprod. Sci. 2012, 134, 84–94. [Google Scholar] [CrossRef]
- Klohonatz, K.M.; Coleman, S.J.; Islas-Trejo, A.D.; Medrano, J.F.; Hess, A.M.; Kalbfleisch, T.; Thomas, M.G.; Bouma, G.J.; Bruemmer, J.E. Coding RNA Sequencing of Equine Endometrium during Maternal Recognition of Pregnancy. Genes 2019, 10, 749. [Google Scholar] [CrossRef] [PubMed]
- Swegen, A.; Grupen, C.G.; Gibb, Z.; Baker, M.A.; de Ruijter-Villani, M.; Smith, N.D.; Stout, T.A.E.; Aitken, R.J. From Peptide Masses to Pregnancy Maintenance: A Comprehensive Proteomic Analysis of The Early Equine Embryo Secretome, Blastocoel Fluid, and Capsule. Proteomics 2017, 17, 1600433. [Google Scholar] [CrossRef] [PubMed]
- Smits, K.; Willems, S.; Van Steendam, K.; Van De Velde, M.; De Lange, V.; Ververs, C.; Roels, K.; Govaere, J.; Van Nieuwerburgh, F.; Peelman, L.; et al. Proteins involved in embryo-maternal interaction around the signalling of maternal recognition of pregnancy in the horse. Sci. Rep. 2018, 8, 5249. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- Hayder, H.; O’Brien, J.; Nadeem, U.; Peng, C. MicroRNAs: Crucial regulators of placental development. Reproduction 2018, 155, R259–R271. [Google Scholar] [CrossRef]
- Paul, A.B.M.; Sadek, S.T.; Mahesan, A.M. The role of microRNAs in human embryo implantation: A review. J. Assist. Reprod. Genet. 2019, 36, 179–187. [Google Scholar] [CrossRef]
- Hossain, M.M.; Tesfaye, D.; Salilew-Wondim, D.; Held, E.; Proll, M.J.; Rings, F.; Kirfel, G.; Looft, C.; Tholen, E.; Uddin, J.; et al. Massive deregulation of miRNAs from nuclear reprogramming errors during trophoblast differentiation for placentogenesis in cloned pregnancy. BMC Genom. 2014, 15, 43. [Google Scholar] [CrossRef][Green Version]
- da Silveira, J.C.; Veeramachaneni, D.N.; Winger, Q.A.; Carnevale, E.M.; Bouma, G.J. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: A possible new form of cell communication within the ovarian follicle. Biol. Reprod. 2012, 86, 71. [Google Scholar] [CrossRef] [PubMed]
- Loux, S.C.; Scoggin, K.E.; Bruemmer, J.E.; Canisso, I.F.; Troedsson, M.H.; Squires, E.L.; Ball, B.A. Evaluation of circulating miRNAs during late pregnancy in the mare. PLoS ONE 2017, 12, e0175045. [Google Scholar] [CrossRef] [PubMed]
- Dini, P.; Daels, P.; Loux, S.C.; Esteller-Vico, A.; Carossino, M.; Scoggin, K.E.; Ball, B.A. Kinetics of the chromosome 14 microRNA cluster ortholog and its potential role during placental development in the pregnant mare. BMC Genom. 2018, 19, 954. [Google Scholar] [CrossRef] [PubMed]
- Klohonatz, K.M.; Cameron, A.D.; Hergenreder, J.R.; da Silveira, J.C.; Belk, A.D.; Veeramachaneni, D.N.; Bouma, G.J.; Bruemmer, J.E. Circulating miRNAs as Potential Alternative Cell Signaling Associated with Maternal Recognition of Pregnancy in the Mare. Biol. Reprod. 2016, 95, 124. [Google Scholar] [CrossRef]
- Klohonatz, K.M.; Coleman, S.J.; Cameron, A.D.; Hess, A.M.; Reed, K.J.; Canovas, A.; Medrano, J.F.; Islas-Trejo, A.D.; Kalbfleisch, T.; Bouma, G.J.; et al. Non-Coding RNA Sequencing of Equine Endometrium During Maternal Recognition of Pregnancy. Genes 2019, 10, 749. [Google Scholar] [CrossRef]
- Klohonatz, K.M.; Hess, A.M.; Hansen, T.R.; Squires, E.L.; Bouma, G.J.; Bruemmer, J.E. Equine endometrial gene expression changes during and after maternal recognition of pregnancy. J. Anim. Sci. 2015, 93, 3364–3376. [Google Scholar] [CrossRef]
- Peter Durairaj, R.R.; Aberkane, A.; Polanski, L.; Maruyama, Y.; Baumgarten, M.; Lucas, E.S.; Quenby, S.; Chan, J.K.Y.; Raine-Fenning, N.; Brosens, J.J.; et al. Deregulation of the endometrial stromal cell secretome precedes embryo implantation failure. Mol. Hum. Reprod. 2017, 23, 582. [Google Scholar] [CrossRef][Green Version]
- Sucurovic, S.; Nikolic, T.; Brosens, J.J.; Mulac-Jericevic, B. Spatial and Temporal Analyses of FGF9 Expression During Early Pregnancy. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 42, 2318–2329. [Google Scholar] [CrossRef]
- Hulboy, D.L.; Rudolph, L.A.; Matrisian, L.M. Matrix metalloproteinases as mediators of reproductive function. Mol. Hum. Reprod. 1997, 3, 27–45. [Google Scholar] [CrossRef]
- Tabibzadeh, S. Molecular control of the implantation window. Hum. Reprod. Update 1998, 4, 465–471. [Google Scholar] [CrossRef][Green Version]
- Mirkin, S.; Arslan, M.; Churikov, D.; Corica, A.; Diaz, J.I.; Williams, S.; Bocca, S.; Oehninger, S. In search of candidate genes critically expressed in the human endometrium during the window of implantation. Hum. Reprod. (Oxf. Engl.) 2005, 20, 2104–2117. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Chakraborty, I.; Paria, B.C.; Wang, X.N.; Plowman, G.; Dey, S.K. Amphiregulin is an implantation-specific and progesterone-regulated gene in the mouse uterus. Mol. Endocrinol. 1995, 9, 691–705. [Google Scholar] [PubMed]
- Kim, T.H.; Lee, D.K.; Franco, H.L.; Lydon, J.P.; Jeong, J.W. ERBB receptor feedback inhibitor 1 regulation of estrogen receptor activity is critical for uterine implantation in mice. Biol. Reprod. 2010, 82, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, I.; Lee, D.K.; Petit, F.G.; Jeong, J.; Lee, K.; Lydon, J.P.; DeMayo, F.J.; Tsai, M.J.; Tsai, S.Y. COUP-TFII mediates progesterone regulation of uterine implantation by controlling ER activity. PLoS Genet. 2007, 3, e102. [Google Scholar] [CrossRef]
- Yang, Z.M.; Das, S.K.; Wang, J.; Sugimoto, Y.; Ichikawa, A.; Dey, S.K. Potential sites of prostaglandin actions in the periimplantation mouse uterus: Differential expression and regulation of prostaglandin receptor genes. Biol. Reprod. 1997, 56, 368–379. [Google Scholar] [CrossRef][Green Version]
- McDowell, K.J.; Adams, M.H.; Adam, C.Y.; Simpson, K.S. Changes in equine endometrial oestrogen receptor alpha and progesterone receptor mRNAs during the oestrous cycle, early pregnancy and after treatment with exogenous steroids. J. Reprod. Fertil. 1999, 117, 135–142. [Google Scholar] [CrossRef]
- Chuang, P.C.; Sun, H.S.; Chen, T.M.; Tsai, S.J. Prostaglandin E2 induces fibroblast growth factor 9 via EP3-dependent protein kinase Cdelta and Elk-1 signaling. Mol. Cell. Biol. 2006, 26, 8281–8292. [Google Scholar] [CrossRef]
- Assou, S.; Haouzi, D.; Dechaud, H.; Gala, A.; Ferrieres, A.; Hamamah, S. Comparative gene expression profiling in human cumulus cells according to ovarian gonadotropin treatments. BioMed. Res. Int. 2013, 2013, 354582. [Google Scholar] [CrossRef]
- Yang, Q.; Gu, W.W.; Gu, Y.; Yan, N.N.; Mao, Y.Y.; Zhen, X.X.; Wang, J.M.; Yang, J.; Shi, H.J.; Zhang, X.; et al. Association of the peripheral blood levels of circulating microRNAs with both recurrent miscarriage and the outcomes of embryo transfer in an in vitro fertilization process. J. Transl. Med. 2018, 16, 186. [Google Scholar] [CrossRef]
- Shi, L.; Liu, S.; Zhao, W.; Shi, J. miR-483-5p and miR-486-5p are down-regulated in cumulus cells of metaphase II oocytes from women with polycystic ovary syndrome. Reprod. Biomed. Online 2015, 31, 565–572. [Google Scholar] [CrossRef]
- Lin, X.; Beckers, E.; Mc Cafferty, S.; Gansemans, Y.; Joanna Szymanska, K.; Chaitanya Pavani, K.; Catani, J.P.; Van Nieuwerburgh, F.; Deforce, D.; De Sutter, P.; et al. Bovine Embryo-Secreted microRNA-30c Is a Potential Non-invasive Biomarker for Hampered Preimplantation Developmental Competence. Front. Genet. 2019, 10, 315. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Niu, Z.; Li, Q.; Pang, R.T.; Chiu, P.C.; Yeung, W.S. MicroRNA and Embryo Implantation. Am. J. Reprod. Immunol. 2016, 75, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Shen, H.; Fan, L.J.; Guan, J.; Zheng, X.B.; Chen, X.; Liang, R.; Zhang, X.W.; Cui, Q.H.; Sun, K.K.; et al. Endometrial MicroRNA Signature during the Window of Implantation Changed in Patients with Repeated Implantation Failure. Chin. Med. J. 2017, 130, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, T.; Wu, L.; Liu, X.; Xue, S.; Lei, M. Identification of non-coding and coding RNAs in porcine endometrium. Genomics 2017, 109, 43–50. [Google Scholar] [CrossRef]
- Wu, S.; Sun, H.; Zhang, Q.; Jiang, Y.; Fang, T.; Cui, I.; Yan, G.; Hu, Y. MicroRNA-132 promotes estradiol synthesis in ovarian granulosa cells via translational repression of Nurr1. Reprod. Boil. Endocrinol. 2015, 13, 94. [Google Scholar] [CrossRef]
- Sang, Q.; Yao, Z.; Wang, H.; Feng, R.; Wang, H.; Zhao, X.; Xing, Q.; Jin, L.; He, L.; Wu, L.; et al. Identification of MicroRNAs in Human Follicular Fluid: Characterization of MicroRNAs That Govern Steroidogenesis in Vitro and Are Associated With Polycystic Ovary Syndrome in Vivo. J. Clin. Endocrinol. Metab. 2013, 98, 3068–3079. [Google Scholar] [CrossRef]
- Machtinger, R.; Rodosthenous, R.S.; Adir, M.; Mansour, A.; Racowsky, C.; Baccarelli, A.A.; Hauser, R. Extracellular microRNAs in follicular fluid and their potential association with oocyte fertilization and embryo quality: An exploratory study. J. Assist. Reprod. Genet. 2017, 34, 525–533. [Google Scholar] [CrossRef]
- Reliszko, Z.P.; Gajewski, Z.; Kaczmarek, M.M. Signs of embryo-maternal communication: miRNAs in the maternal serum of pregnant pigs. Reprod. (Camb. Engl.) 2017, 154, 217–228. [Google Scholar] [CrossRef]
- Yang, S.; Li, H.; Ge, Q.; Guo, L.; Chen, F. Deregulated microRNA species in the plasma and placenta of patients with preeclampsia. Mol. Med. Rep. 2015, 12, 527–534. [Google Scholar] [CrossRef]
- Wang, N.; Feng, Y.; Xu, J.; Zou, J.; Chen, M.; He, Y.; Liu, H.; Xue, M.; Gu, Y. miR-362-3p regulates cell proliferation, migration and invasion of trophoblastic cells under hypoxia through targeting Pax3. Biomed. Pharmacother. 2018, 99, 462–468. [Google Scholar] [CrossRef]
- Yang, Y.; Li, H.; Ma, Y.; Zhu, X.; Zhang, S.; Li, J. MiR-221-3p is down-regulated in preeclampsia and affects trophoblast growth, invasion and migration partly via targeting thrombospondin 2. Biomed. Pharmacother. Biomed. Pharmacother. 2019, 109, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Tao, T.; Yin, Y.; Zhao, L.; Yang, L.; Hu, L. miR-144 may regulate the proliferation, migration and invasion of trophoblastic cells through targeting PTEN in preeclampsia. Biomed. Pharmacother. 2017, 94, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lei, B.; Li, H.; Zhu, L.; Wang, L.; Tao, H.; Mei, S.; Li, F. MicroRNA-144 is regulated by CP2 and decreases COX-2 expression and PGE2 production in mouse ovarian granulosa cells. Cell Death Dis. 2017, 8, e2597. [Google Scholar] [CrossRef] [PubMed]
- Wolf, C.A.; Maslchitzky, E.; Gregory, R.M.; Jobim, M.I.; Mattos, R.C. Effect of corticotherapy on proteomics of endometrial fluid from mares susceptible to persistent postbreeding endometritis. Theriogenology 2012, 77, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Shi, J.; Dong, M.; Li, L.; Liu, L.; Luz-Madrigal, A.; Tsonis, P.A.; Del Rio-Tsonis, K.; Liang, C. mirPRo—A novel standalone program for differential expression and variation analysis of miRNAs. Sci. Rep. 2015, 5, 14617. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2007, 36 (Suppl. S1), D154–D158. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2-and edgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Chou, C.-H.; Chang, N.-W.; Shrestha, S.; Hsu, S.-D.; Lin, Y.-L.; Lee, W.-H.; Yang, C.-D.; Hong, H.-C.; Wei, T.-Y.; Tu, S.-J. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2015, 44, D239–D247. [Google Scholar] [CrossRef]
| miRNA B | miRNA P vs. C miRNA P vs. C | mRNA B | mRNA P vs. C | Gene Description |
| eca-miR-30c | Down | HDAC4 | Down | histone deacetylase 4 |
| eca-miR-30c | Down | DLL4 | Down | delta like canonical Notch ligand 4 |
| eca-miR-30c | Down | LRRC8D | Up | leucine rich repeat containing 8 VRAC subunit D |
| eca-miR-30c | Down | LRRC8B | Up | leucine rich repeat containing 8 VRAC subunit B |
| eca-miR-130a | Up | ELL2 | Down | elongation factor for RNA polymerase II 2 |
| eca-miR-130a | Up | TGFBR1 | Down | transforming growth factor beta receptor 1 |
| eca-miR-130a | Up | DLL4 | Down | delta like canonical Notch ligand 4 |
| eca-miR-130a | Up | BTBD3 | Up | BTB domain containing 3 |
| eca-miR-130a | Up | CDCA4 | Up | cell division cycle associated 4 |
| eca-miR-130a | Up | EGLN3 | Up | egl-9 family hypoxia inducible factor 3 |
| eca-miR-130a | Up | MET | Up | MET proto-oncogene, receptor tyrosine kinase |
| eca-miR-130a | Up | LIPA | Up | lipase A, lysosomal acid type |
| eca-miR-130a | Up | MB21D2 | Up | Mab-21 domain containing 2 |
| eca-miR-132 | Up | CD55 | Up | CD55 molecule (Cromer blood group) |
| eca-miR-132 | Up | GCNT1 | Up | glucosaminyl (N-acetyl) transferase 1 |
| eca-miR-144 | Down | SLC7A2 | Down | solute carrier family 7 member 2 |
| eca-miR-144 | Down | ELL2 | Down | elongation factor for RNA polymerase II 2 |
| eca-miR-144 | Down | SAMD12 | Down | sterile alpha motif domain containing 12 |
| eca-miR-144 | Down | NR2F2 | Up | nuclear receptor subfamily 2 group F member 2 |
| eca-miR-144 | Down | MET | Up | MET proto-oncogene, receptor tyrosine kinase |
| eca-miR-221 | Up | FN1 | Down | fibronectin 1 |
| eca-miR-221 | Up | NHSL1 | Down | NHS like 1 |
| eca-miR-221 | Up | USP18 | Up | ubiquitin specific peptidase 18 |
| eca-miR-221 | Up | TNFSF10 | Up | TNF superfamily member 10 |
| eca-novel-miR-132 | Up | LIPG | Up | lipase G, endothelial type |
| eca-novel-miR-132 | Up | BACE2 | Up | beta-secretase 2 |
| eca-novel-miR-146 | Up | SPTLC3 | Up | serine palmitoyltransferase long chain base subunit 3 |
| miRNA B | miRNA P vs. C | Protein UF | Protein P vs. C | Gene Description |
| eca-miR-30c | Down | TKT | Up | Transketolase |
| eca-miR-30c | Down | RAC1 | Up | Rac family small GTPase 1 |
| eca-miR-30c | Down | PFN1 | Up | profilin 1 |
| eca-miR-130a | Up | TPP1 | Down | tripeptidyl peptidase 1 |
| eca-miR-132 | Up | SH3BGRL | Up | SH3 domain-binding glutamic acid-rich-like protein |
| eca-miR-144 | Down | MSN | Down | moesin (membrane-organizing extension spike protein) |
| eca-miR-144 | Down | PLS1 | Down | plastin 1 |
| eca-miR-144 | Down | RAC1 | Up | Rac family small GTPase 1 |
| eca-miR-221 | Up | YWHAE | Up | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon |
| eca-miR-221 | Up | CLIC1 | Up | Chloride intracellular channel protein |
| eca-miR-221 | Up | YWHAB | Up | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta |
| miRNA UF | miRNA P vs. C | mRNA B | mRNA P vs. C | Gene Description |
| eca-miR-23b | Up | CA2 | Down | carbonic anhydrase 2 |
| eca-miR-23b | Up | ABCC1 | Up | ATP binding cassette subfamily C member 1 |
| eca-miR-23b | Up | MET | Up | MET proto-oncogene, receptor tyrosine kinase |
| miRNA UF | miRNA P vs. C | Protein UF | Protein P vs. C | Gene Description |
| eca-miR-23b | Up | CAP1 | Down | adenylyl cyclase-associated protein |
| eca-miR-23b | Up | NOL11 | down | nucleolar protein 11 |
| eca-miR-23b | Up | CCT7 | up | chaperonin containing TCP1 subunit 7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smits, K.; Gansemans, Y.; Tilleman, L.; Van Nieuwerburgh, F.; Van De Velde, M.; Gerits, I.; Ververs, C.; Roels, K.; Govaere, J.; Peelman, L.; et al. Maternal Recognition of Pregnancy in the Horse: Are MicroRNAs the Secret Messengers? Int. J. Mol. Sci. 2020, 21, 419. https://doi.org/10.3390/ijms21020419
Smits K, Gansemans Y, Tilleman L, Van Nieuwerburgh F, Van De Velde M, Gerits I, Ververs C, Roels K, Govaere J, Peelman L, et al. Maternal Recognition of Pregnancy in the Horse: Are MicroRNAs the Secret Messengers? International Journal of Molecular Sciences. 2020; 21(2):419. https://doi.org/10.3390/ijms21020419
Chicago/Turabian StyleSmits, Katrien, Yannick Gansemans, Laurentijn Tilleman, Filip Van Nieuwerburgh, Margot Van De Velde, Ilse Gerits, Cyrillus Ververs, Kim Roels, Jan Govaere, Luc Peelman, and et al. 2020. "Maternal Recognition of Pregnancy in the Horse: Are MicroRNAs the Secret Messengers?" International Journal of Molecular Sciences 21, no. 2: 419. https://doi.org/10.3390/ijms21020419
APA StyleSmits, K., Gansemans, Y., Tilleman, L., Van Nieuwerburgh, F., Van De Velde, M., Gerits, I., Ververs, C., Roels, K., Govaere, J., Peelman, L., Deforce, D., & Van Soom, A. (2020). Maternal Recognition of Pregnancy in the Horse: Are MicroRNAs the Secret Messengers? International Journal of Molecular Sciences, 21(2), 419. https://doi.org/10.3390/ijms21020419





