Overexpression of Rice Expansin7 (Osexpa7) Confers Enhanced Tolerance to Salt Stress in Rice
Abstract
1. Introduction
2. Results
2.1. Production and Analysis of OsEXPA7-OX Rice Plants
2.2. OsEXPA7 Overexpressing Lines Showed Strong Tolerance to Saline Stress
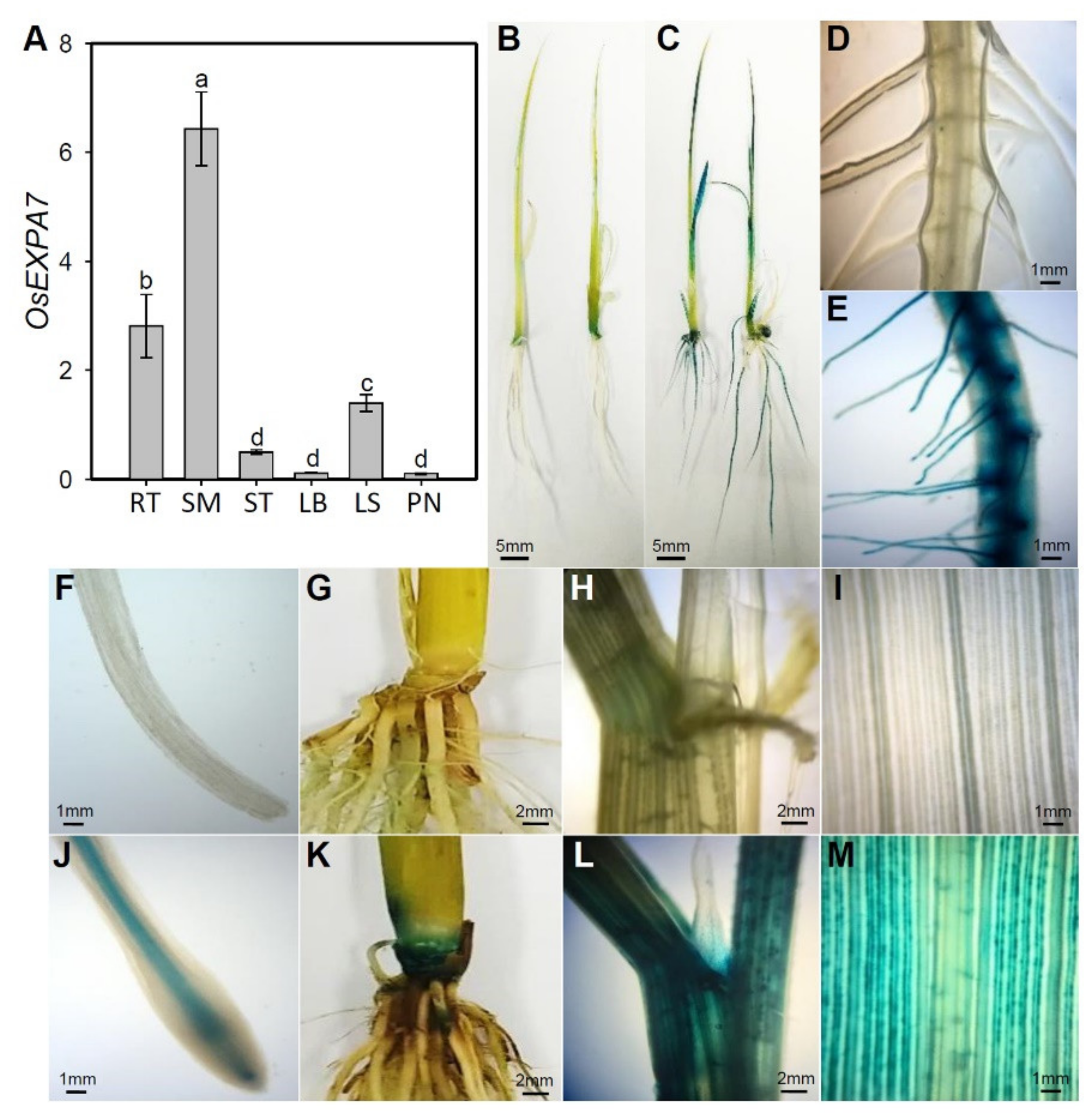
2.3. OsEXPA7 Was Mainly Expressed in Shoot Apical Meristem (SAM), Root, and Leaf Sheath
2.4. Morphological Changes in Leaves and Roots Due to OsEXPA7 Overexpression in Rice
2.5. Improved Ion Homeostasis in OsEXPA7-OX Plants Under Salt Stress
2.6. Overexpression of OsEXPA7 Improved the Antioxidant Capacity of Transgenic Rice Lines
2.7. DEGs in OsEXPA7-OX and WT Plants
2.8. Functional Classification of DEGs Using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
2.9. Genes Involved in Salt Stress Tolerance were Significantly Altered in OsEXPA7-OX Compared to WT
3. Discussion
3.1. OsEXPA7 Overexpression Improved Salt Stress Tolerance in Rice
3.2. OsEXPA7 Overexpression Increased Cell Size and Enhanced Plant Growth
3.3. Improvement of Na+ and K+ Balance in OsEXPA7-OX Plants Under Salt Stress
3.4. Overexpression of OsEXPA7 Improved Oxidative Stress Tolerance
3.5. Possible Mechanisms Underlying Salt Tolerance in OsEXPA7 Overexpressing Rice Plants
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Morphological Analysis
4.3. Measurement of Electrical Conductivity and Chlorophyll Concentration
4.4. Histochemical Assay
4.5. Determination of Na+ and K+ Content
4.6. Determination of ROS and Enzyme Activity
4.7. MDA and Proline Content
4.8. Real-Time PCR Analysis and cDNA Synthesis
4.9. RNA Isolation and RNA-Seq Analysis
4.10. Data Analyses
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CaMV | Cauliflower mosaic virus |
| PGD | Phosphogluconate dehydrogenase |
| GUS | b-glucuronidase |
| WT | Wild type |
| ROS | Reactive oxygen species |
| MDA | Malondialdehyde |
| OX | Overexpression |
| GO | Gene ontology |
| LV | Large vein |
| BS | Bundle sheath |
| Co | Collenchyma |
| MX | Metaxylem |
| ARF | Auxin response factor |
| SOS1 | Salt overly sensitive 1 |
| NHX4 | Sodium/hydrogen exchanger 4 |
| PM | Plasma membrane |
| NaCl | Sodium chloride |
| DAB | 3,3-diaminobenzidine |
| NBT | Nitro-blue tetrazolium |
| DEG | Differentially expressed genes |
| FDR | False discovery rate |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| Aux/IAA | Auxin/indole-3-acetic acid |
| SAM | Shoot apical meristem |
| POD | Peroxidase |
| SOD | Superoxide dismutase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| WAK | Wall-associated kinase |
| ELR | Electrolyte leakage ratio |
References
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef] [PubMed]
- Maas, E.V. Salt tolerance of plants. Appl. Agric. Res. 1986, 1, 12–26. [Google Scholar]
- Ashraf, M.; Akram, N.A. Improving salinity tolerance of plants through conventional breeding and genetic engineering: An analytical comparison. Biotechnol. Adv. 2009, 27, 744–752. [Google Scholar] [CrossRef]
- Bohnert, H.J.; Ayoubi, P.; Borchert, C.; Bressan, R.A.; Burnap, R.L.; Cushman, J.C.; Cushman, M.A.; Deyholos, M.; Fischer, R.; Galbraith, D.W.; et al. A genomics approach towards salt stress tolerance. Plant Physiol. Biochem. 2001, 39, 295–311. [Google Scholar] [CrossRef]
- You, J.; Chan, Z. ROS Regulation During Abiotic Stress Responses in Crop Plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell Signaling during Cold, Drought, and Salt Stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef] [PubMed]
- Galvan-Ampudia, C.S.; Julkowska, M.M.; Darwish, E.; Gandullo, J.; Korver, R.A.; Brunoud, G.; Haring, M.A.; Munnik, T.; Vernoux, T.; Testerink, C. Halotropism Is a Response of Plant Roots to Avoid a Saline Environment. Curr. Biol. 2013, 23, 2044–2050. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Tang, Z.; Su, W.; Sun, W. Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics 2005, 5, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Blumwald, E. Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 2000, 12, 431–434. [Google Scholar] [CrossRef]
- Louise, J.; Simon, M.-M. A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum. FEBS Lett. 2004, 559, 61–65. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Kader, M.A.; Seidel, T.; Golldack, D.; Lindberg, S. Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa L.) cultivars. J. Exp. Bot. 2006, 57, 4257–4268. [Google Scholar] [CrossRef] [PubMed]
- White, P.J.; Karley, A.J. Plant Cell Monographs; Springer: Heidelberg, Germany, 2010; Volume 17, pp. 199–224. [Google Scholar]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 18. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Kumar, M.; Kim, S.-R.; Ryu, H.; Cho, Y.-G. Insights into genomics of salt stress response in rice. Rice 2013, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K.; Liu, J.; Xiong, L. Genetic Analysis of Salt Tolerance in Arabidopsis: Evidence for a Critical Role of Potassium Nutrition. Plant Cell 1998, 10, 1181–1191. [Google Scholar] [CrossRef]
- De Lourdes Oliveira Otoch, M.; Menezes Sobreira, A.C.; Farias de Aragão, M.E.; Orellano, E.G.; da Guia Silva Lima, M.; Fernandes de Melo, D. Salt modulation of vacuolar H+-ATPase and H+-Pyrophosphatase activities in Vigna unguiculata. J. Plant Physiol. 2001, 158, 545–551. [Google Scholar] [CrossRef]
- Wang, B.; Lüttge, U.; Ratajczak, R. Effects of salt treatment and osmotic stress on V-ATPase and V-PPase in leaves of the halophyte Suaeda salsa. J. Exp. Bot. 2001, 52, 2355–2365. [Google Scholar] [CrossRef]
- Barragán, V.; Leidi, E.O.; Andrés, Z.; Rubio, L.; De Luca, A.; Fernández, J.A.; Cubero, B.; Pardo, J.M. Ion Exchangers NHX1 and NHX2 Mediate Active Potassium Uptake into Vacuoles to Regulate Cell Turgor and Stomatal Function in Arabidopsis. Plant Cell 2012, 24, 1127–1142. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef][Green Version]
- Bae, J.M.; Kwak, M.S.; Noh, S.A.; Oh, M.-J.; Kim, Y.-S.; Shin, J.S. Overexpression of sweetpotato expansin cDNA (IbEXP1) increases seed yield in Arabidopsis. Transgenic Res. 2014, 23, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Wenwen, Y.; Zang, G.; Kang, Z.; Zhang, Z.; Huang, J.; Wang, G. OsEXPB2, a β-expansin gene, is involved in rice root system architecture. Mol. Breed. 2015, 35, 41. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, N.; Qiu, S.; Zou, H.; Zang, G.; Kang, Z.; Wang, G.; Huang, J. Regulation of the α-expansin gene OsEXPA8 expression affects root system architecture in transgenic rice plants. Mol. Breed. 2014, 34, 47–57. [Google Scholar] [CrossRef]
- He, X.; Zeng, J.; Cao, F.; Ahmed, I.M.; Zhang, G.; Vincze, E.; Wu, F. HvEXPB7, a novel β-expansin gene revealed by the root hair transcriptome of Tibetan wild barley, improves root hair growth under drought stress. J. Exp. Bot. 2015, 66, 7405–7419. [Google Scholar] [CrossRef]
- Kuluev, B.R.; Safiullina, M.G.; Knyazev, A.V.; Chemeris, A.V. Effect of ectopic expression of NtEXPA5 gene on cell size and growth of organs of transgenic tobacco plants. Russ. J. Dev. Biol. 2013, 44, 28–34. [Google Scholar] [CrossRef]
- Goh, H.-H.; Sloan, J.; Dorca-Fornell, C.; Fleming, A. Inducible repression of multiple expansin genes leads to growth suppression during leaf development. Plant Physiol. 2012, 159, 1759–1770. [Google Scholar] [CrossRef]
- Palapol, Y.; Kunyamee, S.; Thongkhum, M.; Ketsa, S.; Ferguson, I.B.; van Doorn, W.G. Expression of expansin genes in the pulp and the dehiscence zone of ripening durian (Durio zibethinus) fruit. J. Plant Physiol. 2015, 182, 33–39. [Google Scholar] [CrossRef]
- Le Gall, H.; Philippe, F.; Domon, J.-M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell Wall Metabolism in Response to Abiotic Stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef]
- Wu, Y.; Thorne, E.T.; Sharp, R.E.; Cosgrove, D.J. Modification of expansin transcript levels in the maize primary root at low water potentials. Plant Physiol. 2001, 126, 1471–1479. [Google Scholar] [CrossRef]
- Han, Y.; Chen, Y.; Yin, S.; Zhang, M.; Wang, W. Over-expression of TaEXPB23, a wheat expansin gene, improves oxidative stress tolerance in transgenic tobacco plants. J. Plant Physiol. 2015, 173, 62–71. [Google Scholar] [CrossRef]
- Guo, W.; Zhao, J.; Li, X.; Qin, L.; Yan, X.; Liao, H. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011, 66, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Noh, S.A.; Park, S.H.; Huh, G.H.; Paek, K.-H.; Shin, J.S.; Bae, J.M. Growth retardation and differential regulation of expansin genes in chilling-stressed sweetpotato. Plant Biotechnol. Rep. 2009, 3, 75–85. [Google Scholar] [CrossRef]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2015, 5, 771. [Google Scholar] [CrossRef]
- Feng, X.; Xu, Y.; Peng, L.; Yu, X.; Zhao, Q.; Feng, S.; Zhao, Z.; Li, F.; Hu, B. TaEXPB7-B, a β-expansin gene involved in low-temperature stress and abscisic acid responses, promotes growth and cold resistance in Arabidopsis thaliana. J. Plant Physiol. 2019, 240, 153004. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tian, J.; Belanger, F.C.; Huang, B. Identification and characterization of an expansin gene AsEXP1 associated with heat tolerance in C3Agrostis grass species. J. Exp. Bot. 2007, 58, 3789–3796. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Han, Y.; Kong, X.; Kang, H.; Ren, Y.; Wang, W. Ectopic expression of wheat expansin gene TaEXPA2 improved the salt tolerance of transgenic tobacco by regulating Na+/K+ and antioxidant competence. Physiol. Plant. 2017, 159, 161–177. [Google Scholar] [CrossRef]
- Geilfus, C.-M.; Ober, D.; Eichacker, L.; Mühling, K.; Zörb, C. Down regulation of ZmEXPB6 is correlated with salt mediated growth reduction in leaves of Zea mays L. J. Biol. Chem. 2015, 290, 11235–11245. [Google Scholar] [CrossRef]
- Qiu, S.; Ma, N.; Che, S.; Wang, Y.; Peng, X.; Zhang, G.; Wang, G.; Huang, J. Repression of OsEXPA3 Expression Leads to Root System Growth Suppression in Rice. Crop Sci. 2014, 54, 2201–2213. [Google Scholar] [CrossRef]
- Lasanthi-Kudahettige, R.; Magneschi, L.; Loreti, E.; Gonzali, S.; Licausi, F.; Novi, G.; Beretta, O.; Vitulli, F.; Alpi, A.; Perata, P. Transcript Profiling of the Anoxic Rice Coleoptile. Plant Physiol. 2007, 144, 218–231. [Google Scholar] [CrossRef]
- Bevilacqua, C.B.; Basu, S.; Pereira, A.; Tseng, T.-M.; Zimmer, P.D.; Burgos, N.R. Analysis of Stress-Responsive Gene Expression in Cultivated and Weedy Rice Differing in Cold Stress Tolerance. PLoS ONE 2015, 10, e0132100. [Google Scholar] [CrossRef][Green Version]
- Chen, Z.; Pottosin, I.I.; Cuin, T.A.; Fuglsang, A.T.; Tester, M.; Jha, D.; Zepeda-Jazo, I.; Zhou, M.; Palmgren, M.G.; Newman, I.A.; et al. Root Plasma Membrane Transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiol. 2007, 145, 1714–1725. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.; Ahmad, S. Influence of sodium chloride on ion accumulation, yield components and fibre characteristics in salt-tolerant and salt-sensitive lines of cotton (Gossypium hirsutum L.). Field Crops Res. 2000, 66, 115–127. [Google Scholar] [CrossRef]
- Li, J.; Guo, X.; Zhang, M.; Wang, X.; Zhao, Y.; Yin, Z.; Zhang, Z.; Wang, Y.; Xiong, H.; Zhang, H.; et al. OsERF71 confers drought tolerance via modulating ABA signaling and proline biosynthesis. Plant Sci. 2018, 270, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Hamamoto, S.; Uozumi, N. Sodium transport system in plant cells. Front. Plant Sci. 2013, 4, 410. [Google Scholar] [CrossRef]
- Fukuda, A.; Nakamura, A.; Hara, N.; Toki, S.; Tanaka, Y. Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 2011, 233, 175–188. [Google Scholar] [CrossRef]
- Majda, M.; Robert, S. The Role of Auxin in Cell Wall Expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef]
- Li, G.; Liang, W.; Zhang, X.; Ren, H.; Hu, J.; Bennett, M.; Zhang, D. Rice actin-binding protein RMD is a key link in the auxin-actin regulatory loop that controls cell growth. Proc. Natl. Acad. Sci. USA 2014, 111, 10377–10382. [Google Scholar] [CrossRef]
- Decreux, A.; Messiaen, J. Wall-associated Kinase WAK1 Interacts with Cell Wall Pectins in a Calcium-induced Conformation. Plant Cell Physiol. 2005, 46, 268–278. [Google Scholar] [CrossRef]
- Kanneganti, V.; Gupta, A. RNAi mediated silencing of a wall associated kinase, 5 OsiWAK1 in Oryza sativa results in impaired root development and sterility due to anther indehiscence. Physiol. Mol. Biol. Plants 2011, 17, 65–77. [Google Scholar] [CrossRef]
- Bakshi, M.; Oelmüller, R. WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, L.; Chen, J.; Liu, Z.; Park, C.-M.; Xiang, F. WRKY71 Acts Antagonistically Against Salt-Delayed Flowering in Arabidopsis thaliana. Plant Cell Physiol. 2017, 59, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Gho, Y.-S.; Jung, K.-H.; Kim, S.-R. Genome-Wide Identification and Analysis of Genes, conserved between japonica and indica Rice Cultivars, that Respond to Low-Temperature Stress at the Vegetative Growth Stage. Front. Plant Sci. 2017, 8, 1120. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.-D.; Zhang, F.; Huang, L.-Y.; Zhuo, D.-L.; Zhou, Y.; Shi, Y.-Y.; Li, Z. Stress-activated Protein Kinase OsSAPK2 Involved in Regulating Resistant Response to Xanthomonas oryzae pv. oryzae in Rice. Acta Agron. Sin. 2015, 41, 1191. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Enzymes and Other Agents That Enhance Cell Wall Extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 391–417. [Google Scholar] [CrossRef]
- Taiz, L. Plant Cell Expansion: Regulation of Cell Wall Mechanical Properties. Annu. Rev. Plant Physiol. 1984, 35, 585–657. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Cell Wall Loosening by Expansins. Plant Physiol. 1998, 118, 333–339. [Google Scholar] [CrossRef]
- Wu, Y.; Sharp, R.E.; Durachko, D.M.; Cosgrove, D.J. Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiol. 1996, 111, 765–772. [Google Scholar] [CrossRef]
- Lee, D.-K.; Ahn, J.H.; Song, S.-K.; Choi, Y.D.; Lee, J.S. Expression of an expansin gene is correlated with root elongation in soybean. Plant Physiol. 2003, 131, 985–997. [Google Scholar] [CrossRef]
- Michael, A.J. A cDNA from pea petals with sequence similarity to pollen allergen, cytokinin-induced and genetic tumour-specific genes: Identification of a new family of related sequences. Plant Mol. Biol. 1996, 30, 219–224. [Google Scholar] [CrossRef]
- Ma, N.; Wang, Y.; Qiu, S.; Kang, Z.; Che, S.; Wang, G.; Huang, J. Overexpression of OsEXPA8, a Root-Specific Gene, Improves Rice Growth and Root System Architecture by Facilitating Cell Extension. PLoS ONE 2013, 8, e75997. [Google Scholar] [CrossRef]
- Neumann, P.M.; Azaizeh, H.; Leon, D. Hardening of root cell walls: A growth inhibitory response to salinity stress. Plant Cell Environ. 1994, 17, 303–309. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Relaxation in a high-stress environment: The molecular bases of extensible cell walls and cell enlargement. Plant Cell 1997, 9, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.T.; Cosgrove, D.J. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 9783–9788. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, D.; Wittwer, F.; Mandel, T.; Kuhlemeier, C. Localized Upregulation of a New Expansin Gene Predicts the Site of Leaf Formation in the Tomato Meristem. Plant Cell 1998, 10, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Hodge, A.; Berta, G.; Doussan, C.; Merchan, F.; Crespi, M. Plant root growth, architecture and function. Plant Soil 2009, 321, 153–187. [Google Scholar] [CrossRef]
- Kaashyap, M.; Ford, R.; Kudapa, H.; Jain, M.; Edwards, D.; Varshney, R.; Mantri, N. Differential Regulation of Genes Involved in Root Morphogenesis and Cell Wall Modification is Associated with Salinity Tolerance in Chickpea. Sci. Rep. 2018, 8, 4855. [Google Scholar] [CrossRef] [PubMed]
- Kubo, F.C.; Yasui, Y.; Kumamaru, T.; Sato, Y.; Hirano, H.-Y. Genetic analysis of rice mutants responsible for narrow leaf phenotype and reduced vein number. Genes Genet. Syst. 2016, 91, 235–240. [Google Scholar] [CrossRef][Green Version]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.-K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef]
- Apse, M.P.; Aharon, G.S.; Snedden, W.A.; Blumwald, E. Salt Tolerance Conferred by Overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 1999, 285, 1256–1258. [Google Scholar] [CrossRef]
- Qiu, Q.-S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.-K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef]
- Halfter, U.; Ishitani, M.; Zhu, J.K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 2000, 97, 3735–3740. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.-K.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Shaukat, M.; Ashraf, M.; Zhu, C.; Jin, Q.; Zhang, J. Climate Change and Agriculture; Intechopen: London, UK, 2019; pp. 1–26. [Google Scholar]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Pang, C.-H.; Wang, B.-S. Progress in Botany; Springer: Berlin, Germany, 2008; Volume 69, pp. 231–245. [Google Scholar]
- Ahmad, P.; Alyemeni, M.; Abass, M.; Wijaya, L.; Alam, P.; Kumar, A.; Ashraf, M. Upregulation of antioxidant and glyoxalase systems mitigates NaCl stress in Brassica juncea by supplementation of zinc and calcium. J. Plant Interact. 2018, 13, 151–162. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xing, S.; Guo, Q.; Zhao, M.; Zhang, J.; Gao, Q.; Wang, G.; Wang, W. Drought tolerance through over-expression of the expansin gene TaEXPB23 in transgenic tobacco. J. Plant Physiol. 2011, 168, 960–966. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, H.; Yang, R.; Xu, X.; Liu, X.; Xu, J. Over-expression of PttEXPA8 gene showed various resistances to diverse stresses. Int. J. Biol. Macromol. 2019, 130, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Masuta, Y.; Saito, K.; Murayama, S.; Ozawa, K. Enhanced chilling tolerance at the booting stage in rice by transgenic overexpression of the ascorbate peroxidase gene, OsAPXa. Plant Cell Rep. 2011, 30, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, S.R.; Sadhasivam, V.; Parida, A. Over expression of cytosolic copper/zinc superoxide dismutase from a mangrove plant Avicennia marina in indica Rice var Pusa Basmati-1 confers abiotic stress tolerance. Transgenic Res. 2008, 17, 281–291. [Google Scholar] [CrossRef]
- Naser, V.; Shani, E. Auxin response under osmotic stress. Plant Mol. Biol. 2016, 91, 661–672. [Google Scholar] [CrossRef]
- Iqbal, N.; Umar, S.; Khan, N.A.; Khan, M.I.R. A new perspective of phytohormones in salinity tolerance: Regulation of proline metabolism. Environ. Exp. Bot. 2014, 100, 34–42. [Google Scholar] [CrossRef]
- Sahi, C.; Singh, A.; Kumar, K.; Blumwald, E.; Grover, A. Salt stress response in rice: Genetics, molecular biology, and comparative genomics. Funct. Integr. Genom. 2006, 6, 263–284. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhong, H.; Deng, X.; Gautam, M.; Gong, Z.; Zhang, Y.; Zhao, G.; Liu, C.; Li, Y. Evolutionary Analysis of GH3 Genes in Six Oryza Species/Subspecies and Their Expression under Salinity Stress in Oryza sativa ssp. japonica. Plants 2019, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Quint, M.; Gray, W.M. Auxin signaling. Curr. Opin. Plant Biol. 2006, 9, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Gray, W.M. SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Perrot-Rechenmann, C. Cellular Responses to Auxin: Division versus Expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.-I.; Lim, H.-M.; Siddiqui, Z.S.; Park, S.-H.; Kim, A.R.; Kwon, T.-R.; Lee, S.-K.; Park, S.-C.; Jeong, M.-J.; Lee, G.-S. Over-expression of PsGPD, a mushroom glyceraldehyde-3-phosphate dehydrogenase gene, enhances salt tolerance in rice plants. Biotechnol. Lett. 2014, 36, 1641–1648. [Google Scholar] [CrossRef]
- Keisham, M.; Mukherjee, S.; Bhatla, S.C. Mechanisms of Sodium Transport in Plants-Progresses and Challenges. Int. J. Mol. Sci. 2018, 19, 647. [Google Scholar] [CrossRef]
- Park, S.-H.; Yi, N.; Kim, Y.S.; Jeong, M.-H.; Bang, S.-W.; Choi, Y.D.; Kim, J.-K. Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J. Exp. Bot. 2010, 61, 2459–2467. [Google Scholar] [CrossRef]
- Park, S.-H.; Bang, S.W.; Jeong, J.S.; Jung, H.; Redillas, M.C.F.R.; Kim, H.I.; Lee, K.H.; Kim, Y.S.; Kim, J.-K. Analysis of the APX, PGD1 and R1G1B constitutive gene promoters in various organs over three homozygous generations of transgenic rice plants. Planta 2012, 235, 1397–1408. [Google Scholar] [CrossRef] [PubMed]
- Gregorio, G.; Senadhira, D.; Mendoza, R. Screening Rice for Salinity Tolerance; IRRI Discussion Paper Series; International Rice Research Institute: Manila, Phillipines, 1997; Volume 22. [Google Scholar]
- Bado, S.; Forster, B.P.; Ghanim, A.M.A.; Jankowicz-Cieslak, J.; Berthold, G.; Luxiang, L. Protocol for Screening for Salt Tolerance in Rice. In Protocols for Pre-Field Screening of Mutants for Salt Tolerance in Rice, Wheat and Barley; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- Periasamy, K. A technique of staining sections of paraffin-embedded plant materials without employing a graded ethanol series. J. R. Microsc. Soc. 1967, 87, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Schichnes, D.; Nemson, J.; Ruzin, S. Microwave Paraffin Techniques for Botanical Tissues. In Microwave Techniques and Protocols; Springer: Berlin, Germany, 2008. [Google Scholar]
- Wang, Y.; Jiang, J.; Zhao, X.; Liu, G.; Yang, C.; Zhan, L. A novel LEA gene from Tamarix androssowii confers drought tolerance in transgenic tobacco. Plant Sci. 2006, 171, 655–662. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. In Methods in Enzymology; Academic Press: Cambridge, UK, 1987; Volume 148, pp. 350–382. [Google Scholar]
- Poosakkannu, A.; Loganathan, A. Beta glucuronidase activity in early stages of rice seedlings and callus: A comparison with Escherichia coli beta glucuronidase expressed in the transgenic rice. Int. J. Biotechnol. Mol. Biol. Res. 2013, 4, 52–59. [Google Scholar]
- Shen, Y.; Shen, L.; Shen, Z.; Jing, W.; Ge, H.; Zhao, J.; Zhang, W. The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ. 2015, 38, 2766–2779. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Yusuf, M.A.; Singh, P.; Sardar, M.; Sarin, N.B. Histochemical Detection of Superoxide and H2O2 Accumulation in Brassica juncea Seedlings. Bio Protoc. 2014, 4, e1108. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide Dismutases: I. Occurrence in Higher Plants 1977, 59, 309–314. [Google Scholar]
- Chance, B.; Maehly, A.C. Assay of catalases and peroxidases. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1955; Volume 2, pp. 764–775. [Google Scholar]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef]








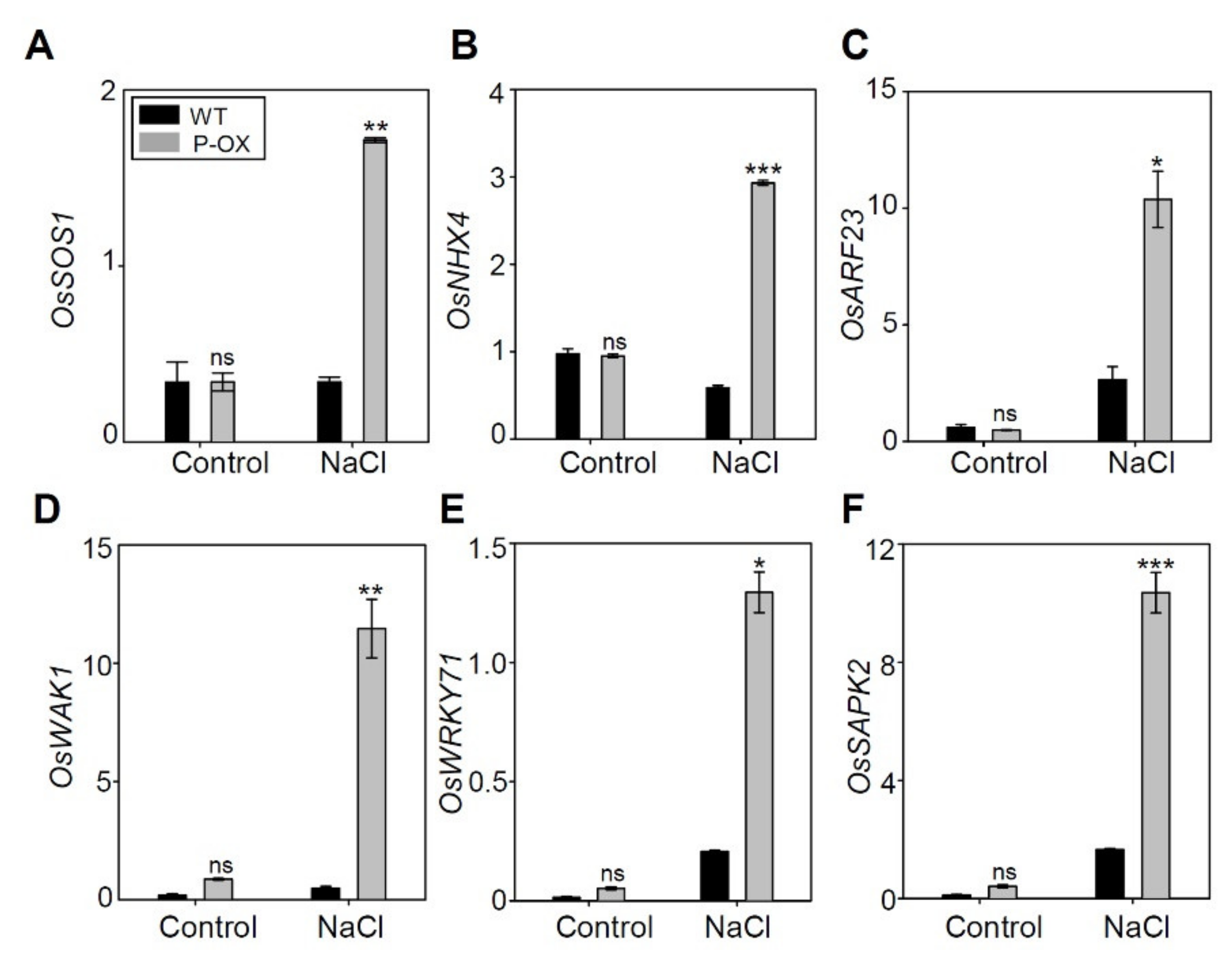
| Gene Name | Gene ID | Up or Down Ratio (Log2-Fold Change) | Function |
|---|---|---|---|
| OsSOS1 | LOC_Os12g44360.1 | up (2.35) | Sodium/hydrogen exchanger 7, putative, expressed |
| OsARF23 | LOC_Os11g32110.3 | up (25.30) | auxin response factor, putative, expressed |
| OsWAK1 | LOC_Os11g46860.1 | up (3.93) | wall-associated receptor kinase 3 precursor |
| OsMSRA4.1 | LOC_Os10g41400.2 | up (4.64) | response to salt stress, cellular response to oxidative stress |
| OsWRKY71 | LOC_Os02g08440.2 | up (6.00) | Response to salt stress, response to ethylene, response to abscisic acid |
| OsSAPK2 | LOC_Os07g42940.6 | up (3.30) | serine/threonine-protein kinase SAPK2 |
| OsGH3.12 | LOC_Os11g08340.1 | up (2.15) | Auxin-responsive Glycoside Hydrolase 3 (GH3) family member |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jadamba, C.; Kang, K.; Paek, N.-C.; Lee, S.I.; Yoo, S.-C. Overexpression of Rice Expansin7 (Osexpa7) Confers Enhanced Tolerance to Salt Stress in Rice. Int. J. Mol. Sci. 2020, 21, 454. https://doi.org/10.3390/ijms21020454
Jadamba C, Kang K, Paek N-C, Lee SI, Yoo S-C. Overexpression of Rice Expansin7 (Osexpa7) Confers Enhanced Tolerance to Salt Stress in Rice. International Journal of Molecular Sciences. 2020; 21(2):454. https://doi.org/10.3390/ijms21020454
Chicago/Turabian StyleJadamba, Chuluuntsetseg, Kiyoon Kang, Nam-Chon Paek, Soo In Lee, and Soo-Cheul Yoo. 2020. "Overexpression of Rice Expansin7 (Osexpa7) Confers Enhanced Tolerance to Salt Stress in Rice" International Journal of Molecular Sciences 21, no. 2: 454. https://doi.org/10.3390/ijms21020454
APA StyleJadamba, C., Kang, K., Paek, N.-C., Lee, S. I., & Yoo, S.-C. (2020). Overexpression of Rice Expansin7 (Osexpa7) Confers Enhanced Tolerance to Salt Stress in Rice. International Journal of Molecular Sciences, 21(2), 454. https://doi.org/10.3390/ijms21020454





